Abstract
Rationale: Independent replication of genetic associations in complex diseases, particularly in whole-genome association studies, is critical to confirm the association.
Objectives: A whole-genome association study identified ORMDL3 as a promising candidate gene for asthma in white populations. Here, we attempted to confirm the role of ORMDL3 genetic variants in asthma in three ethnically diverse populations: Mexican, Puerto Rican, and African American.
Methods: We used family-based analyses to test for association between seven candidate single-nucleotide polymorphisms (SNPs) in and around the ORMDL3 gene and asthma and related phenotypes in 701 Puerto Rican and Mexican parent–child trios. We also evaluated these seven SNPs and an additional ORMDL3 SNP in 264 African American subjects with asthma and 176 healthy control subjects.
Measurements and Main Results: We found significant associations between two SNPs within ORMDL3 (rs4378650 and rs12603332) and asthma in Mexicans and African Americans (P = 0.028 and 0.001 for rs4378650 and P = 0.021 and 0.001 for rs12603332, respectively), and a trend toward association in Puerto Ricans (P = 0.076 and 0.080 for SNPs rs4378650 and rs12603332, respectively). These associations became stronger among Mexican and Puerto Rican subjects with asthma with IgE levels greater than 100 IU/ml. We did not find any association between ORMDL3 SNPs and baseline lung function or response to the bronchodilator albuterol.
Conclusions: Our results confirm that the ORMDL3 locus is a risk factor for asthma in ethnically diverse populations. However, inconsistent SNP-level results suggest that further studies will be needed to determine the mechanism by which ORMDL3 predisposes to asthma.
Keywords: asthma, genetics, ORMDL3, Latinos, African Americans
AT A GLANCE COMMENTARY
Scientific Knowledge on This Subject
A whole-genome association study identified the gene ORMDL3 as a promising candidate for asthma in white populations. This association has not yet been independently replicated in nonwhite populations.
What This Study Adds to the Field
This study confirms the association between ORMDL3 and asthma. The association in three genetically diverse populations provides strong support for its role in asthma susceptibility.
Asthma is a complex disease with both genetic and environmental risk factors (1, 2). More than 100 gene candidates have previously been associated with asthma, but fewer than half of the associations have been successfully replicated in multiple populations (3). Moffatt and colleagues performed a whole-genome association study on a cohort of 994 British and German white patients with childhood asthma and 1,243 control subjects, using both family and case–control cohorts (4). They identified a region on chromosome 17q21 that was strongly associated with asthma and replicated the results in two large cohorts (also of German and British origin). ORMDL3 was identified as a candidate gene for asthma by evaluating the association between genetic markers in the region and gene transcript levels in Epstein-Barr virus–transformed lymphoblastoid cell lines from the children with asthma. The study found that ORMDL3 transcription levels were strongly correlated with several of the genetic markers near the ORMDL3 gene.
The ORMDL3 gene is 6,560 base pairs long and includes three exons, which encode a 153–amino acid protein with four putative transmembrane domains whose function is unknown. ORMDL3 and two other members of its gene family were first identified by protein sequence comparisons by Hjelmqvist and coworkers, who showed significant sequence conservation to homologs in Drosophila and Saccharomyces cerevisiae (5). ORMDL3 is expressed ubiquitously and its protein product is localized to the endoplasmic reticulum membrane. Double knockouts of the homologs found in Saccharomyces cerevisiae led to impaired growth and higher sensitivity to toxins. The knockouts could be rescued by transformation with vectors bearing ORMDL3 under the control of a constitutive promoter. It is hypothesized that the ORMDL3 gene product has a role in protein folding, but no further studies have been performed to confirm this hypothesis.
There is great variability in the burden of asthma between populations. In the United States, the morbidity and mortality of asthma are highest in African Americans and Puerto Ricans and lowest in Mexicans (6, 7). The genetic risk factors for asthma have been shown to vary within and between populations (8). To determine whether the ORMDL3 gene is a risk factor for asthma in nonwhite populations, we sought to replicate the association between the ORMDL3 gene and asthma in three independent cohorts of Mexican, Puerto Rican, and African American individuals with asthma. The Genetics of Asthma in Latino Americans (GALA) study is an ongoing, multicenter, international, collaborative effort to identify novel clinical and genetic risk factors associated with asthma, and related phenotypes in Mexicans and Puerto Ricans, the two largest Latino populations in the United States. The study includes 701 family trios consisting of a proband with asthma and both biological parents (9). The Study of African Americans, Asthma, Genes, and Environments (SAGE) likewise seeks to identify genetic and environmental risk factors associated with asthma, asthma severity, and drug response in African Americans and consists of 264 cases with asthma and 176 healthy control subjects (10).
METHODS
Subject Recruitment
Mexican and Puerto Rican subjects with asthma (probands) and their biological parents were recruited for the GALA study. Three hundred and ninety-nine Puerto Rican children and their parents were recruited from Puerto Rico (241) and New York City (158), and Mexican children were recruited in Mexico City (100) and the San Francisco Bay Area (201). Probands were enrolled if they had a physician diagnosis of asthma and were either taking a medication for asthma or had two or more asthma-related symptoms (wheezing, coughing, and/or dyspnea). African American subjects with asthma (261) between 8 and 40 years of age with physician-diagnosed asthma and two or more asthma symptoms (wheezing, coughing, and/or dyspnea) in the previous 2 years were recruited from three San Francisco Bay Area asthma specialty clinics. One hundred and seventy-six healthy African Americans between 8 and 40 years of age without symptoms of asthma (wheezing, cough, or dyspnea) or a diagnosis of asthma or other lung disease or chronic illness, and not taking any medications, were enrolled as control subjects. Patients enrolled in both studies were included on the basis of self-identified ethnicity, and only if all four biological grandparents were of the same ethnicity. Detailed recruitment criteria and subject characteristics are described elsewhere (9–12). Local institutional review boards approved all the studies, and all subjects provided written, age-appropriate informed consent or assent. Trained interviewers administered all cases and probands a modified version of the 1978 American Thoracic Society–Division of Lung Diseases Epidemiology Questionnaire (13). Blood samples were collected from each participant and were processed, anonymously labeled, and stored at the University of California, San Francisco DNA Bank (http://www.genomics.ucsf.edu/DNA_Bank/index.aspx).
Pulmonary Function Tests and IgE Measurements
Spirometry was performed according to American Thoracic Society guidelines (14). Pulmonary function test results are shown in Table 1 and are expressed as a percentage of the predicted normal value, using race- and age-adjusted prediction equations from Hankinson and coworkers (15). A quantitative measure of bronchodilator responsiveness was calculated as ΔFEV1, which is relative percent change in pre-FEV1 after administration of 180 μg of albuterol. Total plasma IgE was measured in duplicate, using ImmunoCAP (Phadia US, Portage, MI [formerly UniCAP, Pharmacia/Upjohn, Kalamazoo, MI]).
TABLE 1.
BASELINE CLINICAL CHARACTERISTICS AND SPIROMETRIC VALUES FOR PUERTO RICAN AND MEXICAN PROBANDS WITH ASTHMA AND FOR AFRICAN AMERICAN SUBJECTS WITH ASTHMA
| Characteristic | Puerto Ricans (n = 399: 241 from Puerto Rico, 158 from New York) | Mexicans (n =301: 100 from Mexico, 201 from San Francisco) | African Americans (n = 261) | P Value for Comparison |
|---|---|---|---|---|
| Age, yr | 12.0 (10–15) | 13.1 (11–19) | 16.1 (12–24) | 0.0001 |
| Sex, % male | 55.9 | 54.1 | 40.2 | 0.0001 |
| BMI, kg/m2 | 21.2 (17–26) | 23.8 (20–28) | 25.5 (21–32) | 0.0001 |
| Serum IgE, IU/ml | 258 (92–627) | 270 (99–611) | 118 (40–360) | 0.0007 |
| Spirometry | ||||
| Pre-FEV1, % predicted | 83.1 (74–93) | 90.0 (77–100) | 93.0 (83–102) | 0.0001 |
| Pre-FEV1 < 80% predicted, % | 40.5 | 30.8 | 21.5 | <0.0001 |
| FVC, % predicted | 93.8 (83–105) | 97.7 (87–109) | 98.4 (89–109) | 0.0026 |
| FEV1/FVC, % predicted | 90.3 (82–97) | 91.8 (86–99) | 94.4 (86–100) | 0.0010 |
| ΔFEV1, relative % predicted | 5.0 (0.6–10) | 7.4 (4–12) | 7.0 (4–14) | 0.0001 |
Definition of abbreviation: BMI = body mass index.
Data are given as medians and interquartile ranges.
Single-Nucleotide Polymorphism Selection
A panel of single-nucleotide polymorphisms (SNPs) was selected from the publicly available HapMap database (see http://www.hapmap.org/) for genotyping. SNPs were selected on the basis of the following criteria: (1) They were either within the ORMDL3 gene or within 500 base pairs upstream or downstream of the gene; and (2) they had a reported minor allele frequency of greater than 5% in white (CEU), African (YRI), or combined Asian (JPT + HCB) populations. Seven SNPs were identified for further screening on the basis of these criteria. In addition to these seven SNPs, three SNPs found associated with asthma in the Moffat and coworkers study (rs7216389, rs11650680, and rs3859192) were also included.
Because the HapMap project does not have genotype data on Latino or African American samples, we genotyped the 10 selected SNPs in a panel of 72 unrelated subjects with asthma, 24 of each ethnic group (Mexican, Puerto Rican, and African American), to determine their allele frequency and linkage disequilibrium pattern in our populations (see Table E1 in the online supplement). Pairwise linkage disequilibrium between the SNPs was estimated using the r2 statistic. An SNP was included for further genotyping in the entire GALA and SAGE cohort if its minor allele frequency was greater than 5% and it was not in tight (r2 > 0.8) linkage disequilibrium with another SNP.
Genotyping
Eight ORMDL3 SNPs were selected for further genotyping on the basis of the above-described criteria. Seven SNPs (rs7216389, rs4378650, rs8076131, rs12603332, rs3744246, rs11650680, and rs3859192) were genotyped in Mexican, Puerto Rican, and African American samples. In addition, SNP rs9894164 was genotyped in African Americans only, as the minor allele frequency in the combined Mexican and Puerto Rican cohorts was less than 5%.
All SNPs were genotyped through allele-specific polymerase chain reaction (PCR) followed by detection with the AcycloPrime-FP SNP detection kit (PerkinElmer Life and Analytical Sciences, Waltham, MA) (16). Allele-specific PCR primers were obtained from Integrated DNA Technologies (Coralville, IA).
Statistical Analysis
See the online supplement for detailed information on genetic and statistical analyses. Hardy-Weinberg equilibrium (HWE) was calculated separately for each population by means of χ2 goodness-of-fit tests. In the GALA trios (n = 701), Mendelian inconsistencies were identified with the program PedCheck (17) and were excluded from further analysis. All the SNPs showed no more than 12 Mendelian inconsistencies (see Table E2). FBAT (18) and HaploFBAT (19) were used to assess the association between individual SNPs and haplotypes, respectively, with asthma and quantitative and qualitative measures of asthma-related phenotypes in the GALA trios assuming an additive model. Quantitative phenotypes included the following: asthma severity (defined by baseline FEV1), bronchodilator responsiveness (defined by change in FEV1 after administration of albuterol, relative percentage of the predicted value), and IgE levels, log10 transformed for normal distribution. Qualitative traits included the following: asthma severity defined by a composite, four-point criterion that included the presence of daily symptoms, nocturnal symptoms, prebronchodilator FEV1, and medication use; pre-FEV1 greater or less than 80%; ΔFEV1 greater or less than 12%; and IgE level greater or less than 100 IU/ml. The odds ratios for association between asthma and individual SNPs in GALA trios were estimated with the program UNPHASED (20, 21).
The association tests between genotypes and asthma and qualitative phenotypes in African American subjects were performed with logistic regression models. Association between genotypes and quantitative phenotypes was tested by multiple linear regressions. HAPSTAT (22–24) was used to determine haplotype frequencies and to test their association with asthma, and qualitative and quantitative phenotypes in African Americans. All the case–control analyses were adjusted for potential confounders such as sex, age, and log-transformed body mass index (log10 body mass index). Analysis of response to albuterol was also adjusted for asthma duration, steroid use, and regular or as needed bronchodilator use. Case-controlled genetic association analyses may be confounded by population stratification in admixed populations (25). Therefore, all case–control and cross-sectional analyses were also adjusted for individual admixture estimated from 104 unlinked ancestry informative markers, using a method described previously (26). All case-control and cross-sectional analyses were performed with the statistical software package STATA/SE 9.0 (Stata Corp., College Station, TX). Integration of the case–control and transmission disequilibrium test statistics was performed according to the method described by Kazeem and Farrall (27).
RESULTS
Characteristics of the Subjects
Demographic, clinical, and spirometric characteristics of all subjects with asthma who were analyzed for this study are shown in Table 1. There were a total of 885 subjects with complete spirometric data (Mexican, n = 273; Puerto Rican, n = 336; and African American, n = 261). The median age of the Mexican, Puerto Rican, and African American subjects with asthma was 13, 12, and 19 years, respectively. The median pre-FEV1 and ΔFEV1 were 89 and 7.4%; 83 and 4.9%; and 93 and 7.5% in Mexican, Puerto Rican, and African American subjects with asthma, respectively. The demographic, clinical, and spirometric characteristics of our GALA and SAGE cohorts have been discussed in detail by Naqvi and coworkers (28).
SNP Selection
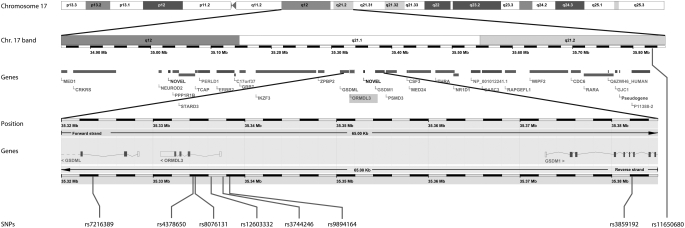
The genotype results for the 10 SNPs selected for screening in the panel of 24 unrelated subjects with asthma from each population are shown in the online supplement (see Figure E1). All of the SNPs were in partial linkage disequilibrium (LD) with each other, but none were in tight LD (r2 > 0.8) (data not shown). Of these 10 SNPs, 7 met the criteria (minor allele frequency > 5% and r2 < 0.8) for genotyping in the Mexican and Puerto Rican populations and 8 met the criteria for genotyping in the African American population. Figure 1 shows the ORMDL3 gene location on chromosome 17, and the location of the eight SNPs that were selected for further genotyping in the entire GALA and SAGE cohorts.
Figure 1.
Schematic of the location of the ORMDL3 gene on chromosome 17, and the position of the eight genotyped single-nucleotide polymorphisms (SNPs) in relation to the gene.
Association between ORMDL3 Genetic Variants and Asthma
All SNPs had a high call rate (92%) and were in Hardy-Weinberg equilibrium in all three populations, separately in probands and their parents in Mexican and Puerto Rican samples, and separately in African American cases and control subjects (see Tables E2 and E3). None of the SNPs were in tight linkage disequilibrium in any of the three populations (see Table E4). The frequency of each SNP and common haplotypes are shown in Table 2.
TABLE 2.
SINGLE-NUCLEOTIDE POLYMORPHISM ALLELE AND HAPLOTYPE FREQUENCIES IN MEXICAN AND PUERTO RICAN PROBANDS WITH ASTHMA AND THEIR PARENTS IN GALA STUDY, AND IN AFRICAN AMERICAN CASES AND CONTROL SUBJECTS IN SAGE
| Frequency (%)
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Puerto Ricans
|
Mexicans
|
African Americans
|
Germans
|
British
|
||||||||
| SNP/ Haplotype | Alleles/ Haplotype | Allele | Probands (n = 399) | Parents (n = 798) | Probands (n = 301) | Parents (n = 602) | Cases (n = 261) | Control Subjects (n = 176) | Cases (n = 728) | Control Subjects (n = 694) | Cases (n = 306) | Control Subjects (n = 1,041) |
| rs7216389 | T/C | T | 66.8 | 63.1 | 73.2 | 70.1 | 83.8 | 78.4 | 57.0 | 47.3 | 61.7 | 49.7 |
| rs4378650 | T/C | C | 58.7 | 56.3 | 73.0 | 68.8 | 70.3 | 57.7 | — | — | — | — |
| rs8076131 | G/A | A | 67.0 | 64.2 | 76.5 | 73.7 | 86.9 | 82.2 | — | — | — | — |
| rs12603332 | T/C | C | 53.6 | 51.5 | 71.5 | 68.2 | 45.8 | 35.6 | — | — | — | — |
| rs3744246 | T/C | C | 74.2 | 73.6 | 82.1 | 80.1 | 73.3 | 69.3 | 83.3 | 80.3 | 82.7 | 79.3 |
| rs9894164 | T/C | T | — | — | — | — | 84.2 | 83.0 | — | — | — | — |
| rs3859192 | T/C | C | 62.3 | 61.6 | 60.0 | 61.8 | 62.5 | 67.9 | 49.7 | 59.1 | 46.2 | 53.6 |
| rs11650680 | T/C | C | 81.4 | 80.7 | 78.1 | 76.5 | 95.6 | 98.0 | 75.9 | 80.1 | 74.8 | 78.8 |
| Haplotype 1 | C/A/C | — | 52.0 | 50.5 | 71.0 | 67.3 | 44.4 | 33.7 | — | — | — | — |
| Haplotype 2 | T/G/T | — | 24.0 | 29.5 | 14.7 | 19.4 | 11.5 | 15.5 | — | — | — | — |
| Haplotype 3 | T/A/T | — | 16.3 | 14.0 | 12.2 | 10.4 | 17.8 | 26.7 | — | — | — | — |
| Haplotype 4 | C/A/T | — | 6.2 | 4.9 | 1.9 | 1.7 | 24.9 | 21.7 | — | — | — | — |
Definition of abbreviation: SNP = single-nucleotide polymorphism.
Dashes indicate unavailable data.
Data on German and British cases and control subjects, where available, were obtained from Moffatt and coworkers (4) and are provided for comparison. Haplotypes incorporated SNPs rs4378650, rs8076131, and rs12603332 and were included if their frequency was greater than 5% in any of the three populations.
Single SNP family–based analyses using the programs FBAT and UNPHASED demonstrated that several ORMDL3 SNPs were significantly associated with increased risk for asthma in Mexican and Puerto Rican populations (Table 3). Logistic regression analysis, adjusting for population stratification as well as other potential confounders, found several SNPs associated with asthma in the African American population (Tables 4 and 5). Removing adjustments for potential confounders did not significantly alter the findings.
TABLE 3.
FAMILY-BASED ANALYSIS OF ASSOCIATION BETWEEN ASTHMA AND QUALITATIVE ASTHMA TRAITS WITH ORMDL3 SINGLE-NUCLEOTIDE POLYMORPHISMS IN PUERTO RICANS AND MEXICANS
| SNP/Haplotype: | rs7216389 | rs4378650 | rs8076131 | rs12603332 | rs3744246 | rs3859192 | rs11650680 |
|---|---|---|---|---|---|---|---|
| Allele: | T | C | A | C | C | C | C |
| Asthma | |||||||
| Mexican | 1.26 | 1.33* | 1.38* | 1.36* | 1.35* | 0.797 | 1.185 |
| (0.95–1.65) | (1.01–1.76) | (1.05–1.81) | (1.04–1.78) | (1.01–1.81) | (0.60–1.05) | (0.86–1.63) | |
| Puerto Rican | 1.35* | 1.23 | 1.29* | 1.22 | 1.10 | 0.96 | 1.03 |
| (1.07–1.70) | (0.98–1.54) | (1.03–1.62) | (0.98–1.52) | (0.86–1.41) | (0.76–1.23) | (0.79–1.35) | |
| Qualitative traits | |||||||
| Severe asthma | |||||||
| Mexican | 1.45 | 1.69* | 1.57* | 1.67* | 1.41 | 0.91 | 1.25 |
| (0.98–2.14) | (1.12–2.56) | (1.03–2.40) | (1.11–2.50) | (0.89–2.25) | (0.61–1.37) | (0.79–1.80) | |
| Puerto Rican | 1.54* | 1.61* | 1.56* | 1.71* | 1.16 | 0.86 | 1.04 |
| (1.10–2.16) | (1.15–2.26) | (1.11–2.18) | (1.21–2.40) | (0.78–1.71) | (0.59–1.25) | (0.72–1.51) | |
| ΔFEV1 > 12% | |||||||
| Mexican | 1.00 | 1.43 | 1.15 | 1.36 | 1.08 | 1.05 | 1.36 |
| (0.52–1.93) | (0.73–2.81) | (0.67–2.34) | (0.68–2.64) | (0.52–2.27) | (0.51–2.16) | (0.61–3.06) | |
| Puerto Rican | 2.17 | 1.5 | 2.40 | 2.00 | 2.00 | 0.70 | 1.14 |
| (0.89–5.30) | (0.49–4.59) | (0.96–6.03) | (0.67–5.94) | (0.59–6.78) | (0.24–2.05) | (0.43–3.02) | |
| IgE > 100 IU/ml | |||||||
| Mexican | 1.46 | 1.45 | 1.50* | 1.56* | 1.26 | 0.82 | 1.15 |
| (0.98–2.18) | (0.97–2.16) | (1.02–2.20) | (1.06–2.29) | (0.82–1.94) | (0.53–1.26) | (0.73–1.80) | |
| Puerto Rican | 1.41* | 1.36* | 1.34* | 1.43* | 1.06 | 0.95 | 0.96 |
| (1.07–1.85) | (1.05–1.77) | (1.03–1.75) | (1.10–1.86) | (0.79–1.41) | (0.70–1.28) | (0.72–1.34) |
Odds ratios are indicated, with 95% confidence intervals in parentheses.
P < 0.05.
TABLE 4.
CASE-CONTROL–BASED ASSOCIATION ANALYSIS OF QUALITATIVE AND QUANTITATIVE ASTHMA TRAITS WITH ORMDL3 SINGLE-NUCLEOTIDE POLYMORPHISMS IN AFRICAN AMERICANS
| SNP: | rs7216389 | rs4378650 | rs8076131 | rs12603332 | rs3744246 | rs9894164 | rs3859192 | rs11650680 |
|---|---|---|---|---|---|---|---|---|
| Allele: | T | C | A | C | C | T | C | C |
| Asthma | 1.21 | 1.79* | 1.20 | 1.74* | 1.40 | 1.25 | 0.78 | 0.32* |
| (0.79–1.84) | (1.27–2.50) | (0.76–1.90) | (1.24–2.44) | (0.96–2.04) | (0.80–1.96) | (0.55–1.09) | (0.12–0.85) | |
| Qualitative traits | ||||||||
| Severe asthma | 1.20 | 1.84 | 1.28 | 1.89* | 1.35 | 1.39 | 0.78 | 0.23* |
| (0.75–1.93) | (1.26–2.69) | (0.76–2.15) | (1.30–2.77) | (0.88–2.06) | (0.83–2.33) | (0.53–1.14) | (0.08–0.62) | |
| ΔFEV > 12% | 1.72 | 2.36* | 1.80 | 1.98* | 1.26 | 1.19 | 0.72 | 0.28 |
| (0.89–3.33) | (1.40–3.96) | (0.87–3.75) | (1.23–3.21) | (0.74–2.15) | (0.62–2.29) | (0.45–1.14) | (0.08–1.04) | |
| IgE > 100 IU/ml | 1.34 | 1.67* | 1.16 | 1.82* | 1.30 | 1.27 | 0.75 | 0.23* |
| (0.81–2.22) | (1.12–2.50) | (0.69–1.98) | (1.21–2.73) | (0.82–2.05) | (0.73–2.22) | (0.50–1.13) | (0.08–0.67) |
Values are adjusted for race, sex, and percentage of African ancestry.
P < 0.05.
TABLE 5.
CASE–CONTROL-BASED ASSOCIATION ANALYSIS OF QUALITATIVE AND QUANTITATIVE ASTHMA TRAITS WITH ORMDL3 SINGLE-NUCLEOTIDE POLYMORPHISMS AND HAPLOTYPES IN AFRICAN AMERICANS
| Haplotype 1 | Haplotype 2 | Haplotype 3 | Haplotype 4 | |
|---|---|---|---|---|
| (C/A/C) | (T/G/T) | (T/A/T) | (C/A/T) | |
| Asthma | 1.56* | 0.69 | 0.59* | 1.18 |
| (1.17–2.07) | (0.46–1.03) | (0.42–0.82) | (0.85–1.63) | |
| Qualitative traits | ||||
| Severe asthma | 1.16 | 0.59* | 0.69 | 1.19 |
| (0.82–1.66) | (0.37–0.94) | (0.43–1.13) | (0.82–1.74) | |
| ΔFEV > 12% | 1.21 | 1.11 | 0.58 | 1.07 |
| (0.11–13.35) | (0.58–2.14) | (0.26–1.27) | (0.63–1.80) | |
| IgE > 100 IU/ml | 1.60* | 0.69 | 0.56* | 1.19 |
| (1.14–2.23) | (0.43–1.13) | (0.37–0.84) | (0.82–1.74) |
Values are adjusted for race, sex, and percentage of African ancestry. (+) or (–) represents the direction of associations.
P < 0.051.
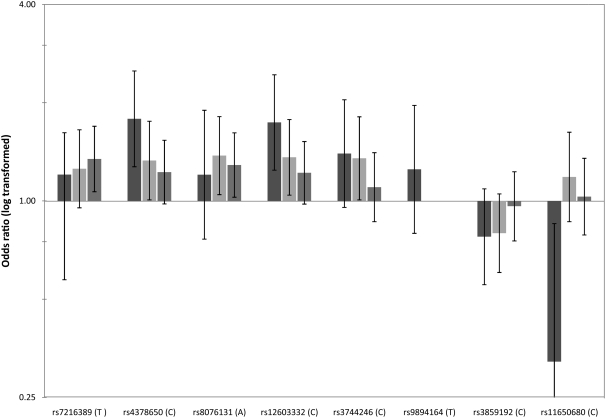
Two individual SNPs, both within intronic regions of the ORMDL3 gene, were significantly associated with asthma in African Americans and Mexicans, and there was a trend toward association for both SNPs in Puerto Ricans. The C allele of SNP rs4378650 carries an odds ratio of 1.79 (95% confidence interval [CI], 1.27 to 2.50; P = 0.001) in African Americans, 1.33 (95% CI, 1.01 to 1.76; P = 0.028) in Mexicans, and 1.23 (95% CI, 0.98 to 1.54; P = 0.076) in Puerto Ricans. The odds ratio for asthma of the C allele of SNP rs12603332 was 1.74 (95% CI, 1.24 to 2.44; P = 0.001), 1.36 (95% CI, 1.042 to 1.78; P = 0.021), and 1.22 (95% CI, 0.98 to 1.52; P = 0.080) in African Americans, Mexicans, and Puerto Ricans, respectively. The data are compared graphically in Figure 2. A joint analysis of the three populations, using the methods for combining data from transmission disequilibrium tests and case–control studies, produced a combined odds ratio of 1.39 (95% CI, 1.21 to 1.61; P = 5.7 × 10−6) for SNP rs4378650 and 1.38 (95% CI, 1.18 to 1.57; P = 2.4 × 10−5) for SNP rs12603332. This analysis showed no significant heterogeneity in the odds ratios in the transmission disequilibrium test and case-control studies for these two SNPs (P = 0.08 and P = 0.35 for SNPs rs4378650 and rs12603332, respectively) (27).
Figure 2.
Plot of odds ratio of association analysis of ORMDL3 single-nucleotide polymorphisms (SNPs) and haplotypes with asthma in African Americans (solid bars), Mexicans (light shaded bars), and Puerto Ricans (dark shaded bars). Error bars indicate 95% confidence intervals.
SNP rs8076131 was significantly associated with asthma in Mexican and Puerto Rican subjects (odds ratio [OR], 1.38; 95% CI, 1.05 to 1.81; P = 0.02 for Mexicans and OR, 1.29; 95% CI, 1.03 to 1.62; P = 0.04 for Puerto Ricans), but not in African Americans (OR, 1.20; 95% CI, 0.76 to 1.90; P = 0.424). SNP rs7216389, which showed the strongest association with asthma in the Moffatt and coworkers study, showed a significant association with asthma in Puerto Ricans (OR, 1.35; 95% CI, 1.07 to 1.70; P = 0.017) and a trend toward association in Mexicans (OR, 1.26; 95% CI, 0.95 to 1.65; P = 0.099). SNPs rs3744246 and rs11650680 showed a statistically significant association with asthma only in Mexicans (OR, 1.35; 95% CI, 1.01 to 1.81; P = 0.05), and African Americans (OR, 0.32; 95% CI, 0.12 to 0.85; P = 0.023), respectively.
There were no statistically significant associations between any of the SNPs and baseline FEV1 or change in FEV1 in response to albuterol. Except for one significant association between SNP rs12603332 and log10 IgE levels in Puerto Ricans (P = 0.015), we did not find any other association between any of the SNPs and log10 IgE levels.
Subgroup analysis showed important associations between SNPs rs4378650 and rs12603332 in patients with allergic asthma (IgE > 100 IU/ml). The association between these SNPs and asthma became stronger in patients with IgE levels exceeding 100 IU/ml in all three ethnic groups. The association between the two SNPs and asthma also became stronger among individuals with severe asthma, defined by a four-point criterion including the use of medications, presence of daily symptoms, presence of nocturnal symptoms, and low (<80% predicted) prebronchodilator FEV1, in all three ethnic groups. There was a significant association between both SNPs and asthma in African Americans who responded to bronchodilators (as defined by a change in FEV1 in response to albuterol of >12%), but not in Puerto Ricans or Mexicans.
The SNPs located within 500 base pairs of the ORMDL3 gene (five in the African American sample and four in Mexican and Puerto Rican cohorts) were selected for haplotype analysis by HaploFBAT (for the family-based Puerto Rican and Mexican cohorts) and HAPSTAT (for the African American case-control cohort). Because SNPs rs9894164 and rs3744246 did not contribute significantly to the analysis, we reduced the five SNP haplotypes to three SNP haplotypes located within the ORMDL3 gene (rs4378650, rs8076131, and rs12603332) for further haplotype testing. There was a significant association between the C/A/C haplotype and asthma in Mexicans and African Americans (P = 0.015 and P = 0.002, respectively). This haplotype was the most common in all three populations, with an observed parental frequency of 51 and 67% in Puerto Ricans and Mexicans, respectively, and a frequency of 34% in African American control subjects. The T/G/T haplotype offered a protective effect against asthma in Puerto Ricans (P = 0.003) and showed a trend toward protection in Mexicans and African Americans (P = 0.07 in both groups). This haplotype was the second most common haplotype in Puerto Ricans and Mexicans, with an observed parental frequency of 19 and 30%, respectively, and a frequency of 16% in African American control subjects. The T/A/T haplotype was associated with reduced risk for asthma in African Americans (P = 0.002), and was the second most common haplotype (frequency of 27%) in this population.
DISCUSSION
This study has replicated the association between ORMDL3 gene and asthma first identified by Moffatt and coworkers (4). The association is reproducible in three distinct populations: Mexicans, Puerto Ricans, and African Americans. Thus, ORMDL3 has now been implicated as a risk factor for asthma in two white populations (of German and British ancestry), and three admixed populations: two Latino populations (Mexicans and Puerto Ricans), and an African American population. This suggests that despite its unknown function, ORMDL3 is a risk factor for asthma, which is shared across ethnically and racially distinct populations. Thus, ORMDL3 joins a short list of genes associated with asthma in multiple populations, and replicated at the gene level, such as IL4, IL13, CD14, ADRB2, FcER1B, and IL4RA (29).
However, our findings suggest that the specific SNPs identified in the Moffatt and coworkers study (rs7216389, rs11650680, and rs3859192) as independently contributing to the risk of asthma are not likely to be the causative SNPs for asthma in the ORMDL3 gene. As the three SNPs fell within a large block of LD in the populations studied by Moffatt and coworkers, it is likely that these SNPs were in LD with one or more of the causative SNPs. rs11650680 and rs3859192 were in an intron in the upstream GSDM1 gene and in the intergenic region upstream from GSDM1, respectively, so it was not surprising that they are associated with asthma through linkage with another SNP. However, rs7216389 was the single SNP most strongly associated with disease in the genome-wide association study (P = 3 × 10−9), and was most strongly associated with ORMDL3 gene expression (P < 10−22). In addition, this SNP lies within a highly conserved element across species and was highly homologous to the proinflammatory transcription factor C/EBPb (CCAAT/enhancer-binding protein, β). Linkage disequilibrium patterns found in and around the ORMDL3 gene differed significantly between populations in this study and also from those found by Moffatt and coworkers in their samples from white subjects. This may explain why several of the SNPs found to be significant in the Moffatt and coworkers study were not associated with asthma in the present study, or were associated with asthma in only one of the three populations studied. The magnitudes of the odds ratios observed by the Moffatt and coworkers study were moderate (between 1.25 and 1.7, depending on the SNP) and are comparable to those seen in this study.
Of the two SNPs most strongly associated with asthma in the present study, SNP rs4378650 was genotyped in the supplementary study by Moffatt and coworkers and was found to have a significant association with asthma (P < 10−9 in the combined U.K. and German populations). Although both rs4378650 and rs12603332 had the most consistent association with asthma across our three populations, these SNPs showed only a trend toward association in Puerto Ricans. On the other hand, SNPs rs7216389 and rs8076131 showed significant associations with asthma in Puerto Ricans, but not in African Americans. The differing patterns of association suggest that there may be a yet undiscovered SNP in linkage disequilibrium with SNPs genotyped in this study. The differing patterns of LD between the populations may explain the differences in associations observed. However, our finding that multiple SNPs within the ORMDL3 gene are associated with asthma in multiple populations and the strong association between these SNPs and asthma in the combined population analysis support a role for ORMDL3 gene in asthma pathogenesis.
The analysis of the ORMDL3 gene in various populations in the present study and the differences in LD structure in these populations have significantly narrowed the region that may carry the causative SNP. Although none of the SNPs associated with asthma in the current study is located in the coding region of the ORMDL3 gene, several studies (30–32) have raised the possibility that polymorphisms within introns may affect splicing efficiency or transcriptional regulation of the gene. In particular, SNP rs12603332 lies in a highly conserved element across species with high correlation to the consensus target sequence for the transcription factor E47 (33), with a transcription factor score of 88.5 (http://www.cbrc.jp/research/db/TFSEARCH.html) (34, 35). E47, a member of the “E protein” family, a subset of helix–loop–helix proteins, has been strongly linked to B- and T-cell development and survival (36, 37). Notably, the substitution of a C allele in the position of rs12603332 reduces the transcription factor score of the region to 77.9. Although speculative, this is a putative explanation for the association seen in our study. Further studies will be necessary to identify the causative SNP in the ORMDL3 gene and to establish a mechanistic link between the gene and asthma.
Supplementary Material
Acknowledgments
The authors thank the families and the patients for their participation. The authors also thank the numerous health care providers and community clinics for their support and participation in the GALA and SAGE Studies. Finally, to the authors especially thank Jeffrey M. Drazen, M.D., Scott Weiss, M.D., Ed Silverman, M.D., Ph.D., Homer A. Boushey, M.D., Jean G. Ford, M.D., and Dean Sheppard for all their effort toward the creation of the GALA Study.
Supported by the National Institutes of Health (HL078885), American Lung Association of California, RWJF Amos Medical Faculty Development Award, NCMHD Health Disparities Scholar, Extramural Clinical Research Loan Repayment Program for Individuals from Disadvantaged Backgrounds, 2001–2003, to E.G.B., by an American Thoracic Society Breakthrough Opportunities in Lung Disease (BOLD) Award and Tobacco-related Disease Research Program New Investigator Award (15KT-0008) to S.C., by the Ernest S. Bazley Trust to P.C.A., and by the Sandler Center for Basic Research in Asthma and the Sandler Family Supporting Foundation.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.200711-1644OC on February 28, 2008
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Becklake MR, Ernst P. Environmental factors. Lancet 1997;350:SII10–SII13. [DOI] [PubMed] [Google Scholar]
- 2.Newman-Taylor A. Environmental determinants of asthma. Lancet 1995;345:296–299. [DOI] [PubMed] [Google Scholar]
- 3.Ober C, Hoffjan S. Asthma genetics 2006: the long and winding road to gene discovery. Genes Immun 2006;7:95–100. [DOI] [PubMed] [Google Scholar]
- 4.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, Depner M, von Berg A, Bufe A, Rietschel E, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 2007;448:470–473. [DOI] [PubMed] [Google Scholar]
- 5.Hjelmqvist L, Tuson M, Marfany G, Herrero E, Balcells S, Gonzalez-Duarte R. ORMDL proteins are a conserved new family of endoplasmic reticulum membrane proteins. Genome Biol 2002;3:research0027.1–research0027.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Homa DM, Mannino DM, Lara M. Asthma mortality in US Hispanics of Mexican, Puerto Rican, and Cuban heritage, 1990–1995. Am J Respir Crit Care Med 2000;161:504–509. [DOI] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. Asthma mortality and hospitalization among children and young adults—United States, 1980–1993. MMWR Morb Mortal Wkly Rep 1996;45:350–353. [PubMed] [Google Scholar]
- 8.Burchard EG, Ziv E, Coyle N, Gomez SL, Tang H, Karter AJ, Mountain JL, Perez-Stable EJ, Sheppard D, Risch N. The importance of race and ethnic background in biomedical research and clinical practice. N Engl J Med 2003;348:1170–1175. [DOI] [PubMed] [Google Scholar]
- 9.Burchard EG, Avila PC, Nazario S, Casal J, Torres A, Rodriguez-Santana JR, Toscano M, Sylvia JS, Alioto M, Salazar M, et al. Lower bronchodilator responsiveness in Puerto Rican than in Mexican subjects with asthma. Am J Respir Crit Care Med 2004;169:386–392. [DOI] [PubMed] [Google Scholar]
- 10.Tsai HJ, Shaikh N, Kho JY, Battle N, Naqvi M, Navarro D, Matallana H, Lilly CM, Eng CS, Kumar G, et al. β2-adrenergic receptor polymorphisms: pharmacogenetic response to bronchodilator among African American asthmatics. Hum Genet 2006;119:547–557. [DOI] [PubMed] [Google Scholar]
- 11.Lind DL, Choudhry S, Ung N, Ziv E, Avila PC, Salari K, Ha C, Lovins EG, Coyle NE, Nazario S, et al. Adam33 is not associated with asthma in Puerto Rican or Mexican populations. Am J Respir Crit Care Med 2003;168:1312–1316. [DOI] [PubMed] [Google Scholar]
- 12.Choudhry S, Ung N, Avila PC, Ziv E, Nazario S, Casal J, Torres A, Gorman JD, Salari K, Rodriguez-Santana JR, et al. Pharmacogenetic differences in response to albuterol between Puerto Ricans and Mexicans with asthma. Am J Respir Crit Care Med 2005;171:563–570. [DOI] [PubMed] [Google Scholar]
- 13.Ferris B. Epidemiology standardization project (American Thoracic Society). Am Rev Respir Dis 1978;118:1–120. [PubMed] [Google Scholar]
- 14.Miller MR, Hankinson J, Brusasco V, Burgos F, Casaburi R, Coates A, Crapo R, Enright P, van der Grinten CP, Gustafsson P, et al.; ATS/ERS Task Force. Standardisation of spirometry. Eur Respir J 2005;26:319–338. [DOI] [PubMed] [Google Scholar]
- 15.Hankinson JL, Odencrantz JR, Fedan KB. Spirometric reference values from a sample of the general US population. Am J Respir Crit Care Med 1999;159:179–187. [DOI] [PubMed] [Google Scholar]
- 16.Chen X, Levine L, Kwok P-Y. Fluorescence polarization in homogeneous nucleic acid analysis. Genome Res 1999;9:492–498. [PMC free article] [PubMed] [Google Scholar]
- 17.O'Connell JR, Weeks DE. Pedcheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 1998;63:259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laird NM, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genet Epidemiol 2000;19:S36–S42. [DOI] [PubMed] [Google Scholar]
- 19.Horvath S, Xu X, Laird NM. The family based association test method: strategies for studying general genotype–phenotype associations. Eur J Hum Genet 2001;9:301–306. [DOI] [PubMed] [Google Scholar]
- 20.Dudbridge F. Pedigree disequilibrium tests for multilocus haplotypes. Genet Epidemiol 2003;25:115–121. [DOI] [PubMed] [Google Scholar]
- 21.Dudbridge F. UNPHASED user guide. Technical Report 2006/5. Cambridge, UK: MRC Biostatistics Unit; 2006.
- 22.Lin DY. Likelihood-based inference on haplotype effects in genetic association studies. J Am Stat Assoc 2006;101:89–118. [Google Scholar]
- 23.Lin DY, Zeng D, Millikan R. Maximum likelihood estimation of haplotype effects and haplotype–environment interactions in association studies. Genet Epidemiol 2005;29:299–312. [DOI] [PubMed] [Google Scholar]
- 24.Zeng D, Lin DY, Avery CL, North KE, Bray MS. Efficient semiparametric estimation of haplotype–disease associations in case–cohort and nested case–control studies. Biostatistics 2006;7:486–502. [DOI] [PubMed] [Google Scholar]
- 25.Cardon LR, Palmer LJ. Population stratification and spurious allelic association. Lancet 2003;361:598–604. [DOI] [PubMed] [Google Scholar]
- 26.Choudhry S, Coyle NE, Tang H, Salari K, Lind D, Clark SL, Tsai HJ, Naqvi M, Phong A, Ung N, et al. Population stratification confounds genetic association studies among Latinos. Hum Genet 2006;118:652–664. [DOI] [PubMed] [Google Scholar]
- 27.Kazeem GR, Farrall M. Integrating case–control and TDT studies. Ann Hum Genet 2005;69:329–335. [DOI] [PubMed] [Google Scholar]
- 28.Naqvi M, Thyne S, Choudhry S, Tsai H, Navarro D, Castro R, Nazario S, Rodriguez-Santana J, Casal J, Torres A, et al. Ethnic-specific differences in bronchodilator responsiveness among African Americans, Puerto Ricans, and Mexicans with asthma. J Asthma 2007;44:639–648. [DOI] [PubMed] [Google Scholar]
- 29.Hoffjan S, Ober C. Present status on the genetic studies of asthma. Curr Opin Immunol 2002;14:709–717. [DOI] [PubMed] [Google Scholar]
- 30.Hull J, Campino S, Rowlands K, Chan MS, Copley RR, Taylor MS, Rockett K, Elvidge G, Keating B, Knight J, et al. Identification of common genetic variation that modulates alternative splicing. PLoS Genet 2007;3:e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laitinen T, Polvi A, Rydman P, Vendelin J, Pulkkinen V, Salmikangas P, Makela S, Rehn M, Pirskanen A, Rautanen A, et al. Characterization of a common susceptibility locus for asthma-related traits. Science 2004;304:300–304. [DOI] [PubMed] [Google Scholar]
- 32.Ueda H, Howson JM, Esposito L, Heward J, Snook H, Chamberlain G, Rainbow DB, Hunter KM, Smith AN, Di Genova G, et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature 2003;423:506–511. [DOI] [PubMed] [Google Scholar]
- 33.Ellenberger T, Fass D, Arnaud M, Harrison SC. Crystal structure of transcription factor e47: E-box recognition by a basic region helix–loop–helix dimer. Genes Dev 1994;8:970–980. [DOI] [PubMed] [Google Scholar]
- 34.Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA, Kolpakov FA, et al. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res 1998;26:362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res 2003;31:374–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murre C. Helix–loop–helix proteins and lymphocyte development. Nat Immunol 2005;6:1079–1086. [DOI] [PubMed] [Google Scholar]
- 37.Quong MW, Romanow WJ, Murre C. E protein function in lymphocyte development. Annu Rev Immunol 2002;20:301–322. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.