The α-carbonic anhydrases (CAs) are zinc metalloenzymes that catalyze the reversible hydration of CO2 in forming HCO3-. The active site of an α-CA contains a catalytically essential Zn2+ coordinated by three histidine residues at the bottom of a 15 Å deep cleft, and the tightest binding CA inhibitors developed to date contain a sulfonamide moiety that coordinates to Zn2+ as a sulfonamidate anion.1 Notably, human isozyme II (CAII) is an ideal model system for exploring new inhibitor designs, some of which can be exploited in biosensing applications.2-4 Here, CAII is utilized for the structure-based design of a xenon (129Xe) biosensor for potential use as a magnetic resonance imaging (MRI) contrast agent.
The 129Xe isotope has a spin-1/2 nucleus, a >200-ppm chemical shift window in water, and a natural isotopic abundance of 26% (commercially available up to 86%), which makes it an appealing biomolecular probe for MRI. Moreover, 129Xe can be laser polarized to enhance MRI signals ∼10,000-fold.5 Although current in vivo MRI applications are limited to functional lung imaging through the diffusion of Xe gas,6 the encapsulation of 129 Xe within a cryptophane cage (KD ≈ 30 μM at 37 °C in phosphate-buffered solution)7 facilitates its use as a biosensor that can be targeted to specific proteins using an appropriate affinity tag.8,9 For example, racemic biosensor 1 (Figure 1a) has been designed to bind to the CA isozymes (KD = 60 ± 20 nM against CAII in solution), and yields a distinctive 129Xe-MRI spectrum when bound to CAII.10 Here, we report the X-ray crystal structure of the CAII-1-Xe complex at 1.70 Å resolution.
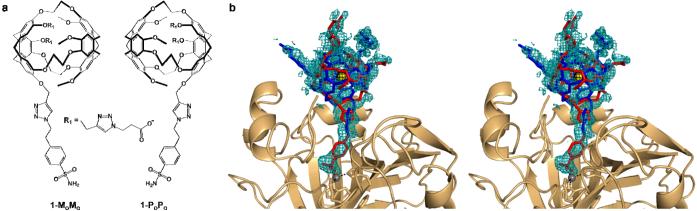
Figure 1.
(a) The MoMo and PoPo enantiomers of the cryptophane-A-derived CA biosensor. The benzenesulfonamide moiety serves as an affinity tag that targets the Zn2+ ion, and the R1 substituents contain triazole propionate moieties that enhance aqueous solubility. (b) Stereoview of a simulated annealing omit map showing 1-MoMo (blue) and 1-PoPo (red) bound in the active site (1.9 σ contour, teal). A Bijvoet difference Fourier map (2.0 σ, black) confirms the encapsulation of Xe (yellow). Coordination interactions with Zn2+ (grey sphere) are indicated by dotted lines.
For structure determination, CAII was overexpressed in E. coli and purified as described,11 then incubated with a two-fold excess of 1, concentrated to 10 mg/mL, and crystallized by the hanging drop vapor diffusion method. Crystals were cryoprotected in 15% glycerol and subsequently pressurized under 20 atm Xe for 30 min prior to flash cooling and X-ray data collection. The structure was refined to final Rwork and Rfree values of 0.23 and 0.25, respectively.
Biosensor 1 coordinates to the active site Zn2+ ion as the sulfonamidate anion, displacing the zinc-bound hydroxide ion of the native enzyme as previously observed in other complexes of CAII with benzenesulfonamide derivatives.1,2,12 The crystallographic occupancies of 1 and Zn2+ are refined at 0.5. It is unusual to observe diminished Zn2+ occupancy in a CAII-inhibitor complex, but the molecular origins of this effect are not clear.
The encapsulation of Xe within the cryptophane cage of 1 is confirmed by inspection of the Bijvoet difference Fourier map calculated from anomalous scattering data (Figures 1b and S1.) X-ray diffraction data was collected at a wavelength λ = 0.9795 Å, which is far from the Xe LI edge of 2.27 Å.13 Nevertheless, the anomalous scattering component f” is 3.4 e- for Xe, so the anomalous signal is still prominent at the wavelength of data collection. A second Xe binding site is observed in a hydrophobic pocket defined by A116, L148, V218, L157, V223 and F226 (Figure S2). The crystallographic occupancies of these Xe sites refine to 0.50 and 0.37, respectively. Anomalous scattering peaks are absent from crystals not subject to Xe pressurization.
Notably, 1 contains a chiral axis and the electron density map reveals the binding of equal populations of both enantiomers, (each refined with an occupancy of 0.25; Figure 1)14-16 Overall, the binding of 1 does not cause any significant structural changes in the active site, and the root-mean-square deviation is 0.34 Å for 256 Cα atoms between the current structure and the unliganded enzyme (PDB 2CBA).17
The total surface area of 1 is ∼1500 Å2, of which ∼500 Å2 becomes solvent inaccessible due to contacts of 1 within the active site cleft of CAII designated molecule I in Figure 2. The surrounding CAII molecules in the unit cell (molecules II-IV), sequester an additional ∼540 Å2 of the surface area of 1 from solvent. Some structural changes are observed near the outer rim of the active site cleft where the cryptophane binds. The most notable change is observed for Q136, which rotates ∼180° to make van der Waals contacts with the cryptophane and the symmetry-related cryptophane bound to molecule III in the crystal lattice. Other residues at the active site rim of molecule I that make close contacts with the cryptophane are G132 and P202. Additional structural changes in the crystal lattice result from the binding of 1 to molecule I: in molecule II, H36 rotates ∼90° to make a van der Waals contact with the cage, and Q137 of molecule III rotates ∼90° to donate a hydrogen bond to an ether oxygen atom of 1.
Figure 2.
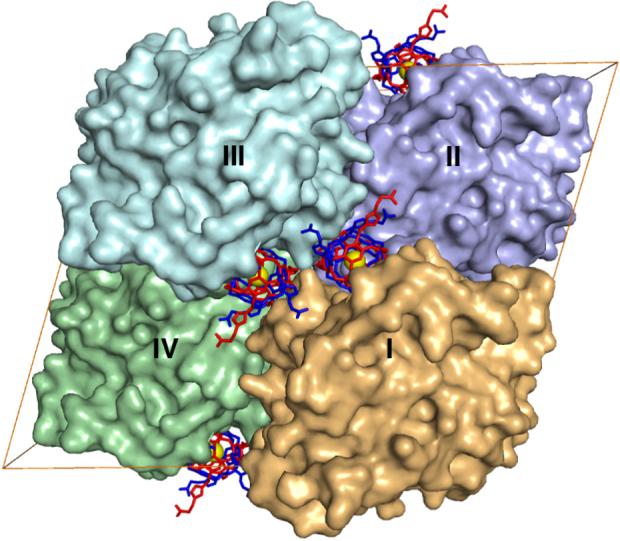
The unit cell of CAII crystals in space group C2 contains four molecules: I (x,y,z), II (x+1/2,y+1/2,z), III (-x,y,-z) and IV (-x+1/2,y+1/2,-z). The binding of 1 in the active site cleft of molecule I buries ∼500 Å2. Crystal contacts bury an additional 540 Å2 of the surface of 1 as follows: 270 Å2 with molecule III, and 240 Å2 and 30 Å2 with the front and back faces of molecule II, respectively. Molecule IV does not contact 1 bound to molecule I.
Although the pendant propionates appear to be more disordered than the cryptophane and are characterized by correspondingly weaker electron density, a hydrogen bond between a propionate moiety and Q53 of molecule II is observed. The relative dearth of strong cryptophane-protein interactions may explain why the affinity of 1 measured by ITC is only slightly better than that measured for the parent triazole-benzenesulfonamide lacking the cryptophane (KD = 100 ± 10 nM).10
Limited hydrogen bond interactions between CAII and the cryptophane moiety of 1 may be advantageous for the use of cryptophanes as 129Xe biosensors. Translational and rotational freedom, the consequence of a flexible linker between the cryptophane and the benzensulfonamide, could allow the cage to reorient rapidly in situ, independently of the protein, to result in decreased correlation times and narrower line widths that increase the sensitivity of 129Xe NMR measurements in solution.8
In conclusion, this work reveals the first experimentally determined structure showing how an encapsulated 129Xe atom can be specifically directed to a biomedically relevant protein target. The possible implications for cancer diagnosis are profound, given that CA isozymes IX and XII are overexpressed on the surface of certain cancer cells.18 Moreover, a search of the Protein Data Bank reveals that with its molecular mass of 1554, the 1-Xe complex is one of the largest synthetic organic ligands ever cocrystallized with a protein. Thus, this work demonstrates the feasibility of preparing crystalline complexes between proteins and nonbiological, nanometer-scale ligands.19
Supplementary Material
Acknowledgement
We thank the Advanced Light Source and the Cornell High Energy Synchrotron Source beamline F-2 for access to X-ray crystallographic data collection facilities and Ulrich Englich for support with the Xe chamber experiments. We also thank P.A. Hill for helpful discussions. I.J.D. is a Camille and Henry Dreyfus Teacher-Scholar and thanks DOD for grant W81XWH-04-1-0657. Finally, we thank the NIH for a Chemical Biology Interface training grant (to J.A.A.), 1R21CA110104 (to I.J.D.) and GM49758 (to D.W.C.). D.W.C. also thanks the BBSRC for the Underwood Fellowship.
References
- (1).Supuran CT, Scozzafava A. Bioorg. Med. Chem. 2007;15:4336–4350. doi: 10.1016/j.bmc.2007.04.020. [DOI] [PubMed] [Google Scholar]
- (2).Elbaum D, Nair SK, Patchan MW, Thompson RB, Christianson DWJ. Am. Chem. Soc. 1996;118:8381–8387. [Google Scholar]
- (3).Krishnamurthy VM, Kaufman GK, Urbach AR, Gitlin I, Gudiksen KL, Weibel DB, Whitesides GM. Chem. Rev. 2008;108:946–1051. doi: 10.1021/cr050262p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Bozym RA, Thompson RB, Stoddard AK, Fierke CA. ACS Chem. Biol. 2006;1:103–111. doi: 10.1021/cb500043a. [DOI] [PubMed] [Google Scholar]
- (5).Cherubini A, Bifone A. Prog. Nucl. Magn. Reson. Spectrosc. 2003;42:1–30. [Google Scholar]
- (6).Fain SB, Korosec FR, Holmes JH, O’Halloran R, Sorkness RL, Grist TM. J. Magn. Reson. Imaging. 2007;25:910–923. doi: 10.1002/jmri.20876. [DOI] [PubMed] [Google Scholar]
- (7).Hill PA, Wei Q, Eckenhoff RG, Dmochowski IJ. J. Am. Chem. Soc. 2007;129:9262–9263. doi: 10.1021/ja072965p. [DOI] [PubMed] [Google Scholar]
- (8).Lowery TJ, Garcia S, Chavez L, Ruiz EJ, Wu T, Brotin T, Dutasta JP, King DS, Schultz PG, Pines A, Wemmer DE. ChemBioChem. 2006;7:65–73. doi: 10.1002/cbic.200500327. [DOI] [PubMed] [Google Scholar]
- (9).Schröder L, Lowery TJ, Hilty C, Wemmer DE, Pines A. Science. 2006;314:446–449. doi: 10.1126/science.1131847. [DOI] [PubMed] [Google Scholar]
- (10).Chambers JM, Hill PA, Aaron JA, Han Z, Christianson DW, Kuzma NN, Dmochowski IJ. Manuscript in preparation. [DOI] [PMC free article] [PubMed]
- (11).Alexander RS, Kiefer LL, Fierke CA, Christianson DW. Biochemistry. 1993;32:1510–1518. doi: 10.1021/bi00057a015. [DOI] [PubMed] [Google Scholar]
- (12).Eriksson AE, Kylsten PM, Jones TA, Liljas A. Proteins: Struct. Funct. Gen. 1988;4:283–293. doi: 10.1002/prot.340040407. [DOI] [PubMed] [Google Scholar]
- (13).Watanabe T. Phys. Rev. 1965;137:1380–1382. [Google Scholar]
- (14).Collet A. In: Comprehensive Supramolecular Chemistry. Atwood JL, Davis JED, MacNicol DD, Vogtle F, editors. Vol. 2. Pergamon; New York: 1996. pp. 325–365. Chapter 11. [Google Scholar]
- (15).Ruiz EJ, Sears DN, Pines A, Jameson CJ. J. Am. Chem Soc. 2006;128:16980–16988. doi: 10.1021/ja066661z. [DOI] [PubMed] [Google Scholar]
- (16).Eliel EL, Wilen SH. Stereochemistry of Organic Compounds. John Wiley & Sons, Inc.; New York: 1994. pp. 1119–1190. [Google Scholar]
- (17).Håkansson K, Carlsson M, Svensson LA, Liljas A. J. Mol. Biol. 1992;227:1192–1204. doi: 10.1016/0022-2836(92)90531-n. [DOI] [PubMed] [Google Scholar]
- (18).Pastorekova S, Parkkila S, Zavada J. Adv. Clin. Chem. 2006;42:167–216. [PubMed] [Google Scholar]
- (19). The atomic coordinates of the human carbonic anhydrase II-sulfonamide cryptophane-A complex have been deposited in the Protein Data Bank with accession code 3CYU.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.