Abstract
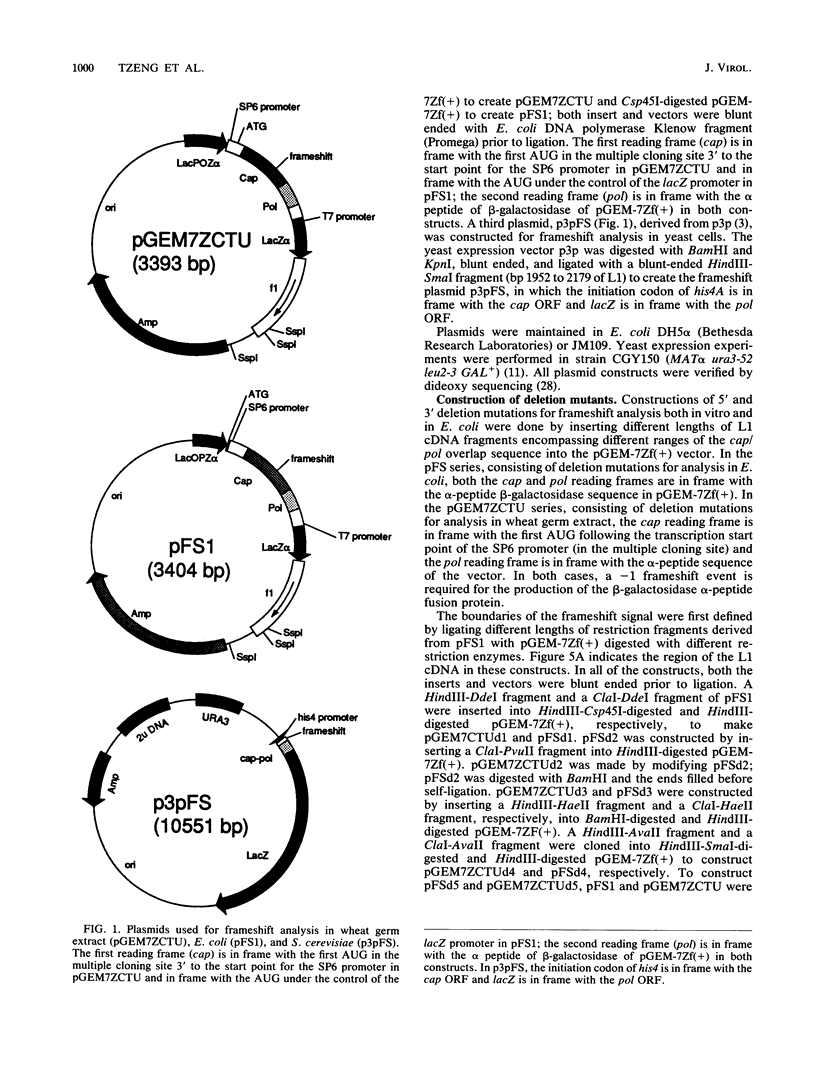
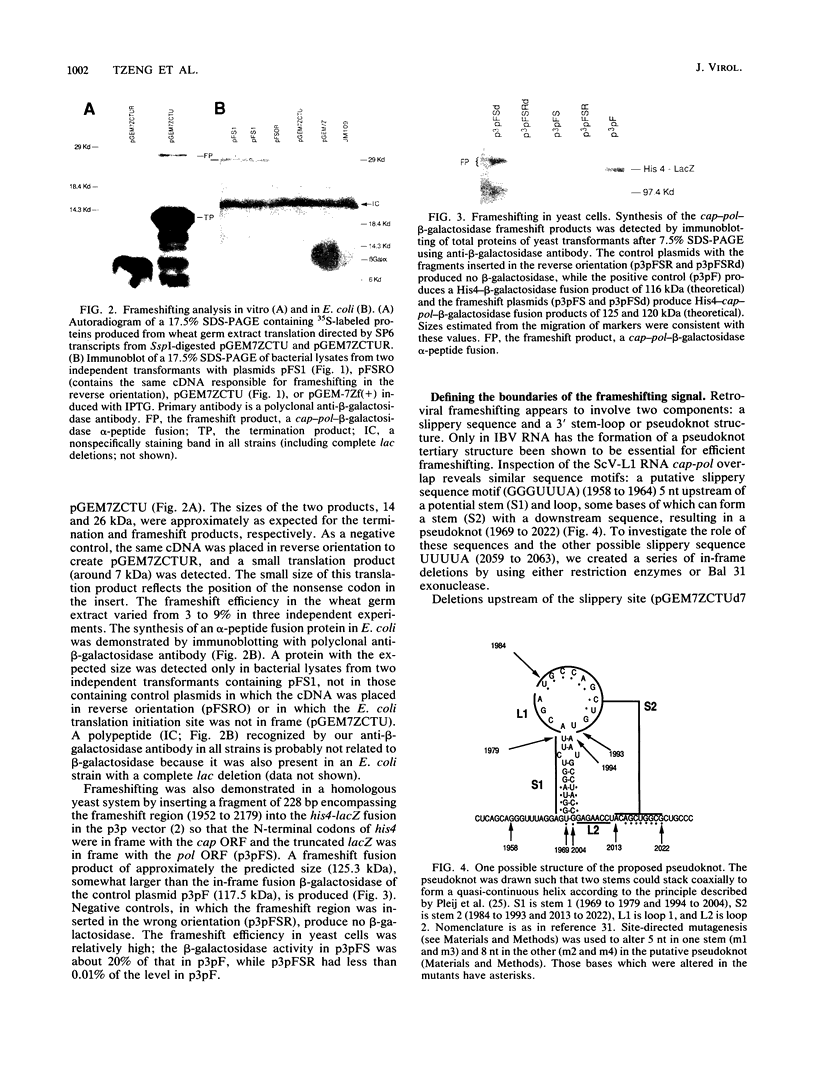
The large double-stranded RNA of the Saccharomyces cerevisiae (yeast) virus has two large overlapping open reading frames on the plus strand, one of which is translated via a -1 ribosomal frameshift. Sequences including the overlapping region, placed in novel contexts, can direct ribosomes to make a -1 frameshift in wheat germ extract, Escherichia coli and S. cerevisiae. This sequence includes a consensus slippery sequence, GGGUUUA, and has the potential to form a pseudoknot 3' to the putative frameshift site. Based on deletion analysis, a region of 71 nucleotides including the potential pseudoknot and the putative slippery sequence is sufficient for frameshifting. Site-directed mutagenesis demonstrates that the pseudoknot is essential for frameshifting.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson C. W., Straus J. W., Dudock B. S. Preparation of a cell-free protein-synthesizing system from wheat germ. Methods Enzymol. 1983;101:635–644. doi: 10.1016/0076-6879(83)01044-7. [DOI] [PubMed] [Google Scholar]
- Belcourt M. F., Farabaugh P. J. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell. 1990 Jul 27;62(2):339–352. doi: 10.1016/0092-8674(90)90371-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Digard P., Inglis S. C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989 May 19;57(4):537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruenn J. A. Relationships among the positive strand and double-strand RNA viruses as viewed through their RNA-dependent RNA polymerases. Nucleic Acids Res. 1991 Jan 25;19(2):217–226. doi: 10.1093/nar/19.2.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clare J., Farabaugh P. Nucleotide sequence of a yeast Ty element: evidence for an unusual mechanism of gene expression. Proc Natl Acad Sci U S A. 1985 May;82(9):2829–2833. doi: 10.1073/pnas.82.9.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigen W. J., Caskey C. T. Translational frameshifting: where will it stop? Cell. 1987 Jul 3;50(1):1–2. doi: 10.1016/0092-8674(87)90652-0. [DOI] [PubMed] [Google Scholar]
- Diamond M. E., Dowhanick J. J., Nemeroff M. E., Pietras D. F., Tu C. L., Bruenn J. A. Overlapping genes in a yeast double-stranded RNA virus. J Virol. 1989 Sep;63(9):3983–3990. doi: 10.1128/jvi.63.9.3983-3990.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dihanich M., van Tuinen E., Lambris J. D., Marshallsay B. Accumulation of viruslike particles in a yeast mutant lacking a mitochondrial pore protein. Mol Cell Biol. 1989 Mar;9(3):1100–1108. doi: 10.1128/mcb.9.3.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman J. D., Icho T., Wickner R. B. A -1 ribosomal frameshift in a double-stranded RNA virus of yeast forms a gag-pol fusion protein. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):174–178. doi: 10.1073/pnas.88.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura T., Wickner R. B. Gene overlap results in a viral protein having an RNA binding domain and a major coat protein domain. Cell. 1988 Nov 18;55(4):663–671. doi: 10.1016/0092-8674(88)90225-5. [DOI] [PubMed] [Google Scholar]
- Goff C. G., Moir D. T., Kohno T., Gravius T. C., Smith R. A., Yamasaki E., Taunton-Rigby A. Expression of calf prochymosin in Saccharomyces cerevisiae. Gene. 1984 Jan;27(1):35–46. doi: 10.1016/0378-1119(84)90236-1. [DOI] [PubMed] [Google Scholar]
- Hopper J. E., Bostian K. A., Rowe L. B., Tipper D. J. Translation of the L-species dsRNA genome of the killer-associated virus-like particles of Saccharomyces cerevisiae. J Biol Chem. 1977 Dec 25;252(24):9010–9017. [PubMed] [Google Scholar]
- Jacks T., Madhani H. D., Masiarz F. R., Varmus H. E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988 Nov 4;55(3):447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Power M. D., Masiarz F. R., Luciw P. A., Barr P. J., Varmus H. E. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988 Jan 21;331(6153):280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- Jacks T., Townsley K., Varmus H. E., Majors J. Two efficient ribosomal frameshifting events are required for synthesis of mouse mammary tumor virus gag-related polyproteins. Proc Natl Acad Sci U S A. 1987 Jun;84(12):4298–4302. doi: 10.1073/pnas.84.12.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Varmus H. E. Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting. Science. 1985 Dec 13;230(4731):1237–1242. doi: 10.1126/science.2416054. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S. Y., Chen J. H., Maizel J. V. Thermodynamic stability and statistical significance of potential stem-loop structures situated at the frameshift sites of retroviruses. Nucleic Acids Res. 1989 Aug 11;17(15):6143–6152. doi: 10.1093/nar/17.15.6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor J., Fulton S. M., Dobson M. J., Wilson W., Kingsman S. M., Kingsman A. J. A retrovirus-like strategy for expression of a fusion protein encoded by yeast transposon Ty1. Nature. 1985 Jan 17;313(5999):243–246. doi: 10.1038/313243a0. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W. A., Waterhouse P. M., Gerlach W. L. Sequence and organization of barley yellow dwarf virus genomic RNA. Nucleic Acids Res. 1988 Jul 11;16(13):6097–6111. doi: 10.1093/nar/16.13.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras D. F., Diamond M. E., Bruenn J. A. Identification of a putative RNA dependent RNA polymerase encoded by a yeast double stranded RNA virus. Nucleic Acids Res. 1988 Jul 11;16(13):6225–6225. doi: 10.1093/nar/16.13.6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleij C. W., Rietveld K., Bosch L. A new principle of RNA folding based on pseudoknotting. Nucleic Acids Res. 1985 Mar 11;13(5):1717–1731. doi: 10.1093/nar/13.5.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi J. D., Wyatt J. R., Tinoco I., Jr A pseudoknotted RNA oligonucleotide. Nature. 1988 Jan 21;331(6153):283–286. doi: 10.1038/331283a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel P. RNA pseudoknots that interact with components of the translation apparatus. Cell. 1989 Jul 14;58(1):9–12. doi: 10.1016/0092-8674(89)90395-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C. K., Draper D. E. Unusual mRNA pseudoknot structure is recognized by a protein translational repressor. Cell. 1989 May 19;57(4):531–536. doi: 10.1016/0092-8674(89)90123-2. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Atkins J. F., Gesteland R. F. Slippery runs, shifty stops, backward steps, and forward hops: -2, -1, +1, +2, +5, and +6 ribosomal frameshifting. Cold Spring Harb Symp Quant Biol. 1987;52:687–693. doi: 10.1101/sqb.1987.052.01.078. [DOI] [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Dahlberg A. E., Atkins J. F., Gesteland R. F. Reading frame switch caused by base-pair formation between the 3' end of 16S rRNA and the mRNA during elongation of protein synthesis in Escherichia coli. EMBO J. 1988 May;7(5):1503–1507. doi: 10.1002/j.1460-2075.1988.tb02969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Shuh M., Atkins J. F., Gesteland R. F. E. coli ribosomes re-phase on retroviral frameshift signals at rates ranging from 2 to 50 percent. New Biol. 1989 Nov;1(2):159–169. [PubMed] [Google Scholar]
- Weiss R., Lindsley D., Falahee B., Gallant J. On the mechanism of ribosomal frameshifting at hungry codons. J Mol Biol. 1988 Sep 20;203(2):403–410. doi: 10.1016/0022-2836(88)90008-3. [DOI] [PubMed] [Google Scholar]
- Wilson W., Braddock M., Adams S. E., Rathjen P. D., Kingsman S. M., Kingsman A. J. HIV expression strategies: ribosomal frameshifting is directed by a short sequence in both mammalian and yeast systems. Cell. 1988 Dec 23;55(6):1159–1169. doi: 10.1016/0092-8674(88)90260-7. [DOI] [PubMed] [Google Scholar]
- Wyatt J. R., Puglisi J. D., Tinoco I., Jr RNA pseudoknots. Stability and loop size requirements. J Mol Biol. 1990 Jul 20;214(2):455–470. doi: 10.1016/0022-2836(90)90193-P. [DOI] [PubMed] [Google Scholar]
- ten Dam E. B., Pleij C. W., Bosch L. RNA pseudoknots: translational frameshifting and readthrough on viral RNAs. Virus Genes. 1990 Jul;4(2):121–136. doi: 10.1007/BF00678404. [DOI] [PMC free article] [PubMed] [Google Scholar]







