Abstract
We reported previously that endogenous reactive oxygen species (ROS) function as myogenic signaling molecules. It has also been determined that excess ROS induce electrophile-response element (EpRE)-driven gene expression via activation of nuclear factor erythroid 2-related factor 2 (Nrf2). Nonetheless, the relationship between the metabolism of ROS (eg, H2O2) through glutathione (GSH) up-regulation, GSH-dependent reduction of H2O2, and Nrf2-dependent gene regulation is not well established. Therefore, we attempted to determine whether H2O2 controls the intracellular GSH redox state via the Nrf2-glutamate-cysteine ligase (GCL)/glutathione reductase (GR)-GSH signaling pathway. In our experiments, enhanced H2O2 generation was accompanied by an increase in both total GSH levels and the GSH/GSSG ratio during muscle differentiation. Both GCL and GR transcriptional expression levels were markedly increased during muscle differentiation but reduced by catalase treatment. Nrf2 protein expression and nuclear translocation increased during myogenesis. The inhibition of GCL, GR, and Nrf2 both by inhibitors and by RNA interference blocked muscle differentiation. Phosphatidylinositol 3-kinase regulated the expression of the GCL C (a catalytic subunit) and GR genes via the induction of Nrf2 nuclear translocation and expression. In conclusion, endogenous H2O2 generated during muscle differentiation not only functions as a signaling molecule, but also regulates the GSH redox state via activation of the Nrf2-GCL/GR-GSH signaling pathway downstream of phosphatidylinositol 3-kinase.
Muscle wasting (cachexia or atrophy) is a frequently observed syndrome in many chronic diseases, such as chronic obstructive pulmonary disease, chronic heart failure, renal failure, AIDS, and cancer.1 Therefore, investigations into the mechanisms of muscle regeneration and myogenesis are critical in the design of such chronic disorder therapies. To initiate the process of muscle differentiation, proliferating myoblasts irreversibly arrest from the cell cycle, followed by fusion with each other, resulting in the formation of multinucleated myotubes.2 Phosphatidylinositol (PI) 3-kinase is one of the essential signaling molecules for muscle differentiation.3 It stimulates myogenesis via the activation of downstream signaling molecules including Akt/protein kinase B,4,5 Rac,6 p70 S6-kinase,7 PLC-γ1,8,9 mTOR,5 and p38 mitogen-activated protein kinase (MAPK).4,10 In contrast, the Raf/MEK/Erk pathway has been shown to stimulate cell proliferation rather than muscle differentiation.11
The reduced form of glutathione (GSH) is the most abundant of the intracellular low molecular weight thiols. The accumulation of oxidized glutathione (GSSG) and the depletion of intracellular GSH have been shown to markedly impair protein function.12 Therefore, the GSH/GSSG ratio has been identified as an important component in the regulation of the protein thiol-disulfide redox status in cells.13 There are two major pathways of GSH supply by which the GSH/GSSG balance can be maintained: one is glutamate-cysteine ligase (GCL) composed of GCLC (a catalytic subunit) and GCLM (a modifier subunit), which is the rate-limiting enzyme in GSH synthesis14; and the other is glutathione reductase (GR), which catalyzes the reduction of GSSG to GSH using NADPH.15
Nuclear factor erythroid 2-related factor 2 (Nrf2), a redox-sensitive transcription factor, binds to the electrophile-response element (EpRE) and regulates the transcriptional activation of EpRE-harboring genes.16,17,18 The GCLC and GCLM of humans and mice, but not rats, were determined to harbor EpRE regulatory elements.19 It has been recently reported that the level of Nrf2 protein can be increased by the induction or stabilization of the Nrf2 protein,17,20 and several signaling molecules, including PI 3-kinase,21 p38 MAPK,22 and ERK1/2,23 have been shown to participate in the translocation and stabilization of Nrf2 in response to a variety of phase 2 gene inducers.
Whereas reactive oxygen species (ROS), which consist principally of superoxide, hydroxyl radicals, and hydrogen peroxide, transmit signals downstream via induced alterations in protein redox status,24,25 H2O2 is the only one that could actually function directly as a second messenger in a physiologically relevant manner.12 We demonstrated previously that ROS are essential mediators of muscle differentiation.6 In addition, GSH has also been shown to regulate muscle differentiation.26 The underlying mechanism by which the GSH redox system is regulated in response to ROS generation during muscle differentiation, however, was not assessed further. In this study, we provide evidence that H2O2 generated during muscle differentiation contributes to the formation of myotubes from myoblasts via the activation of the Nrf2-GCL/GR-GSH redox signaling pathway.
Materials and Methods
Materials
Dulbecco’s modified Eagle’s medium (DMEM), DMEM/F-12, fetal bovine serum, donor calf serum, and G418 were purchased from Invitrogen Life Technologies (Grand Island, NY). Buthionine-sulfoximine (BSO), 1,3-bis-(2-chloroethyl)-1-nitrosourea (BCNU), catalase, 2′,7′-dichlorofluorescein diacetate (DCF-DA), diethyl maleate (DEM), H2O2, Hoechst 33342, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), metaphosphoric acid (MPA), and actinomycin D were acquired from Sigma (St. Louis, MO). LY294002, LY303511, SB203580, and PD98059 were obtained from Tocris Cookson, Ltd. (Bristol, UK). The oligonucleotide probe of EpRE, which was used for the electrophoretic mobility shift assay (EMSA), was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). [γ-32P]ATP was purchased from Amersham Biosciences (Buckinghamshire, UK). Anti-GCLC antibody was donated by Dr. Henry Jay Forman (University of California at Merced, Atwater, CA). Anti-GR antibody was obtained from Lab Frontier (Seoul, Korea). Antibodies specific to Nrf2, myosin heavy chain (MHC), myogenin, actin, Akt, and lamin B protein were purchased from Santa Cruz Biotechnology. Antibody specific to phospho-Akt (S473) was acquired from Cell Signaling Technology (Beverly, MA).
Cell Culture
H9c2 rat cardiac and C2C12 mouse skeletal myoblasts were grown in 10-cm-diameter dishes in DMEM/F-12 containing 10% (v/v) donor calf serum and DMEM supplemented with 10% fetal bovine serum (growth medium, GM), respectively. Cell differentiation was induced by placing them in DMEM/F-12 containing 1% (v/v) horse serum or DMEM containing 1% (v/v) fetal bovine serum (differentiation medium, DM). Full differentiation was achieved 6 days after the induction of differentiation. Differentiation was evaluated by three ways: the expression levels of differentiation markers, including myogenin and MHC 24 and 72 hours, respectively; the morphological changes in myotube formation 6 days; and muscle creatine kinase (MCK)-dependent luciferase assay 48 hours after the induction of differentiation. Unless specified otherwise, the culture media were replaced daily with fresh media. In an attempt to observe the effects of catalase or inhibitors on muscle differentiation or target molecules, they were dissolved in DM and administered to the differentiating cells for the indicated time periods.
Expression Vectors and Stable Transfection
Stable transfectants with the dominant-negative PI 3-kinase (Δp85) construct were previously described.10 Cells transfected with empty pcDNA3.0 (mock) were used as experimental controls.
RNA Interference
The small interfering RNAs (siRNAs) either specific to Nrf2 (Nrf2-siRNA) or consisting of a scrambled sequence (scrambled-siRNA) were prepared by Intron Biotechnology (Kyungki, Korea). Nrf2-siRNA target sequences were as follows: sense, 5′-CCACCUUGAACACAGAUUUdTdT-3′; antisense, 5′-AA AUCUGUGUUCAAGGUGGdTdT-3′. The siRNAs either specific to GR (GR-siRNA) or FLIP (FLIP-siRNA), or consisting of a scrambled sequence (scrambled-siRNA) were purchased from Santa Cruz. The scrambled-siRNAs were used as negative controls. The short hairpin RNAs (shRNAs) specific to GCLC or GFP were purchased from OriGene Technologies, Inc. (Rockville, MD). GFP-shRNA with a vector harboring a noneffective shRNA cassette against GFP was used as a specific negative control for the down-regulation of the gene. The efficiency of siRNA- and shRNA-based interference of Nrf2, GCLC, and GR was monitored via Western blotting analysis. siRNA (40 nmol/L) and shRNA (0.5 μg/ml) were transfected into cells using GenePORTER transfection reagent (Gene Therapy Systems, Inc., San Diego, CA).
GSH Determination
Aliquots of each medium sample were obtained for the determination of extracellular GSH. The cells were washed in phosphate-buffered saline (PBS) and harvested by scraping in 5% MPA for the determination of intracellular GSH levels. After homogenization, the homogenates were incubated for 30 minutes on ice and centrifuged for 20 minutes at 18,000 × g to discard the precipitated proteins, and the GSH and GSSG contents were assessed via high performance liquid chromatography.27 The levels of GSH and GSSG were determined via comparison with the standards and normalized to the protein contents.
Luciferase Assay
H9c2 cells were transfected with 0.5-μg amounts of the pGL3 enhancer vector, GCLC-Luc construct, pGL2 vector, or MCK-Luc construct, coupled with the internal control plasmid, pSV-β-gal (Promega, Madison, WI). The pGL3 and (−1758/+2) GCLC-Luc plasmids were obtained from Dr. Shelly C. Lu (University of California–Los Angeles, Los Angeles, CA). The MCK-Luc vector was generously donated by Dr. KY. Lee (Chungbuk National University, Chungbuk, Korea). Cells incubated in GM or DM for the indicated time periods were harvested in 150 μl of lysis buffer (1% Triton X-100, 25 mmol/L of Gly-Gly, 15 mmol/L of MgSO4, and 2 mmol/L of EGTA, pH 8.0). Luciferase and β-gal activities (data not shown) were measured with a microplate reader (Bio-Rad, Richmond, CA) using 20 μl of each cell lysate, and the luciferase activity was normalized on the basis of β-gal activity.
Analysis of Cellular ROS Levels
Cellular ROS levels were estimated via DCF fluorescence as an indicator in the following two different methods.28
Confocal Microscopy
Cells with the same cell number were seeded on cover glasses in a 12-well microplate. After being compelled to differentiate for the indicated time periods, the cells were incubated for 15 minutes with 10 μmol/L DCF-DA and 10 μmol/L Hoechst 33342, and washed with PBS. Fluorescence was evaluated with a confocal laser microscope (META 510; Zeiss, Thornwood, NY). The excitation and emission wavelengths for the DCF-DA were 488 nm and 525 nm. The nuclei were stained with Hoechst 33342. The detector gains used on the confocal microscope for both dyes were selected to allow for the discrimination of signals proportional to activity. These gain levels remained unaltered throughout the course of the experiments.
Flow Cytometry
The cells were loaded for 30 minutes with 10 μmol/L of DCF-DA at 37°C and resuspended in 0.5 ml of PBS. Fluorescence was assessed via flow cytometry (FACS Calibur; Becton-Dickinson, Franklin Lakes, NJ). The mean DCF fluorescence intensity was assessed with an excitation wavelength of 488 nm and an emission wavelength of 525 nm. The untreated cells were used as a reference for the ROS levels.
Preparation of Total and Nuclear Extracts
Cells were washed three times with PBS and harvested in 500 μl of lysis buffer (20 mmol/L Tris-HCl, pH 7.5, 137 mmol/L NaCl, 1 mmol/L EGTA, pH 8.0, 1% Triton X-100, 10% glycerol, 1.5 mmol/L MgCl2, 10mmol/L NaF, 1 mmol/L phenylmethyl sulfonyl fluoride, 1 mmol/L Na3VO4, 150 nmol/L aprotinin, 1 μmol/L leupeptin hemisulfate). Cellular debris was removed via 15 minutes of centrifugation at 22,000 × g. Whole cell extracts were snap-frozen in liquid nitrogen or stored at −80°C until use. For Western blotting and EMSA analysis, the nuclear extracts were prepared as previously described.6
EMSA
A synthetic double-strand oligonucleotide probe for the EpRE (5′-TGGGGAACCTGTGCTGGTCACTGGAG-3′) was labeled with [γ-32P] ATP. The nuclear extracts were incubated with radiolabeled oligonucleotide probes for the EpRE and analyzed via autoradiography of polyacrylamide gels as previously described.6
RNA Extraction and Semiquantitative Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
Total RNA was extracted with Trizol reagent (Invitrogen, San Diego, CA). RT-PCR was conducted as described previously.6 In brief, the PCR products were amplified using specific primers as follows: GCLC forward, 5′-ATGTGCCAATATTCAAGGAC-3′; GCLC reverse, 5′-CTCTTCAAAAAGGGTCAGTG-3′; GR forward, 5′-GCTTTCAAAACTGCAAGAGT-3′; GR reverse, 5′-ACTGTGAGAACCTCAACACC-3′; Nrf2 forward, 5′-AGTGTCAAACAGAATGGACCTAAAG-3′; Nrf2 reverse, 5′-CTG AGTAGTTTTTCTCTCTCGTCTT-3′; GAPDH forward, 5′-ACTCAGAAGACTGTGGATGG-3′; GAPDH reverse, 5′-GTCATCATACTTGGCAGGTT-3′. The PCR reactions were conducted for 5 minutes at 95°C, followed by 27 cycles of 94°C for 1 minute, 55°C for 1 minute, and 72°C for 1 minute. The amplified products were visualized on 1% agarose gel. GAPDH was used as a loading control.
Real-Time Quantitative RT-PCR
Expression of mRNA for Nrf2, β-actin, and GAPDH was measured via a TaqMan-based real-time RT-PCR assay. The primer and probe sequences designed by Sigma-Proligo (Sigma, The Woodlands, TX) were as follows: Nrf2 forward, 5′-TTCCCAGCCACGTTGAGAGC-3′; Nrf2 reverse, 5′-CTGTAACTCGGGAATGGAAAATAGC-3′; Nrf2 probe, 5′-(6-Fam) AACTTGCTCCATGTCCTGCTGTATGCTGC-(Tamra)-3′; β-actin forward, 5′-GGTATGGGTCAGAAGGACTCC-3′; β-actin reverse, 5′-TGACAATGCCGTGTTCAATGG-3′; β-actin probe, 5′-(6-Fam) ACTTCAGGGTCAGGATGCCTCTCTTGCTC-(Tamra)-3′; GAPDH forward, 5′-TACCCACGGCAAGTTCAACG-3′; GAPDH reverse, 5′-GTGGTGAAGACGCCAGTAGAC-3′; GAPDH probe, 5′-(6-Fam) CCACGACATACTCAGCACCAGCATCACC-(Tamra)-3′. Real-time PCR amplification was conducted in a 96-well optical tray with a final reaction volume of 20 μl containing 10 μl of TaqMan Universal Mastermix (PE Applied Biosystems, Branchburg, NJ), 0.2 μl of forward (20 pmol) and reverse primers (20 pmol), and 1 μl of probe (1 μmol/L), and 20 ng of reverse-transcribed total RNA. It was conducted in an ABI Prism 7300 (PE Applied Biosystems). The relative expression of Nrf2 was normalized to two reference genes, β-actin and GAPDH.
MTT Assay
Cell viability was assessed using the MTT conversion assay in a 12-well plate. The culture medium was replaced by 1 ml of medium containing 0.5 mg/ml of MTT and incubated for 60 minutes at 37°C. The blue-colored tetrazolium crystals resulting from mitochondrial enzymatic activity on MTT substrate were solubilized with 200 μl of dimethyl sulfoxide. The optical density was read at 595 nm in a microplate reader (Bio-Rad). Cell survival was expressed as the percentage of absorbance relative to that of the untreated or untransfected cells.
Western Blotting Analysis
Cell lysates (20 μg protein) were separated via sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electroblotted onto nitrocellulose paper for 1 hour at 100 V at 4°C. The membranes were blocked with 3% bovine serum albumin in TBST (20 mmol/L Tris-HCl, pH 7.5, 50 mmol/L NaCl, 0.1% Tween 20). After blocking, the membranes were incubated for 1 hour with primary antibodies diluted to 1:1000 in TBST. After washing, the membranes were incubated for 1 hour with horseradish peroxidase-labeled secondary antibody diluted to 1:2000 in TBST, and the labeled proteins were detected with chemiluminescence reagents, as described by the manufacturer (Santa Cruz Biotechnology). Unless specified, actin was immunoblotted to standardize the quantity of sample proteins for all Western blotting analyses.
Densitometry and Statistical Analysis
The density of bands in the Western blotting or semiquantitative RT-PCR analysis was determined via densitometry, using Quantity One software (Bio-Rad). The results were expressed as the means ± SE from at least three independent experiments. Statistical analyses were conducted via Student’s t-test. Unless otherwise indicated, a P value of <0.05 was significant.
Results
ROS Generation and Intracellular GSH Redox Changes during Muscle Differentiation
Previously, on a semiquantitative basis, we demonstrated the enhanced ROS generation in differentiating myoblasts.6 Because ROS measurement in myotube-forming cells by flow cytometry analysis was not possible because of technical limitations, we first measured ROS generation in the myotube-forming cells via confocal microscopy using green fluorescent DCF-DA. The Hoechst staining of the same cells revealed the localization of the nuclei. As compared with the proliferation stage, the enhanced ROS generation began to be observed 1 hour after the induction of differentiation, and then was maintained throughout the 6-day period (Figure 1A).
Figure 1.
ROS generation and intracellular GSH redox changes during muscle differentiation. H9c2 cells were grown in GM or further incubated in DM for the indicated time periods. A: DCF-DA and Hoechst 33342 were added to each group of cells, and ROS generation was observed via confocal microscopy. Nuclei were stained with Hoechst 33342. B–D: Samples were prepared from cells incubated in GM or DM for the indicated time periods, and the intracellular total GSH (B) and GSSG levels (C) and the GSH/GSSG ratio (D) were determined. The data are expressed as the means ± SE of at least three independent experiments. *P < 0.05, versus cells in GM. h and d are hours and days after the induction of differentiation. Scale bars = 20 μm. Original magnifications, ×40.
Because ROS were expected to alter the redox status of cells, we next determined the intracellular levels of total GSH, GSSG, and the GSH/GSSG ratio during myogenesis. The levels of total GSH increased from 12 hours, achieving a maximum of ∼2.5-fold at 24 to 48 hours and then decreased gradually from 72 hours after the switch to DM (Figure 1B). The levels of GSSG decreased gradually, and the GSH/GSSG ratio increased significantly as differentiation progressed (Figure 1, C and D). Identical results were observed during the C2C12 differentiation process (data not shown) as observed in the H9c2 cells.
Inhibition of Glutamate-Cysteine Ligase and GR Impairs Myogenesis
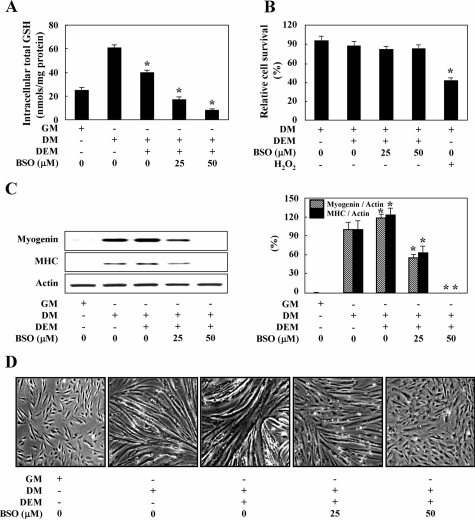
To determine whether GSH depletion impairs muscle differentiation, the cells were initially pretreated for 20 minutes with the thiol alkylating agent, DEM, at a concentration of 50 μmol/L. Furthermore, to prevent GSH synthesis and demonstrate the roles of GCL, we inhibited GCL activity in the myoblasts during the differentiation process, by cultivating the cells in DM with BSO (GCL inhibitor) for 24 hours. DEM treatment alone effected a mild reduction in the intracellular total GSH levels, but BSO treatment for 24 hours in DM dose dependently reduced GSH levels as compared with DM alone (Figure 2A). In addition, to assess the functions of GR in muscle differentiation, we initially inhibited GR activity in myoblasts during the differentiation process by cultivating the cells for 24 hours in DM with BCNU (GR inhibitor). Because the half-life of BCNU in cells was reported to be quite short (∼40 minutes),29 we exchanged the DM with BCNU every 6 hours. BCNU treatment for 24 hours in DM did not detectably reduce the levels of intracellular total GSH even at a relatively high concentration (20 μmol/L) (Figure 3A), but elevated GSSG levels (Figure 3B) and reduced the GSH/GSSG ratio (Figure 3C) in a dose-dependent manner when compared with DM alone. To ascertain whether these results were because of toxic effects of the inhibitors or not, we monitored cell survival 24 hours after BSO or BCNU treatment, using an MTT assay. As shown in Figures 2B and 3D, DEM, BSO, and BCNU at the concentration we used in DM did not significantly affect cell survival, compared with DM alone. The addition of 100 μmol/L H2O2 in DM was used as a positive control, which induced cell damage and abolished the reduction of MTT (Figures 2B and 3D).
Figure 2.
GCL inhibition impairs myogenesis. Cells in DM were treated with 50 μmol/L DEM for 20 minutes followed by the indicated doses of BSO. The intracellular total GSH levels (A) and MTT activity (B) were assessed as described in the Materials and Methods section. The addition of 100 μmol/L H2O2 in DM was used as a positive control. Differentiation was evaluated via the expression of differentiation markers by Western blotting (C, left) and densitometric (C, right) analysis, and morphological changes with photomicrographs (D) after incubation in DM in the absence or presence of DEM and BSO. Actin was used as a loading control. The data are expressed as the means ± SE of at least three independent experiments. *P < 0.05, versus untreated cells in DM. Original magnifications, ×20.
Figure 3.
Inhibition of GR impairs myogenesis. Cells in DM were treated with the indicated doses of BCNU to inhibit GR activity. The intracellular total GSH levels (A), GSSG levels (B), GSH/GSSG ratio (C), and MTT activity assays (D) were assessed. H2O2 in DM was used as a positive control. Differentiation was evaluated via the expression of differentiation markers by Western blotting (E, left) and densitometric (E, right) analysis, and morphological changes with photomicrographs (F) after incubation in DM in the absence or presence of BCNU. Actin was used as a loading control. The data are expressed as the means ± SE of at least three independent experiments. *P < 0.05, versus untreated cells in DM. Original magnifications, ×20.
Treatment with BSO and BCNU in DM dose dependently suppressed the expression of both myogenin and MHC (Figures 2C and 3E) and morphological changes in myotube formation (Figures 2D and 3F), as compared with what was observed with DM alone, suggesting that excessive oxidative stress may impair myogenesis. However, consistent with the results of the previous report,26 the mild depletion of intracellular GSH by treating cells only with DEM in DM induced a slight increase in the expression of both myogenin and MHC and morphological changes in myotube formation, thereby indicating that physiological level of H2O2 indeed accelerates, rather than impairs, myogenesis (Figure 2, A, C, and D).
Next, we suppressed the expression of GCLC and GR by transfecting the cells with shRNA and siRNA specific to them. The cells transfected with GCLC-shRNA and GR-siRNA evidenced an almost complete suppression of GCLC and GR expression during the experiment (Figure 4, A and B). We then assessed their effects on muscle differentiation after myogenic stimulation. As is shown in Figure 4, C and D, the suppression of their expressions resulted in a complete blockage of the expression of both myogenin and MHC and morphological alterations in myotube formation.
Figure 4.
Knockdown of GCLC and GR impairs myogenesis. Cells transfected with shRNA or siRNA specific to GCLC or GR were induced to differentiate in DM. The expression levels of GCLC and GR were determined via Western blotting (A and B, left) and densitometric (A and B, right) analysis. Differentiation was evaluated by the expression of differentiation markers by Western blotting (C, left) and densitometric (C, right) analysis as well as morphological changes with photomicrographs (D) after switching to DM. GFP-shRNA and scrambled-siRNA were used as negative controls for these knockdown experiments. Actin was used as a loading control. The data are expressed as the means ± SE of at least three independent experiments. *P < 0.05, versus cells transfected with GFP-shRNA or scrambled-siRNA in DM. Original magnifications, ×20.
H2O2 Contributes to the Induction of GCLC Subunit and GR during the Myogenic Process
To determine whether GCLC and GR are induced during the myogenic process, we assessed the levels of GCLC and GR protein via Western blotting analysis (Figure 5A) and mRNA levels by semiquantitative RT-PCR analysis (Figure 5B). Rat normal kidney lysates were used as a positive control, because this organ reportedly expressed high levels of GCLC and GR.30 Compared with the proliferation stage in GM, the expression of GCLC protein was shown to have increased markedly during the differentiation process from 12 hours, achieving a maximum level at 24 to 48 hours, and decreasing gradually from 72 to 96 hours. The expression of GR protein apparently increased in a time-dependent manner up to 96 hours after induction of differentiation (Figure 5A). During the normal differentiation process, myogenin and MHC expressions markedly increased (Figure 5A). The mRNA levels of GCLC also increased during differentiation from 12 hours, achieving a peak at 24 to 48 hours, and decreasing gradually from 72 to 96 hours. In contrast, the GR mRNA levels increased gradually in a time-dependent manner up to 96 hours after the induction of differentiation (Figure 5B). Also, we observed identical results during the process of differentiation of C2C12 myoblasts (data not shown), as observed with H9c2.
Figure 5.
Induction of GCLC and GR during muscle differentiation. A and B: Proliferating cells in GM were induced to differentiate for the indicated time periods in DM. The expression patterns of GCLC and GR were determined via Western blotting (A, top), semiquantitative RT-PCR (B, top), and densitometric (A and B, bottom) analysis. Differentiation was evaluated by measuring the expression of myogenin and MHC. The protein sample from rat kidney tissue (3 μg) was loaded as a size marker of GCLC and GR (Con). C: Confluent cells were incubated in GM or further exposed to DM with actinomycin D (5 μg/ml) for the indicated time periods, and semiquantitative RT-PCR analysis was conducted (C, top). Cells incubated in DM for 24 hours were again exposed to GM or DM with actinomycin D, and the mRNA levels of GCLC and GR were determined via semiquantitative RT-PCR analysis (C, bottom). D: Confluent cells were incubated in DM in the absence or presence of catalase and Western blotting analysis was conducted. E and F: Proliferating cells in GM were induced to differentiate for the indicated time periods in DM. The patterns of Mn-SOD expression were determined via Western blotting (E, top), semiquantitative RT-PCR (F, top), and densitometric (E and F, bottom) analysis. Actin and GAPDH were used as loading controls. The data are representative of at least three different experiments and are expressed as the means ± SE. *P < 0.05, versus cells in GM.
We assumed that the increase in the expression of GCLC and GR genes may occur at the transcriptional level by increasing the transcriptional rate, or at the posttranscriptional level via mRNA stabilization. To exclude the possibility of stabilizing mRNA, we used actinomycin D to inhibit de novo mRNA synthesis. In an effort to determine whether GCLC and GR mRNA inductions were inhibited by actinomycin D, we first treated the differentiating cells with 5 μg/ml of actinomycin D for the indicated times, and then assessed their mRNA levels. The results revealed that GCLC and GR mRNA inductions were inhibited completely at 8 hours and 24 hours, respectively, after actinomycin D treatment, which suggested that the half-life of GR might be longer than that of GCLC (Figure 5C, top). We further assessed the decay rates of their mRNAs via treatment with actinomycin D in GM or DM for up to 24 hours after the induction of their mRNAs in DM for 24 hours. As is shown in Figure 5C, bottom, the decay rates of the GCLC and GR mRNAs were shown to be similar in both GM and DM, thereby demonstrating that transcriptional activation induces GCLC and GR during myogenesis.
We also assessed the effects of catalase on the induction of GCLC and GR during myogenesis. As shown, the expressions of GCLC, GR, and myogenin were determined to be reduced 24 hours, and MHC expression was shown to be suppressed 72 hours after the treatment of the cells with catalase in DM, thereby indicating that H2O2 is an essential molecule for muscle differentiation (Figure 5D). In addition, the protein (Figure 5E) and mRNA (Figure 5F) expression levels of MnSOD were up-regulated gradually in a time-dependent manner up to 96 hours after the induction of differentiation, thereby suggesting that increased MnSOD expression may contribute, in part, to the generation of H2O2.
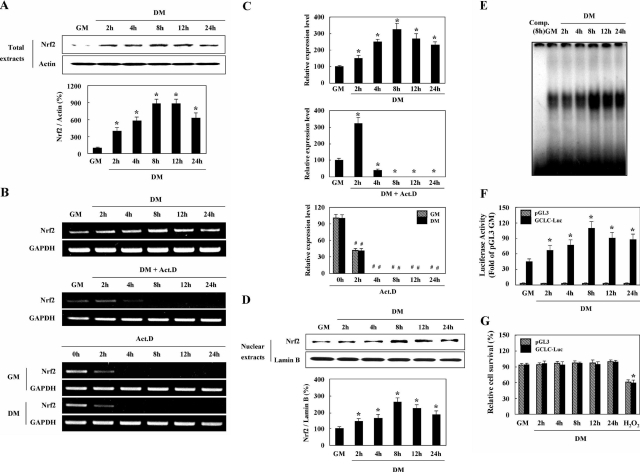
Nrf2 Is Transcriptionally Up-Regulated and Translocated to the Nucleus during Myogenesis
Recently, it was reported that enhanced Nrf2 protein level and its nuclear translocation can be provoked by many inducers, including the phenolic antioxidant tBHQ, and a variety of ROS-producing agents including DEM, sulforaphane, and menadione.31,32 Therefore, we initially assessed whether Nrf2 protein and mRNA expressions are induced during myogenesis via Western blotting (Figure 6A), semiquantitative RT-PCR (Figure 6B, top), and real-time quantitative RT-PCR (Figure 6C, top) analysis. Both Nrf2 protein and mRNA expression levels increased after 2 hours, reached a peak at 8 hours, and then began to decrease from 12 hours until 24 hours after the induction of differentiation. To rule out the possibility of mRNA stabilization, we again treated the cells with actinomycin D, as was done for GCLC and GR. The results of semiquantitative (Figure 6B, middle and bottom) and real-time quantitative (Figure 6C, middle and bottom) RT-PCR analysis indicated that the induction of Nrf2 mRNA was inhibited almost completely after 4 hours (Figure 6, B and C; middle), and the decay rates of Nrf2 mRNA, which was induced for 8 hours in DM, were determined to be similar in both GM and DM (Figure 6, B and C; bottom), thereby indicating that transcriptional activation is the principal cause of the induction of Nrf2 during myogenesis.
Figure 6.
Nrf2 is transcriptionally up-regulated and translocated to the nucleus during myogenesis. A–C: Cells were cultured in GM or DM for the indicated time periods. Expression patterns of total Nrf2 protein were determined via Western blotting (A, top) and densitometric (A, bottom) analysis, and those of Nrf2 mRNA were determined via semiquantitative (B, top) and real-time quantitative (C, top) RT-PCR analysis. In addition, confluent cells were incubated in GM or further exposed to DM with actinomycin D (5 μg/ml) for the indicated time periods, and semiquantitative (B, middle) and real-time quantitative (C, middle) RT-PCR analysis was conducted. Furthermore, cells exposed to DM for 8 hours were incubated in GM or DM with actinomycin D, and the levels of Nrf2 mRNA were determined via semiquantitative (B, bottom) and real-time quantitative (C, bottom) RT-PCR analysis. D: Nuclear extracts were assessed via Western blotting (D, top) and densitometric (D, bottom) analysis. Lamin B was identified as a nuclear protein marker. E: Nuclear fractions were prepared from each sample and EMSA analysis was conducted using the EpRE consensus sequence. The first lane represents competition with 100× unlabeled probe. F and G: Cells were transiently transfected with the pGL3 enhancer vector (pGL3) or the GCLC promoter-luciferase construct (GCLC-Luc) and further incubated in GM or DM for the indicated time periods. The data are expressed in terms of luciferase activity (F) and MTT activity (G), as compared with those of pGL-3 enhancer vector-transfected cells in GM. G: The addition of 100 μmol/L H2O2 in DM was used as a positive control. The data are representative of at least three different experiments and are expressed as the means ± SE. *P < 0.05 versus cells in GM. #P < 0.05 versus untreated cells in GM or DM.
We then determined whether Nrf2 translocates to the nucleus during myogenesis by Western blotting analysis (Figure 6D), EMSA (Figure 6E), and GCLC promoter assay (Figure 6F). Similarly to what was observed with Nrf2 induction, Nrf2 nuclear accumulation, EpRE binding activity, and GCLC promoter activity increased as early in the experiment as 2 hours, achieved a peak level at 8 hours, and began to decrease slightly from 12 hours after the induction of differentiation. The rat GCLC promoter does not contain EpRE. However, Nrf2 was shown to regulate the rat GCLC promoter via the modulation of the expression and trans-activating activity of key nuclear factor (NF)-κB and AP-1 family members.33 Also, NF-κB activation has been suggested to stimulate the process of myogenesis.6,34 Therefore, we suggest that Nrf2 may regulate the transcription of the rat GCLC subunit indirectly via NF-κB during muscle differentiation. To discern whether this result of the luciferase assay was attributable to a toxic effect of the transfection of different constructs, we monitored the survival of the transfected cells after cultivation in GM or DM within 24 hours. As shown, the survival of the differentiating myoblasts was similar, regardless of the transfection of different constructs (Figure 6G).
Role of Nrf2 in the Induction of the Glutamate-Cysteine Ligase Catalytic Subunit and GR during Myogenesis
It was previously reported that Nrf2 transcriptionally regulates GCL subunits and GR, even though putative EpRE sequences have not yet to be identified in the GR promoter.35,36 The cells that were transfected with Nrf2-siRNA evidenced an almost complete suppression of Nrf2 expression after 24 hours of cultivation in GM (Figure 7A, left) or in DM (Figure 7A, right). GCLC and GR expressions in those cells were also reduced markedly. However, no alterations in the levels of phosphorylated Akt (p-Akt) and total Akt in DM (Figure 7A, right) were observed, thereby indicating that PI 3-kinase activity is not affected by Nrf2 knockdown.
Figure 7.
Role of Nrf2 in GCLC and GR induction during muscle differentiation. Cells transfected with siRNA specific to Nrf2 were incubated for 24 hours in GM (A, left) or DM (A, right). A: Total cell lysates were assessed via Western blotting analysis. Differentiation was evaluated by the expression of differentiation markers with Western blotting analysis (B), morphological changes with photomicrographs (C), and an MCK-dependent luciferase assay (D). The intracellular total GSH (E) and ROS (F) levels, as well as the MTT assay (G) were assessed after Nrf2 knockdown. Scrambled-siRNA was used as a negative control for knockdown and FLIP knockdown was used as a positive control for MTT assay. Data represent the means ± SE of at least three independent experiments. *P < 0.05 versus cells transfected with scrambled-siRNA in GM. #P < 0.05 versus cells transfected with scrambled-siRNA in DM. Original magnifications, ×20.
We then attempted to determine whether Nrf2 suppression blocks myogenesis. The knockdown of Nrf2 by siRNA transfection blocked the expression of myogenin and MHC (Figure 7B), as well as morphological changes in myotube formation (Figure 7C). We also verified these results via MCK-dependent luciferase assay using the MCK-responsive luciferase reporter plasmid, MCK-Luc. The knockdown of Nrf2 reduced MCK promoter activity, when measured 48 hours after the induction of differentiation (Figure 7D).
We further observed the effects of Nrf2 on intracellular total GSH content and ROS generation during the processes of proliferation and differentiation 24 hours after the induction of differentiation. Nrf2 knockdown induced a marked reduction in the intracellular total GSH content during both the proliferation and differentiation processes (Figure 7E). Also, Nrf2 knockdown increased DCF fluorescence only in differentiating, but not in proliferating, cells (Figure 7F). Although the reasons for this discrepancy remain unclear, it is possible that even though the intracellular total GSH content is inhibited by the down-regulation of Nrf2 in proliferating cells, the ROS levels of those cells may not increase so apparently as to become detectable by flow cytometry analysis, as is the case in differentiating cells. To rule out the possibility that the decreased viability of the Nrf2-siRNA transfected cells would affect the results, we assessed relative cell survival via MTT assay. As shown in this assay, the survival of the siRNA-transfected cells was similar regardless of Nrf2 knockdown (Figure 7G). Because it has been well documented that the down-regulation of FLIP by chemicals or siRNA sensitizes cells to death receptor-mediated apoptosis,37 we transfected FLIP-siRNA as a positive control in this MTT assay.
Involvement of PI 3-Kinase in the Nrf2 Pathway during Muscle Differentiation
PI 3-kinase has been shown to transmit signals for muscle differentiation.3,4,5,6,7,8,9,10 Therefore, we focused on characterizing the activation of Nrf2, GCLC, and GR downstream of PI 3-kinase. For this purpose, we inhibited PI 3-kinase by LY294002 or via the overexpression of Δp85. To provide further evidence in this regard, we investigated LY303511, a negative analogue of LY294002, which evidences a high degree of structural similarity with LY294002 but completely lacks the ability to inhibit PI 3-kinase. In an effort to assess the effectiveness and specificity of LY294002 treatment, we monitored the phosphorylation of Akt, which was activated by PI 3-kinase during myogenesis. The inhibition of PI 3-kinase by LY294002 for 8 hours after induction of differentiation induced a dose-dependent suppression of Nrf2 nuclear translocation (Figure 8A). In addition, the inhibition of PI 3-kinase by LY294002 for 24 or 72 hours during the differentiation process induced a dose-dependent suppression not only of the expression of phosphorylated Akt, Nrf2, GCLC, GR, and myogenin (24 hours), but also of MHC expression (72 hours) (Figure 8B). Overexpression of Δp85 exerted the same effect on both Nrf2 translocation (Figure 8C) and expression levels of Nrf2, GCLC, and GR (Figure 8D) as LY294002 treatment. Consistent with GCLC suppression, the intracellular total GSH content was reduced by LY294002 treatment for 24 hours in DM in a dose-dependent manner (Figure 8E). To rule out the possibility that the loss of intracellular total GSH is attributable to the increased efflux in the presence of LY294002, we measured the extracellular total GSH content. In parallel with the suppression of GCLC expression and the intracellular total GSH, LY294002 treatment for 24 hours in DM induced a dose-dependent reduction in the extracellular total GSH (Figure 8F), thereby indicating that the intracellular GSH was abrogated principally because LY294002 inhibits GSH synthesis via the suppression of the Nrf2-GCLC pathway downstream of PI 3-kinase. As expected, LY303511 did not show any effects that were demonstrated by LY294002. Although both p38 MAPK and ERK1/2 signaling cascades have been reported to contribute to the activation and stabilization of Nrf2,22,23 we found that SB203580 (inhibitor of p38 MAPK) and PD98059 (inhibitor of ERK1/2) in DM did not suppress the increased nuclear translocation of Nrf2 (Figure 8G, top) and the induction of Nrf2, GCLC, and GR genes (Figure 8G, bottom) during the process of differentiation, thereby indicating that different upstream signaling molecules are involved in the mediation of Nrf2 activation, depending on the cell types and the nature of the stimuli. In addition, as expected, SB203580 in DM inhibited and PD98059 in DM enhanced the expression of myogenin and MHC (Figure 8G, bottom).
Figure 8.
PI 3-kinase is involved in the Nrf2 pathway during muscle differentiation. A: After 8 hours of treatment with LY294002 or LY303511 in DM, the nuclear extracts were assessed via Western blotting (A, top) and densitometric (A, bottom) analysis. B: After treatment with the indicated doses of LY294002 or LY303511 in DM, the total extracts were assessed via Western blotting analysis. Actin was used as a loading control. C and D: In the two clones (1 and 2) of mock and Δp85 transfectants, nuclear extracts were analyzed via Western blotting analysis to observe Nrf2 translocation in GM or in DM for 8 hours (C). After mock and Δp85 transfectants were cultivated in GM or DM for 24 hours, Western blotting analysis was conducted (D). Mock transfectants were used as negative controls. E and F: Cells were treated for 24 hours with LY294002 or LY303511 in DM and the intracellular (E) and extracellular (F) total GSH content was measured. The data are expressed as the means ± SE of at least three independent experiments. *P < 0.05, versus untreated cells. G: After treatment with 20 μmol/L of SB203580 or PD98059 for 8 (top) or 24 (bottom) hours in DM, the nuclear or total extracts were assessed via Western blotting analysis.
Discussion
In this study, we first noted that H2O2 generated during muscle differentiation performs dual functions as a signal messenger molecule for muscle differentiation, and as a mediator for antioxidant GSH synthesis via Nrf2 activation, followed by the increased expression of GCLC and GR. We also noted that PI 3-kinase activity is required to increase Nrf2 nuclear translocation and expression.
H2O2 has also been implicated as an essential signaling mediator in the fields of the differentiation process as well as cellular proliferation and apoptosis. For example, the elevation of H2O2 generation performs a crucial function in cardiomyogenesis in the embryoid bodies,38 NGF-induced neuronal cell differentiation in PC 12 cells,39 and smooth muscle cell differentiation.40 Also, NADPH oxidase (Nox) 4-derived ROS have been demonstrated to drive cardiac differentiation.41 Furthermore, we previously reported that a rise in ROS generation by Nox 2 is crucial to muscle differentiation.6,42 In the present study, we have again confirmed the enhanced ROS generation from the very early stage to the fully differentiated myotube formation stage via confocal microscopy, as compared with the proliferation stage. We also observed that catalase blocks muscle differentiation. In addition, the protein and mRNA levels of MnSOD and mitochondrial ROS (data not shown) were increased markedly during the myogenic process. Therefore, we suggest that although it is not easy to determine which specific component of ROS increased in the DCF experiment, H2O2 may be a major signaling mediator for muscle differentiation. It seems that the superoxide generated by Nox 2 and mitochondria would be a precursor for H2O2 and would be pulled rapidly forward to H2O2 by SOD. However, the possibility cannot be dismissed that other ROS members function as signaling molecules by influencing the redox state in the cells. Interestingly, there have been many contrary reports that ROS generated by TNF-α or exogenous H2O2 inhibits muscle differentiation,43,44,45 and ROS released from the cardiac myocyte are linked to the progression of heart failure.46 This probably reflects differences in the site of H2O2 production and/or the quantity of H2O2.
Exogenous H2O2 has been demonstrated to induce Nrf2 activation and regulate the expression of protective genes in response to oxidative stress.18 In contrast, endogenous H2O2, functioning as a signaling molecule, acts locally and does not exist with a sufficiently high quantity to affect Nrf2 activation and total GSH levels. Interestingly, our results conclusively demonstrated that H2O2 generated during muscle differentiation not only stimulates the process of muscle differentiation, but also increases the levels of intracellular total GSH and the GSH/GSSG ratio via the activation of the Nrf2-GCL/GR pathway. Our conclusion is based on the following findings. First, the levels of intracellular total GSH and the GSH/GSSG ratio increased during differentiation, as compared with proliferating myoblasts, in both H9c2 and C2C12 (data not shown) cells. Second, the differentiating myoblasts increased GCLC and GR expressions at the transcriptional level. Third, catalase suppressed the induction of the GCLC and GR genes. Fourth, Nrf2 was induced by transcriptional activation to express higher amounts of protein. Also, Nrf2 was translocated to the nucleus from the cytoplasm to regulate the gene expressions of GCLC and GR during muscle differentiation. Finally, the pharmacological interventions of GCLC and GR using BSO and BCNU, or the specific knockdown of Nrf2, GCLC, or GR by target-specific RNA interference blocked muscle differentiation by affecting the cellular redox state. Consistent with our results, GSH depletion was reported to suppress muscle differentiation.26
The increased protein and mRNA levels of GCLC peaked at 24 to 48 hours and then decreased slightly from 72 hours to 96 hours after the induction of differentiation. In contrast, the protein and mRNA levels of GR increased steadily up to 96 hours after the initiation of differentiation (Figure 5, A and B). Consistently, the levels of intracellular total GSH achieved a maximum value at 24 to 48 hours and then decreased gradually after 72 hours in DM, whereas the GSH/GSSG ratio increased continuously until 96 hours during myogenesis (Figure 1D). Considering that the level of Nrf2 gene expression reached a maximum level at 8 hours and its maximal translocation to the nucleus from the cytoplasm occurred 8 hours after the induction of differentiation, the continuous increase in the protein and mRNA levels of GR suggests that additional factors may be involved. Although the mechanisms remain to be explored, we speculate that the greater mRNA stability of GR (as compared to GCLC) may be involved. In fact, our results demonstrated that GR mRNA was sustained for a longer time than was GCLC mRNA after actinomycin D treatment (Figure 5C). Further studies will also be required to explain how Nrf2 gene induction or nuclear translocation does not persist even though ROS generation is persistent during muscle differentiation.
PI 3-kinase is an essential molecule for muscle differentiation.3,4,5,6,7,8,9,10 It was also shown to be pivotal for cellular redox homeostasis via the synthesis of GSH in myocytes.47 PI 3-kinase inhibitor LY294002 inhibited the nuclear translocation of Nrf2 and EpRE-mediated transactivation.36,48 Furthermore, Nrf2 has been shown to autoregulate its own expression via EpRE-like cis-elements in the promoter.20 Therefore, we thought that inhibition of PI 3-kinase may suppress Nrf2 expression via blocking its nuclear translocation and subsequent autoregulation. Consistent with this hypothesis, our results also indicated that LY294002 and the dominant-negative form of PI 3-kinase, Δp85, reduced Nrf2 nuclear translocation as well as its induction, followed by reduced expression of both the GCLC and GR genes. Therefore, we concluded that the PI 3-kinase-dependent signaling pathway performs a key function in the induction of GCLC and GR via Nrf2 during muscle differentiation.
In conclusion, we demonstrated that H2O2 functions as a stimulator of GSH synthesis during muscle differentiation. We also demonstrated that the Nrf2-GCLC/GR pathway downstream of PI 3-kinase is a principal regulating system for cellular redox homeostasis during muscle differentiation. Our findings will provide some critical insights into the mechanisms inherent to muscle regeneration and myogenesis, which will prove valuable for the design of therapeutic modalities for chronic muscle disorders, including muscle wasting.
Acknowledgments
We thank Dr. K.Y. Lee for providing us with the MCK-Luc vectors and Dr. Shelly C. Lu for the generous gift of GCLC-Luc vectors.
Footnotes
Address reprint requests to Kyung-Sik Yoon, M.D., Ph.D., or Sung Soo Kim, M.D., Ph.D., Department of Biochemistry and Molecular Biology, School of Medicine, Kyung Hee University, #1, Hoegi-dong, Dongdaemoon-gu, Seoul 130-701, Korea. E-mail: sky9999@khu.ac.kr (K.-S.Y.) or sgskim@khu.ac.kr (S.S.K.).
Supported by the Korea Science and Engineering Foundation (grant no. R13-2002-020-02001-0 2007 to S.S.K.) and the National Institutes of Health (grant ES05511 to H.J.F.).
References
- Lynch GS, Schertzer JD, Ryall JG. Therapeutic approaches for muscle wasting disorders. Pharmacol Ther. 2007;113:461–487. doi: 10.1016/j.pharmthera.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Seale P, Rudnicki MA. A new look at the origin, function, and “stem-cell” status of muscle satellite cells. Dev Biol. 2000;218:115–124. doi: 10.1006/dbio.1999.9565. [DOI] [PubMed] [Google Scholar]
- Lim MJ, Choi KJ, Ding Y, Kim JH, Kim BS, Kim YH, Lee J, Choe W, Kang I, Ha J, Yoon KS, Kim SS. RhoA/Rho kinase blocks muscle differentiation via serine phosphorylation of insulin receptor substrate-1 and -2. Mol Endocrinol. 2007;21:2282–2293. doi: 10.1210/me.2007-0114. [DOI] [PubMed] [Google Scholar]
- Gonzalez I, Tripathi G, Carter EJ, Cobb LJ, Salih DA, Lovett FA, Holding C, Pell JM. Akt2, a novel functional link between p38 mitogen-activated protein kinase and phosphatidylinositol 3-kinase pathways in myogenesis. Mol Cell Biol. 2004;24:3607–3622. doi: 10.1128/MCB.24.9.3607-3622.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latres E, Amini AR, Amini AA, Griffiths J, Martin FJ, Wei Y, Lin HC, Yancopoulos GD, Glass DJ. Insulin-like growth factor-1 (IGF-1) inversely regulates atrophy-induced genes via the phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin (PI3K/Akt/mTOR) pathway. J Biol Chem. 2005;280:2737–2744. doi: 10.1074/jbc.M407517200. [DOI] [PubMed] [Google Scholar]
- Piao YJ, Seo YH, Hong F, Kim JH, Kim YJ, Kang MH, Kim BS, Jo SA, Jo I, Jue DM, Kang I, Ha J, Kim SS. Nox 2 stimulates muscle differentiation via NF-kappaB/iNOS pathway. Free Radic Biol Med. 2005;38:989–1001. doi: 10.1016/j.freeradbiomed.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Coolican SA, Samuel DS, Ewton DZ, McWade FJ, Florini JR. The mitogenic and myogenic actions of insulin-like growth factors utilize distinct signaling pathways. J Biol Chem. 1997;272:6653–6662. doi: 10.1074/jbc.272.10.6653. [DOI] [PubMed] [Google Scholar]
- Faenza I, Ramazzotti G, Bavelloni A, Fiume R, Gaboardi GC, Follo MY, Gilmour RS, Martelli AM, Ravid K, Cocco L. Inositide-dependent phospholipase C signaling mimics insulin in skeletal muscle differentiation by affecting specific regions of the cyclin D3 promoter. Endocrinology. 2007;148:1108–1117. doi: 10.1210/en.2006-1003. [DOI] [PubMed] [Google Scholar]
- Hong F, Moon K, Kim SS, Kim YS, Choi YK, Bae YS, Suh PG, Ryu SH, Choi EJ, Ha J, Kim SS. Role of phospholipase C-gamma1 in insulin-like growth factor I-induced muscle differentiation of H9c2 cardiac myoblasts. Biochem Biophys Res Commun. 2001;282:816–822. doi: 10.1006/bbrc.2001.4644. [DOI] [PubMed] [Google Scholar]
- Chun YK, Kim J, Kwon S, Choi SH, Hong F, Moon K, Kim JM, Choi SL, Kim BS, Ha J, Kim SS. Phosphatidylinositol 3-kinase stimulates muscle differentiation by activating p38 mitogen-activated protein kinase. Biochem Biophys Res Commun. 2000;276:502–507. doi: 10.1006/bbrc.2000.3486. [DOI] [PubMed] [Google Scholar]
- Rommel C, Clarke BA, Zimmermann S, Nunez L, Rossman R, Reid K, Moelling K, Yancopoulos GD, Glass DJ. Differentiation stage-specific inhibition of the Raf-MEK-ERK pathway by Akt. Science. 1999;286:1738–1741. doi: 10.1126/science.286.5445.1738. [DOI] [PubMed] [Google Scholar]
- Forman HJ, Fukuto JM, Torres M. Redox signaling: thiol chemistry defines which reactive oxygen and nitrogen species can act as second messengers. Am J Physiol. 2004;287:C246–C256. doi: 10.1152/ajpcell.00516.2003. [DOI] [PubMed] [Google Scholar]
- Schafer FQ, Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic Biol Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- Griffith OW. Biologic and pharmacologic regulation of mammalian glutathione synthesis. Free Radic Biol Med. 1999;27:922–935. doi: 10.1016/s0891-5849(99)00176-8. [DOI] [PubMed] [Google Scholar]
- Carlberg I, Mannervik B. Glutathione reductase. Methods Enzymol. 1985;113:484–490. doi: 10.1016/s0076-6879(85)13062-4. [DOI] [PubMed] [Google Scholar]
- Moi P, Chan K, Asunis I, Cao A, Kan YW. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc Natl Acad Sci USA. 1994;91:9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T, Sherratt PJ, Huang HC, Yang CS, Pickett CB. Increased protein stability as a mechanism that enhances Nrf2-mediated transcriptional activation of the antioxidant response element. Degradation of Nrf2 by the 26 S proteasome. J Biol Chem. 2003;278:4536–4541. doi: 10.1074/jbc.M207293200. [DOI] [PubMed] [Google Scholar]
- Purdom-Dickinson SE, Lin Y, Dedek M, Morrissy S, Johnson J, Chen QM. Induction of antioxidant and detoxification response by oxidants in cardiomyocytes: evidence from gene expression profiling and activation of Nrf2 transcription factor. J Mol Cell Cardiol. 2007;42:159–176. doi: 10.1016/j.yjmcc.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moinova HR, Mulcahy RT. An electrophile responsive element (EpRE) regulates beta-naphthoflavone induction of the human gamma-glutamylcysteine synthetase regulatory subunit gene. Constitutive expression is mediated by an adjacent AP-1 site. J Biol Chem. 1998;273:14683–14689. doi: 10.1074/jbc.273.24.14683. [DOI] [PubMed] [Google Scholar]
- Kwak MK, Itoh K, Yamamoto M, Kensler TW. Enhanced expression of the transcription factor Nrf2 by cancer chemopreventive agents: role of antioxidant response element-like sequences in the nrf2 promoter. Mol Cell Biol. 2002;22:2883–2892. doi: 10.1128/MCB.22.9.2883-2892.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaso K, Yano H, Fukuhara Y, Takeshima T, Wada-Isoe K, Nakashima K. PI3K is a key molecule in the Nrf2-mediated regulation of antioxidative proteins by hemin in human neuroblastoma cells. FEBS Lett. 2003;546:181–184. doi: 10.1016/s0014-5793(03)00517-9. [DOI] [PubMed] [Google Scholar]
- Anwar AA, Li FY, Leake DS, Ishii T, Mann GE, Siow RC. Induction of heme oxygenase 1 by moderately oxidized low-density lipoproteins in human vascular smooth muscle cells: role of mitogen-activated protein kinases and Nrf2. Free Radic Biol Med. 2005;39:227–236. doi: 10.1016/j.freeradbiomed.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Xu C, Yuan X, Pan Z, Shen G, Kim JH, Yu S, Khor TO, Li W, Ma J, Kong AN. Mechanism of action of isothiocyanates: the induction of ARE-regulated genes is associated with activation of ERK and JNK and the phosphorylation and nuclear translocation of Nrf2. Mol Cancer Ther. 2006;5:1918–1926. doi: 10.1158/1535-7163.MCT-05-0497. [DOI] [PubMed] [Google Scholar]
- Klatt P, Lamas S. Regulation of protein function by S-glutathiolation in response to oxidative and nitrosative stress. Eur J Biochem. 2000;267:4928–4944. doi: 10.1046/j.1432-1327.2000.01601.x. [DOI] [PubMed] [Google Scholar]
- Zheng M, Aslund F, Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]
- Ardite E, Barbera JA, Roca J, Fernandez-Checa JC. Glutathione depletion impairs myogenic differentiation of murine skeletal muscle C2C12 cells through sustained NF-kappaB activation. Am J Pathol. 2004;165:719–728. doi: 10.1016/s0002-9440(10)63335-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinberg L, Fibach E, Am J, Atlas D. N-acetylcysteine amide, a novel cell-permeating thiol, restores cellular glutathione and protects human red blood cells from oxidative stress. Free Radic Biol Med. 2005;38:136–145. doi: 10.1016/j.freeradbiomed.2004.09.025. [DOI] [PubMed] [Google Scholar]
- Bonini MG, Rota C, Tomasi A, Mason RP. The oxidation of 2′,7′-dichlorofluorescin to reactive oxygen species: a self-fulfilling prophesy? Free Radic Biol Med. 2006;40:968–975. doi: 10.1016/j.freeradbiomed.2005.10.042. [DOI] [PubMed] [Google Scholar]
- Ueda-Kawamitsu H, Lawson TA, Gwilt PR. In vitro pharmacokinetics and pharmacodynamics of 1,3-bis(2-chloroethyl)-1-nitrosourea (BCNU). Biochem Pharmacol. 2002;63:1209–1218. doi: 10.1016/s0006-2952(02)00878-x. [DOI] [PubMed] [Google Scholar]
- Li S, Thompson SA, Kavanagh TJ, Woods JS. Localization by in situ hybridization of gamma-glutamylcysteine synthetase mRNA expression in rat kidney following acute methylmercury treatment. Toxicol Appl Pharmacol. 1996;141:59–67. doi: 10.1006/taap.1996.0260. [DOI] [PubMed] [Google Scholar]
- Nguyen T, Sherratt PJ, Nioi P, Yang CS, Pickett CB. Nrf2 controls constitutive and inducible expression of ARE-driven genes through a dynamic pathway involving nucleocytoplasmic shuttling by Keap1. J Biol Chem. 2005;280:32485–32492. doi: 10.1074/jbc.M503074200. [DOI] [PubMed] [Google Scholar]
- Itoh K, Wakabayashi N, Katoh Y, Ishii T, O'Connor T, Yamamoto M. Keap1 regulates both cytoplasmic-nuclear shuttling and degradation of Nrf2 in response to electrophiles. Genes Cell. 2003;8:379–391. doi: 10.1046/j.1365-2443.2003.00640.x. [DOI] [PubMed] [Google Scholar]
- Yang H, Magilnick N, Lee C, Kalmaz D, Ou X, Chan JY, Lu SC. Nrf1 and Nrf2 regulate rat glutamate-cysteine ligase catalytic subunit transcription indirectly via NF-kappaB and AP-1. Mol Cell Biol. 2005;25:5933–5946. doi: 10.1128/MCB.25.14.5933-5946.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeza-Raja B, Munoz-Canoves P. p38 MAPK-induced nuclear factor-kappaB activity is required for skeletal muscle differentiation: role of interleukin-6. Mol Biol Cell. 2004;15:2013–2026. doi: 10.1091/mbc.E03-08-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Itoh K, Yamamoto M, Zweier JL, Li Y. Role of Nrf2 signaling in regulation of antioxidants and phase 2 enzymes in cardiac fibroblasts: protection against reactive oxygen and nitrogen species-induced cell injury. FEBS Lett. 2005;579:3029–3036. doi: 10.1016/j.febslet.2005.04.058. [DOI] [PubMed] [Google Scholar]
- Lee JM, Johnson JA. An important role of Nrf2-ARE pathway in the cellular defense mechanism. J Biochem Mol Biol. 2004;37:139–143. doi: 10.5483/bmbrep.2004.37.2.139. [DOI] [PubMed] [Google Scholar]
- Wajant H. Targeting the FLICE inhibitory protein (FLIP) in cancer therapy. Mol Interv. 2003;3:124–127. doi: 10.1124/mi.3.3.124. [DOI] [PubMed] [Google Scholar]
- Sauer H, Rahimi G, Hescheler J, Wartenberg M. Role of reactive oxygen species and phosphatidylinositol 3-kinase in cardiomyocyte differentiation of embryonic stem cells. FEBS Lett. 2000;476:218–223. doi: 10.1016/s0014-5793(00)01747-6. [DOI] [PubMed] [Google Scholar]
- Suzukawa K, Miura K, Mitsushita J, Resau J, Hirose K, Crystal R, Kamata T. Nerve growth factor-induced neuronal differentiation requires generation of Rac1-regulated reactive oxygen species. J Biol Chem. 2000;275:13175–13178. doi: 10.1074/jbc.275.18.13175. [DOI] [PubMed] [Google Scholar]
- Su B, Mitra S, Gregg H, Flavahan S, Chotani MA, Clark KR, Goldschmidt-Clermont PJ, Flavahan NA. Redox regulation of vascular smooth muscle cell differentiation. Circ Res. 2001;89:39–46. doi: 10.1161/hh1301.093615. [DOI] [PubMed] [Google Scholar]
- Li J, Stouffs M, Serrander L, Banfi B, Bettiol E, Charnay Y, Steger K, Krause KH, Jaconi ME. The NADPH oxidase NOX4 drives cardiac differentiation: role in regulating cardiac transcription factors and MAP kinase activation. Mol Biol Cell. 2006;17:3978–3988. doi: 10.1091/mbc.E05-06-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong F, Lee J, Song JW, Lee SJ, Ahn H, Cho JJ, Ha J, Kim SS. Cyclosporin A blocks muscle differentiation by inducing oxidative stress and inhibiting the peptidyl-prolyl-cis-trans isomerase activity of cyclophilin A: cyclophilin A protects myoblasts from cyclosporin A-induced cytotoxicity. FASEB J. 2002;16:1633–1635. doi: 10.1096/fj.02-0060fje. [DOI] [PubMed] [Google Scholar]
- Hansen JM, Klass M, Harris C, Csete M. A reducing redox environment promotes C2C12 myogenesis: implications for regeneration in aged muscle. Cell Biol Int. 2007;31:546–553. doi: 10.1016/j.cellbi.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langen RC, Schols AM, Kelders MC, van der Velden JLJ, Wouters EF, Janssen-Heininger YM. Tumor necrosis factor-alpha inhibits myogenesis through redox-dependent and -independent pathways. Am J Physiol. 2002;283:C714–C721. doi: 10.1152/ajpcell.00418.2001. [DOI] [PubMed] [Google Scholar]
- Layne MD, Farmer SR. Tumor necrosis factor-alpha and basic fibroblast growth factor differentially inhibit the insulin-like growth factor-I induced expression of myogenin in C2C12 myoblasts. Exp Cell Res. 1999;249:177–187. doi: 10.1006/excr.1999.4465. [DOI] [PubMed] [Google Scholar]
- Sorescu D, Griendling KK. Reactive oxygen species, mitochondria, and NAD(P)H oxidases in the development and progression of heart failure. Congest Heart Fail. 2002;8:132–140. doi: 10.1111/j.1527-5299.2002.00717.x. [DOI] [PubMed] [Google Scholar]
- Li S, Li X, Rozanski GJ. Regulation of glutathione in cardiac myocytes. J Mol Cell Cardiol. 2003;35:1145–1152. doi: 10.1016/s0022-2828(03)00230-x. [DOI] [PubMed] [Google Scholar]
- Lee JM, Hanson JM, Chu WA, Johnson JA. Phosphatidylinositol 3-kinase, not extracellular signal-regulated kinase, regulates activation of the antioxidant-responsive element in IMR-32 human neuroblastoma cells. J Biol Chem. 2001;276:20011–20016. doi: 10.1074/jbc.M100734200. [DOI] [PubMed] [Google Scholar]