Abstract
Objectives
To compare the alignments of two large anatomical ontologies (the Foundational Model of Anatomy and GALEN) produced by three ontology alignment systems (AOAS, FALCON and PRIOR) in the framework of the Ontology Alignment Evaluation Initiative during its 2006 campaign.
Materials
Number of mappings identified by AOAS: 3,132, FALCON: 2,518 and PRIOR: 2,589.
Methods
Three approaches to analyzing and comparing the results were utilized: computing the overlap among result files, manual review of some 2,000 mappings and structural validation.
Conclusions
The generic systems FALCON and PRIOR identify many false positives and false negatives. With a stricter and domain-specific lexical similarity model, AOAS has a better precision, but is more sensitive to missing synonyms and misspellings.
Keywords: anatomy, ontology alignment, collaborative evaluation, cross-validation
Introduction
A given domain is often represented by multiple ontologies, providing overlapping, yet different coverage of the domain knowledge. Anatomy, for example, is represented in ontologies such as the Foundational Model of Anatomy (FMA) and GALEN. There is a need for creating mappings among such ontologies in order to facilitate the integration of data annotated with these ontologies and reasoning across ontologies. Ontology alignment is the identification of correspondences among entities (i.e., concepts and relationships) across ontologies with overlapping content. Ontology alignment is an active field of research and many approaches to aligning ontologies have been developed in the past decade [1].
Like other research communities, such as information retrieval (TREC1) and information extraction (BioCreAtIvE2), ontology alignment researchers have set up a competitive evaluation: the Ontology Alignment Evaluation Initiative (OAEI3), with the goal of comparing systems and algorithms and gaining insights from the best matching strategies [2].
Evaluating ontology alignment is generally challenging. Except for ontologies of limited size (and significance), no reference alignment (“gold standard”) is usually available for most ontologies, particularly in specialized domains such as anatomy. In the absence of a gold standard, the traditional framework of recall and precision cannot be used as the basis for the evaluation. Instead, the organizers used cross-validation as a surrogate. The assumption here is that mappings identified by several teams have a better chance of being valid.
At the 2006 edition of OAEI, five teams – including ours –presented the results of their alignment of the FMA and GALEN. The objective of this study is to review some of the results of the 2006 OAEI campaign for anatomy and to analyze the strengths and weaknesses of the various approaches.
Background
Anatomical ontologies
The two anatomical ontologies under investigation in the 2006 OAEI campaign are the Foundational Model of Anatomy (FMA) and GALEN.
The Foundational Model of Anatomy4 (FMA) is an evolving ontology that has been under development at the University of Washington since 1994 [3]. Its objective is to conceptualize the physical objects and spaces that constitute the human body. The underlying data model for the FMA is a frame-based structure implemented with Protégé5. Over 70,000 concepts cover the entire range of macroscopic, microscopic and subcellular canonical anatomy. In addition to preferred terms (one per concept), some 50,000 synonyms are provided (up to 6 per concept). For example, there is a concept named Uterine tube, which has two synonyms: Oviduct and Fallopian tube. Because single inheritance is one of the modeling principles used in the FMA, every concept (except for the root) stands in a unique is-a relation to other concepts. Additionally, seven kinds of partitive relationships are used to connect anatomical concepts (e.g., part of, constitutional part of, regional part of, and their inverses part, constitutional part, regional part). Beside hierarchical relationships, there are 81 kinds of associative relationships between concepts in the FMA. While most of them have inverses (e.g., branch of and branch), a few do not (e.g., input from).
The Generalized Architecture for Languages, Encyclopedias and Nomenclatures in medicine6 (GALEN) has been developed as a European Union AIM project led by the University of Manchester since 1991 [4]. The GALEN common reference model is a clinical terminology based on description logics. GALEN contains some 25,000 concepts and intends to represent the biomedical domain, of which canonical anatomy is only one part. Only one name is provided for each non-anonymous concept (e.g., Lobe of thyroid gland). There are over 3,000 anonymous concepts (e.g., SolidStructure which <isPairedOrUnpaired leftRightPaired>). GALEN supports multiple inheritance and every concept in GALEN (except for the root) stands in at least one IS-A relation – and often several – to other concepts. Relationships in GALEN are generally finer-grained than in the FMA. There are 41 kinds of PART-OF relationships (e.g., isStructuralComponentOf, IsDivisionOf), and 536 associative relationships (e.g., isBranchOf, isServedBy). All relationships have inverses (e.g., hasStructuralComponent, HasDivision, hasBranch, serves).
OWL Full representation
As mentioned earlier, the FMA and GALEN were created using different knowledge representation formalisms: frames for the FMA and description logics for GALEN. In order to facilitate the alignment, the organizers converted the FMA and the anatomy subset of GALEN into OWL Full, the most expressive version of the Web Ontology Language. The resulting representation includes the class hierarchy and relations between classes for both ontologies. Additionally, concept names (including synonyms) and textual definitions for classes are represented for the FMA. The datasets provided by the organizers contain 72,560 concepts for the FMA (with 44,597 synonyms), and 9,566 concepts for GALEN (anatomy subset), of which 1,035 are anonymous.
Alignment systems
The three alignment systems analyzed in this study are the Anatomical Ontology Alignment System (AOAS), the Propagation and InfOrmation Retrieval based ontology mapping system (PRIOR) and Falcon-AO (FALCON). Two other systems participating in the 2006 OAEI campaign are not included in this review for the following reasons. Almost all mappings identified by COMA++ were specific to this system and could therefore not contribute to cross-validation. The result files contributed by IsLab were not available when this study was performed. As most alignment systems, the three systems under investigation rely on a combination of lexical and structural methods, based on the assumption that equivalent concepts across ontologies have similar names and similar relations to other concepts. A brief description of the three systems analyzed follows.
AOAS is a domain-specific ontology matching system for anatomical entities. Its lexical component compares concept names using a model of lexical resemblance developed for biomedical terms and exploits additional synonyms from an external resource: the Unified Medical Language System® (UMLS®). The presence of shared hierarchical paths among concepts across ontologies is then used as positive evidence for the mappings identified lexically. AOAS also identifies incompatible concepts, which receive negative structural evidence [5, 6].
PRIOR is a domain-independent, generic ontology matching system, based on an information retrieval approach. The features used to establish the profile of a concept include all lexical information available (concept name, label, comments, property restriction, etc.). Profile propagation is used to integrate structural information. To the profile of a concept is added, with different weights, the profiles of its ancestors, descendants and siblings. A search engine is then used to compare profiles in a vector space model [7].
FALCON is a domain-independent, generic ontology matching system. It combines three alignment methods, evaluating concept similarity based on strings (lexical similarity of concept names), “documents” (concept names and definitions treated as bags of words and compared in a vector space model) and graph structures (structural similarity based on a bipartite graph, exploiting all relations represented in the ontology for a given concept). Other features of FALCON include the partitioning of large ontologies into smaller blocks and the strategy used for combining the three mapping approaches [8].
Noticeably, both PRIOR and FALCON allow partial matches between concept names (e.g., Adductor magnus of thigh matches Adductor magnus), while only minor term variations are allowed between matches by AOAS. Unlike AOAS or FALCON, PRIOR can exploit the anonymous concepts in GALEN. And while AOAS only identifies mappings between concepts, PRIOR and FALCON also find mappings between relationships.
Materials
The result files for the OAEI 2006 campaign for anatomy were downloaded from the participants’ web sites. The reporting format required from the organizers imposes four fields: entity1, entity2, measure (of confidence) and relation. Most mappings identified by the 3 systems are between equivalent concepts (relation: =). Incompatible mappings despite lexical similarity (negative evidence) are also reported by AOAS (relation: !=). A measure of confidence (0–1, continuous) is attached to each mapping and thresholds determined heuristically are used to select valid mappings. All mappings reported by PRIOR have a measure of 1.0, while FALCON also reported mappings with measures between .95 and 1.0. The mappings identified by AOAS are accompanied by a two-valued measure of confidence (1 when supported by positive evidence, 0.5 otherwise). The number of mappings identified by the three systems is summarized in Table 1.
Table 1.
Number of mappings from the three systems
| AOAS | FALCON | PRIOR | ||
|---|---|---|---|---|
| Measure of confidence | 1.0 | 3,029 | 2,115 | 2,583 |
| [.95–1.0] | 0 | 397 | 0 | |
| 0.5 | 81 | 0 | 0 | |
| Incompatible | 22 | 0 | 0 | |
| Relationships | 0 | 6 | 6 | |
| Total | 3,132 | 2,518 | 2,589 | |
Methods and results
Overlap
We first computed the intersection among the lists of mappings obtained by the three systems, thus partitioning the set of all mappings into subsets with respect to their origin, i.e., according to the system or systems that identified them (e.g., mappings identified by AOAS and FALCON, but not by PRIOR). The number of mappings with respect to their origin is summarized in Figure 1. 1,429 matches were identified by all of the three alignments, accounting for about 46%, 57% and 55% of concept matches in AOAS, FALCON and PRIOR, respectively (Figure 2). The proportion of mappings specific to one system varies largely, from 14% for FALCON to 39% for PRIOR, with 27% for AOAS.
Figure 1.
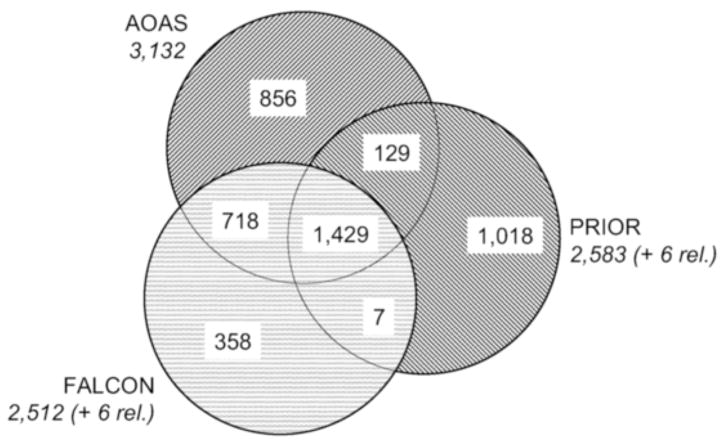
Number of mappings with respect to their origin
Figure 2.
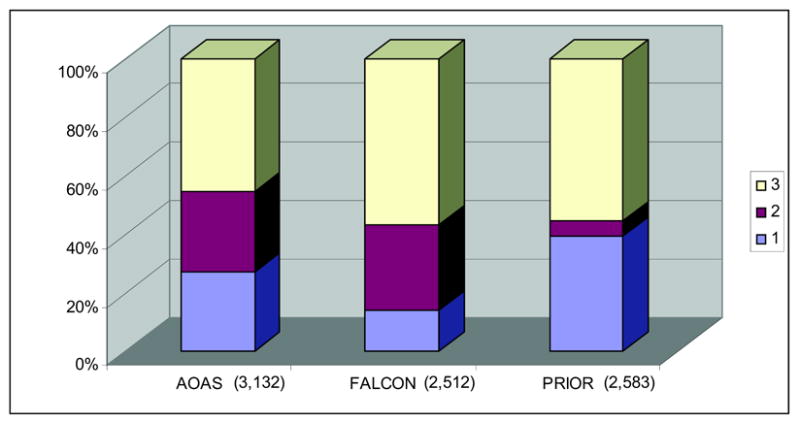
Proportion of mappings identified by 1, 2 and 3 systems
Manual validation
Then, one of us (OB) manually reviewed for accuracy all mappings not identified by AOAS. There are several reasons for explaining our bias towards this system. Unlike the other two systems, AOAS was developed specifically for aligning anatomical ontologies. In previous work, we evaluated it against a gold standard established manually and against other systems. Recall was about .9 and most mappings identified specifically by AOAS were deemed valid [6]. The mappings were classified into the following categories: certain, possible (requires additional domain knowledge) and wrong. The objective of this cursory evaluation is primarily to quantify the false positive for FALCON and PRIOR and the false negatives for AOAS. As shown in Table 2, 1,183 of the 1,383 (86%) mappings not identified by AOAS were deemed invalid. More knowledge is required to establish the validity of half of the remaining 14%.
Table 2.
Manual review of the mappings not identified by AOAS (excluding relations)
| Certain | Possible | Wrong | Total. | |
|---|---|---|---|---|
| FALCON only | 48 | 13 | 297 | 358 |
| PRIOR only | 53 | 80 | 885 | 1,018 |
| FALCON + PRIOR | 5 | 1 | 1 | 7 |
| Total | 106 | 94 | 1,183 | 1,383 |
Structural evidence in AOAS
Another element of validation is provided by the presence of positive structural evidence, i.e., the existence of shared hierarchical paths to other tentative matches (anchors) across systems. We analyze the presence of structural evidence in the various subsets for which no manual review was performed. Overall, 97% of the lexical matches identified by AOAS were supported by positive evidence. Detailed results are reported in Table 3. Except for a larger proportion of conflicts (negative evidence) – 3.1% – in the mappings identified by AOAS and PRIOR, no major differences can be observed in the various subsets.
Table 3.
Structural evidence in AOAS
| Positive | None | Negative | Total. | |
|---|---|---|---|---|
| All 3 systems | 1,382 | 44 | 3 | 1,429 |
| AOAS only | 819 | 30 | 7 | 856 |
| AOAS + FALCON | 705 | 5 | 8 | 718 |
| AOAS + PRIOR | 123 | 2 | 4 | 129 |
| Total | 3,029 | 81 | 22 | 3,132 |
Discussion
Limitations of the evaluation
The evaluation presented in this paper is partial (only the mappings not identified by AOAS were reviewed manually), cursory (some mappings were simply characterized as “possible”, awaiting further review by a domain expert) and non-independent (since the person performing the review was also involved with AOAS). However, for the purpose of cross-validating three alignment systems and in conjunction with other techniques, we believe that it is appropriate in the context of this study.
Strengths and weaknesses of each system
The strengths and weaknesses of each system are summarized in Table 4 and discussed in details below.
Table 4.
Strengths and weaknesses of the three alignment systems
| AOAS | FALCON | PRIOR | |
|---|---|---|---|
| Strengths |
|
|
|
| Weaknesses |
|
|
|
Lexical matching constitutes an important step in ontology alignment. Systems such as PRIOR focusing on bag-of-word matching rather than term matching miss many mappings identified by the other two systems on the basis of exact matches of concept names. Examples include the match of Arm, the match of Eyeball, and the match of Neck of mandible.
Compared to AOAS, FALCON uses a relaxed model of lexical similarity, based on edit distance. AOAS missed some mappings due to improper segmentation of the original GALEN strings. For example, the string “SupraHyoidMuscle” was segmented at points where case changes, leading to the term supra hyoid muscle. However, the proper spelling for this term is suprahyoid muscle and the normalization algorithm used by AOAS could not match the two terms. In contrast, the relaxed approach to string matching employed by FALCON identified the two strings as a match. The analysis of the mappings identified by FALCON and not AOAS revealed about 10 segmentation issues and 15 misspellings in GALEN (e.g., “Mensicus” for “Meniscus”).
Conversely, the relaxed model of lexical resemblance can lead to “egregious” mappings and therefore be extremely detrimental to the alignment. For example, FALCON identified a mapping between Axillary artery (in the armpit) and Maxillary artery (near the mandible).
Both FALCON and PRIOR allow approximate matching to happen, typically resulting in mappings where one term is more specific than the other, because one term contains a modifier while the other does not. These modifiers are often indicative of laterality (left/right) or level (in the vertebrae). For example, FALCON identified a match between Zygomatic process of maxilla and Zygomatic process of left maxilla and PRIOR a match between Spinous process of thoracic vertebra and Spinous process of tenth thoracic vertebra. While related, terms from these pairs should not be identified as equivalent. Of note, in many cases of inaccurate mapping, one term is a proper substring of the other (e.g., Acetabulum and Right acetabulum). Finally, other examples of mismatches involve not a modifier, but the head of the noun phrase or prepositional phrase, as in the mapping between Posterior tibial nerve and Posterior tibial vein (PRIOR) and between Posterior cutaneous nerve of arm and Posterior cutaneous nerve of forearm (FALCON).
AOAS is the only system to fully take advantage of synonymy for the alignment. Some synonyms are provided by the FMA, but others come from the UMLS Metathesaurus. In fact, we verified that most of the 856 mappings identified by AOAS are indeed valid and involve such synonyms. This is the case, for example, of the mapping between Aortic orifice and Ostium of aorta, and between Shoulder joint and Glenohumeral joint. In some cases, however, reliance on synonyms is an issue when the synonyms fail to be represented in the ontologies or external terminologies. For example, the mapping between Heel of foot and Heel and between Cartilage of larynx and Set of cartilages of larynx was missed by AOAS, but identified by PRIOR.
Not relying on lexical similarity, but using an information retrieval paradigm instead makes it possible for PRIOR to identify 56 matches to anonymous concepts in GALEN. Some of them are valid, including the mapping between Artery which <serves Brain> and Set of arteries of brain, and between Bone which <IsDivisionOf (Skull <hasTopology actuallyHollowTopology>)> and Skull bone. Others are not, for example, the mapping between Kidney which <hasLeftRightSelector leftSelection> (i.e., left kidney) and Kidney, or between Potential Cavity which <locativelyContain Pus> and Pus.
The more conservative and linguistically-motivated approach to lexical similarity adopted by AOAS [9] prevents a large number of false positives. As mentioned earlier, however, it is also more sensitive to misspellings and segmentation issues, as well as missing synonyms. Overall, we believe that the benefit of preventing many false positives largely outweighs the few false negatives.
We understand why generic and domain-independent systems such as FALCON and PRIOR have adopted relaxed lexical models. The resources available for biomedicine (UMLS synonyms, domain-specific model of lexical resemblance) are not available for most domains. However, pairs of long terms encountered in anatomy often differing by one qualifier (e.g., for laterality) have an artificially high similarity value when compared with edit distance or in a vector space model. Calibrating the models for a particular domain is an issue that remains to be addressed. Some mappings with a measure of confidence equal to 1 are less than perfect (e.g., between Lamina and Suprachoroid lamina in PRIOR), while near perfect matches fail to have perfect scores (e.g., between Surface Of Calcaneum and Surface of calcaneus in FALCON, with measure=0.962).
Structural validation is specific to AOAS and is designed to operate in combination with a model of lexical resemblance. In fact, structural validation is essentially used to confirm that the terms matching lexically bear some common semantics. Because our model of lexical resemblance is strict, there was no need to calibrate the structural resemblance too strictly. The minimum requirement for positive evidence is that one compatible path to another mapping be shared across ontologies. These requirements are not adapted to the validation of a more relaxed model of lexical similarity. Two terms differing solely by laterality are likely to share paths to many other mappings across systems. For this reason, it would not be sufficient to use our model of structural similarity to validate the mappings identified only by FALCON or PRIOR, which is why we performed a manual review instead.
Consequences for the OAEI Campaign for Anatomy
Evaluating the alignment of large scale, real world ontologies is an interesting, but very challenging endeavor. We showed that the absence of a reference alignment cannot be adequately compensated by the use of cross-validation. A cursory review also leaves many open questions. On the other hand, establishing a gold standard alignment would require the collaboration of domain experts and adequate funding. This study also illustrated that the conversion of the FMA and GALEN into OWL Full and, particularly, different uses of instances, classes and metaclasses by the two models tend to confuse users and impair alignment systems.
Acknowledgments
This research was supported in part by the Intramural Research Program of the National Institutes of Health (NIH), National Library of Medicine (NLM), and by the Natural Science Foundation of China (No.60496324), the National Key Research and Development Program of China (Grant No. 2002CB312004), the Knowledge Innovation Program of the Chinese Academy of Sciences, MADIS of the Chinese Academy of Sciences, and Key Laboratory of Multimedia and Intelligent Software at Beijing University of Technology.
Footnotes
References
- 1.Noy NF. Tools for mapping and merging ontologies. In: Staab S, Studer R, editors. Handbook on Ontologies. Springer-Verlag; 2004. pp. 365–384. [Google Scholar]
- 2.Euzenat J, Mochol M, Shvaiko P, Stuckenschmidt H, Šváb O, Svátek V, et al. First results of the Ontology Alignment Evaluation Initiative 2006. Proceedings of the Ontology Alignment Evaluation Initiative 2006 Campaign (OAEI) 2006;2006:73–90. [Google Scholar]
- 3.Rosse C, Mejino JL., Jr A reference ontology for biomedical informatics: the Foundational Model of Anatomy. J Biomed Inform. 2003;36(6):478–500. doi: 10.1016/j.jbi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 4.Rector AL, Bechhofer S, Goble CA, Horrocks I, Nowlan WA, Solomon WD. The GRAIL concept modelling language for medical terminology. Artif Intell Med. 1997;9(2):139–71. doi: 10.1016/s0933-3657(96)00369-7. [DOI] [PubMed] [Google Scholar]
- 5.Zhang S, Bodenreider O. NLM anatomical ontology alignment system: Results of the 2006 ontology alignment contest. Proceedings of the Ontology Alignment Evaluation Initiative 2006 Campaign (OAEI) 2006;2006:145–156. [Google Scholar]
- 6.Zhang S, Bodenreider O. Experience in aligning anatomical ontologies. International Journal on Semantic Web and Information Systems. 2007 doi: 10.4018/jswis.2007040101. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mao M, Peng Y. PRIOR system: Results for OAEI 2006. Proceedings of the Ontology Alignment Evaluation Initiative 2006 Campaign (OAEI) 2006;2006:165–172. [Google Scholar]
- 8.Hu W, Cheng G, Zheng D, Zhong X, Qu Y. The results of Falcon-AO in the OAEI 2006 Campaign. Proceedings of the Ontology Alignment Evaluation Initiative 2006 Campaign (OAEI) 2006;2006:118–125. [Google Scholar]
- 9.McCray AT, Srinivasan S, Browne AC. Lexical methods for managing variation in biomedical terminologies. Proc Annu Symp Comput Appl Med Care. 1994:235–9. [PMC free article] [PubMed] [Google Scholar]


