Abstract
The third variable region (V3) of the HIV-1 gp120 envelope glycoprotein is immunodominant and contains features essential for coreceptor binding. We determined the structure of V3 in the context of an HIV-1 gp120 core complexed to the CD4 receptor and to the X5 antibody at 3.5 angstrom resolution. Binding of gp120 to cell-surface CD4 would position V3 so that its coreceptor-binding tip protrudes 30 angstroms from the core toward the target cell membrane. The extended nature and antibody accessibility of V3 explain its immunodominance. Together, the results provide a structural rationale for the role of V3 in HIV entry and neutralization.
The HIV envelope spike mediates binding to receptors and virus entry [reviewed in (1)]. The trimeric spike is composed of three gp120 exterior and three gp41 transmembrane envelope glycoproteins. CD4 binding to gp120 in the spike induces conformational changes that allow binding to a coreceptor, either CCR5 or CXCR4, which is required for viral entry (2–6). Snapshots of the gp120 entry mechanism have been visualized through crystal structures of unliganded and CD4-bound states (7, 8). However, an essential component of the coreceptor binding site, the third variable region (V3), has been absent from previous structural characterizations of the gp120 core.
V3 typically consists of 35 amino acids (range 31 to 39) and plays a number of important biological roles [reviewed in (9)]. Not only is it critical for coreceptor binding, but it also determines which coreceptor, CXCR4 or CCR5, will be used for entry (10). In addition, V3 may interact with other elements in the viral spike to control the overall sensitivity of the virus to neutralization (11). Finally, immunization with HIV-1 envelope glycoproteins often elicits neutralizing responses directed primarily against V3 (12, 13).
The structure of V3 in the context of core gp120 bound to CD4, described here, now reveals the entire coreceptor binding site. We propose that V3 acts as a molecular hook, not only for snaring coreceptor but also for modulating subunit associations within the viral spike. Its extended nature is compatible with the elicitation of an immunodominant antibody response.
The extreme glycosylation and conformational flexibility of gp120 inhibit crystallization. We used variational crystallization and various technologies adapted from structural genomics to obtain crystals suitable for x-ray structural analysis (14–16). Constructs of the gp120 core with V3 from three clade B isolates (HXBc2, JR-FL, and YU2) were expressed in Drosophila S2 cells, and the deglycosylated, purified proteins were complexed with CD4 and a CD4-induced antibody (16). A total of 13 different complexes were screened robotically, and crystallization hits were optimized manually. The gp120 core with V3 from JR-FL (17, 18), when complexed to CD4 (two-domain) and the antigen-binding fragment (Fab) of the X5 antibody (19), formed hexagonal crystals that diffracted to approximately 3.5 Å resolution with x-rays provided by an Advanced Photon Source undulator beam line (SER-CAT) (table S1). The structure was solved by molecular replacement (16) and is shown in Fig. 1.
Fig. 1.
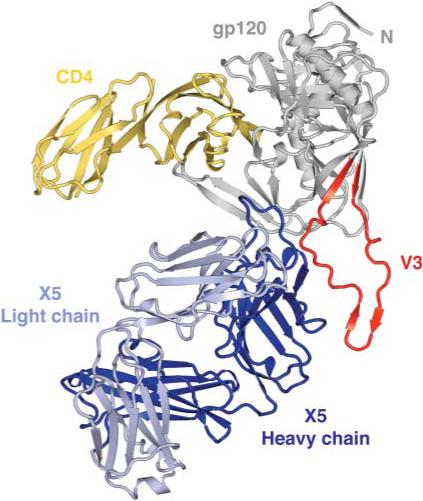
Structure of an HIV-1 gp120 core with V3. The crystal structure of core gp120 (gray) with an intact V3 (red) is shown bound to the membrane-distal two domains of the CD4 receptor (yellow) and the Fab portion of the ×5 antibody (dark and light blue). In this orientation, the viral membrane would be positioned toward the top of the page and the target cell toward the bottom.
As expected, the overall assembly of CD4, X5, and core gp120 resembled the previously determined individual structures of CD4 (20, 21) and of free X5 (22) as well as the complex of core gp120 bound to CD4 (8, 23). For core gp120, some differences were observed in the variable loops and also at the N terminus, regions where variations in gp120 have previously been observed (7, 8, 23, 24). Structural resemblance was maintained around the base of V3, indicating that the previous truncation (7, 8, 23, 24) did not distort this region of the core. In X5, a large structural difference was observed for the third complementarity-determining loop of the X5 heavy chain (CDR H3). Comparison of the refined structures of free X5 (22) and bound X5 showed Ca movements of up to 17 Å, one of the largest induced fits observed for an antibody (fig. S1).
The gp120 envelope protein is composed of inner and outer domains, named for their expected orientation in the oligomeric viral spike (8). V3 emanates from neighboring staves of the stacked double barrel that makes up the outer domain; it is almost 50 Å long from the disulfide bridge at its base to its conserved tip, but is otherwise only 15 Å wide and 5 Å deep (Fig. 2). Overall, it can be subdivided into three structural regions: a conserved base, which forms an integral portion of the core; a flexible stem, which extends away from the core; and a β-hairpin tip. In the crystal structure, the flexibility and position of the V3 tip may be influenced by a lattice contact, in which hydrogen bonds are made to the exposed backbone of the V3 β ribbon between Ile307 and Ile309. Tenuous side-chain contacts are also observed for the returning strand in the V3 stem with X5, as well as with V4 of a symmetry-related gp120 molecule, but these side-chain contacts are unlikely to influence its conformation.
Fig. 2.
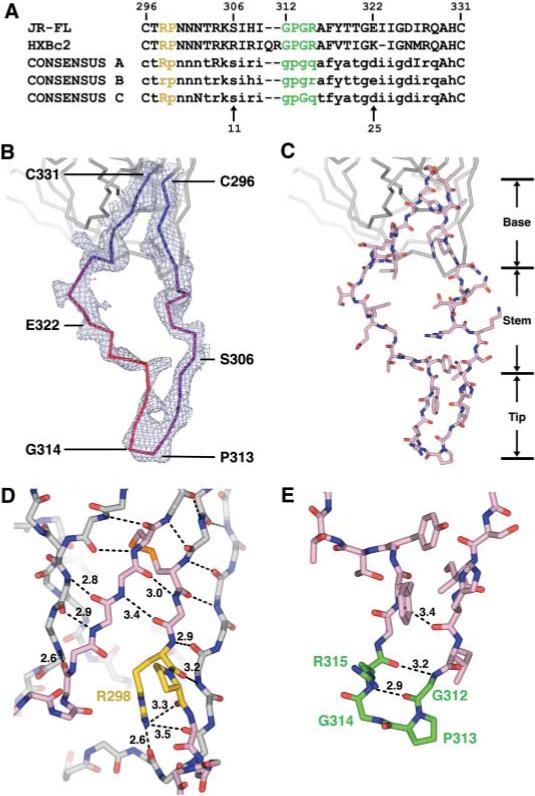
V3 sequence and structure. (A) V3 sequence. The sequences of JR-FL (17) and HXBc2 are shown along with the consensus sequence of clades A, B, and C. For the consensus sequences, absolutely conserved residues are shown in uppercase, with variable residues in lowercase (37). Single-letter aminoacid abbreviations: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; Y, Tyr. The conserved (Arg-Pro) and (Gly-Pro-Gly-Arg) motifs are colored yellow and green, respectively, and are highlighted with the same colors in (D) and (E). (B) V3 electron density and B values. 2Fobs – Fcalc density is shown for the entire V3 region and contoured at 1σ. V3 is color-coded by B value from blue (lower atomic mobility) to red (higher mobility). (C) V3 structure. The entire V3 is shown (color code: salmon, carbon atoms; red, oxygen atoms; dark blue, nitrogen atoms; orange, disulfide bond). Regions corresponding to the fixed base, accordion-like stem, and β-hairpin tip are labeled. (D) Close-up view of the V3 base. From its N terminus (Cys296), V3 extends the antiparallel sheet on the outer domain of gp120. After hydrogen bonding for three residues, additional sheet contacts are interrupted by two conserved residues: Arg298, whose side-chain hydrogen bonds to three carbonyl oxygens, including two on the neighboring outer domain strand; and Pro299, which initiates the separation of outgoing and returning V3 strands. In the returning strand, antiparallel β-sheet interactions with core gp120 recommence with the carbonyl of residue 297 and continue to the disulfide at Cys331. Main-chain atoms are shown for the core and V3 base, colored the same as in (C). Hydrogen bonds are depicted with dashed lines, with select distances in Å. All atoms of the highly conserved Arg298, Pro299, and Cys296-Cys331 disulfide are shown, with Arg and Pro carbons highlighted in yellow and disulfide in orange. (E) Conformation of the V3 tip. From Ser306 to Gly312, the main chain assumes a standard β-conformation, which terminates in a Gly-Pro-Gly-Arg β-turn (residues 312 to 315) (29, 38). After the turn, the returning density is less well defined, indicative of some disorder. All atoms of the tip are colored as in (C), with carbon atoms of the conserved tip highlighted in green. Hydrogen bonds that stabilize the β hairpin are shown as in (D).
Features of gp120 important for coreceptor binding have been mapped by mutagenesis to two regions: (i) the V3 tip, and (ii) the gp120 core around the bridging sheet, the V3 base, and neighboring residues (25–28). Analysis of these two regions on this new structure indicates that they are conserved in both sequence and structure (figs. S2A and S3).
The structural conservation of the V3 tip was surprising here in light of the apparent flexibility of the intervening stem, but we found the V3 tip to be strikingly similar in the context of the core, in antibody–V3 peptide complexes, and as a free peptide; such similarity is consistent with previous reports of recurring conformations for the V3 tip in antibody-peptide complexes (29). The structure shows that conserved regions important for co-receptor binding are separated by 10 to 20 Å and by portions of the V3 stem with moderate to high sequence variation (fig. S2).
Emerging data on the structures of the coreceptors indicate that the regions identified as being important for binding gp120—the coreceptor N terminus and the second extra-cellular loop—may also be spatially separated (30). By integrating the two-site gp120 binding site on the coreceptor with the two-site coreceptor binding site that we observe in the core + V3 gp120 structure, we propose that the N terminus of the coreceptor reaches up and binds to the core and V3 base while the V3 tip of gp120 reaches down to interact with the second extracellular loop of the coreceptor (Fig. 3B). Support for this model comes from several sources: (i) Biochemical studies show that the binding of CCR5 N-terminal peptides to gp120 is affected by gp120 alterations only on the core and around the base of V3 (28); and (ii) small-molecule inhibitors of HIV entry that bind to the second extracellular loop of the coreceptor are observed to no longer affect mutant viruses with V3 truncations (31).
Fig. 3.
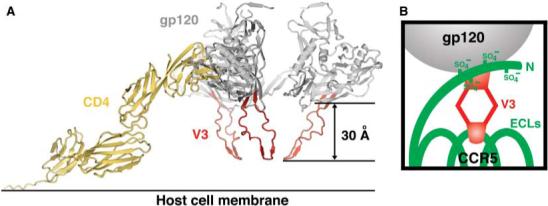
Modeled trimer and co-receptor schematic. (A) V3 in the context of a trimer at the target cell surface. The structure of the CD4-triggered gp120 with V3 was superimposed onto the structure of four-domain CD4 (39) and the trimer model obtained by quantification of surface parameters (32). In this orientation, the target cell membrane and coreceptor are expected to be positioned toward the bottom of the page. (B) Schematic of coreceptor interaction. CCR5 (green) is shown with its tyrosine-sulfated N terminus (at residues 3, 10, 14, and 15) and three extracellular loops (ECLs). V3 (red) is shown with its conserved base interacting with the sulfated CCR5 N terminus and its flexible legs allowing its conserved V3 tip to reach the second ECL of CCR5.
Does binding of the V3 tip to the coreceptor initiate gp41-mediated conformational changes? Despite general tolerance of the V3 stem to changes in sequence, there is less tolerance for insertions or deletions than in other gp120 variable loops. We superimposed the core + V3 structure on the modeled gp120 core trimer that we previously obtained by optimization of quantifiable surface parameters (32). This trimeric model orients gp120 in the context of both cell-surface CD4 and the target cell membrane. Such a superposition projects the highly conserved Pro-Gly of the V3 tip 30 Å toward the target cell membrane (Fig. 3A).
Different coreceptors, primarily CXCR4 or CCR5, can support HIV-1 entry. Sequence analysis has defined an 11/25 rule: If the 11th or 25th positions of V3 are positively charged, viruses will use CXCR4; otherwise they use CCR5 (33). In addition, V3 sequences are more conserved for CCR5-using viruses (fig. S2). The structure shows that positions 11 and 25 (residues 306 and 322) are within the variable stem. They each project about the same distance away from the core but are separated by a Cα distance of 17 Å (fig. S2). This separation suggests that positions 11 and 25 recognize different portions of the coreceptor.
CD4 induces large conformational changes in gp120. Before CD4 binding, V3 may not protrude precisely as observed here for the CD4-triggered coreceptor binding state of gp120 (3, 34). However, structural comparison of unliganded versus CD4-bound conformations of gp120 (7, 8) reveals that the local conformation of the region of the outer domain from which V3 emanates is mostly unchanged. Thus, the extended structure of V3 that we observe here should be generally representative of V3.
Immunization with gp120 or gp120/gp41 in various contexts may elicit an immune response in which virtually all of the neutralizing activity is directed at V3. We examined the crystal and nuclear magnetic resonance structures of V3-reactive antibody-peptide complexes for clues to this immunodominant response (fig. S3). Although the conformation of V3 peptides in these antibody-peptide complexes varies somewhat, the Pro-Gly tip is more conserved. Superimposing the conserved tip in the peptides with the V3 tip in the core + V3 structure permits the V3 peptide-binding antibodies to be placed in the context of the gp120 core. The antibodies completely surround V3 (Fig. 4). Although the accessibility of V3 may be quite different on a primary isolate in its pre-CD4 trimeric state, the extended nature of V3 observed here, when coupled to mechanisms that cloak the rest of the HIV envelope from antibody binding (1, 35, 36), is consistent with its ability to generate an immunodominant response.
Fig. 4.
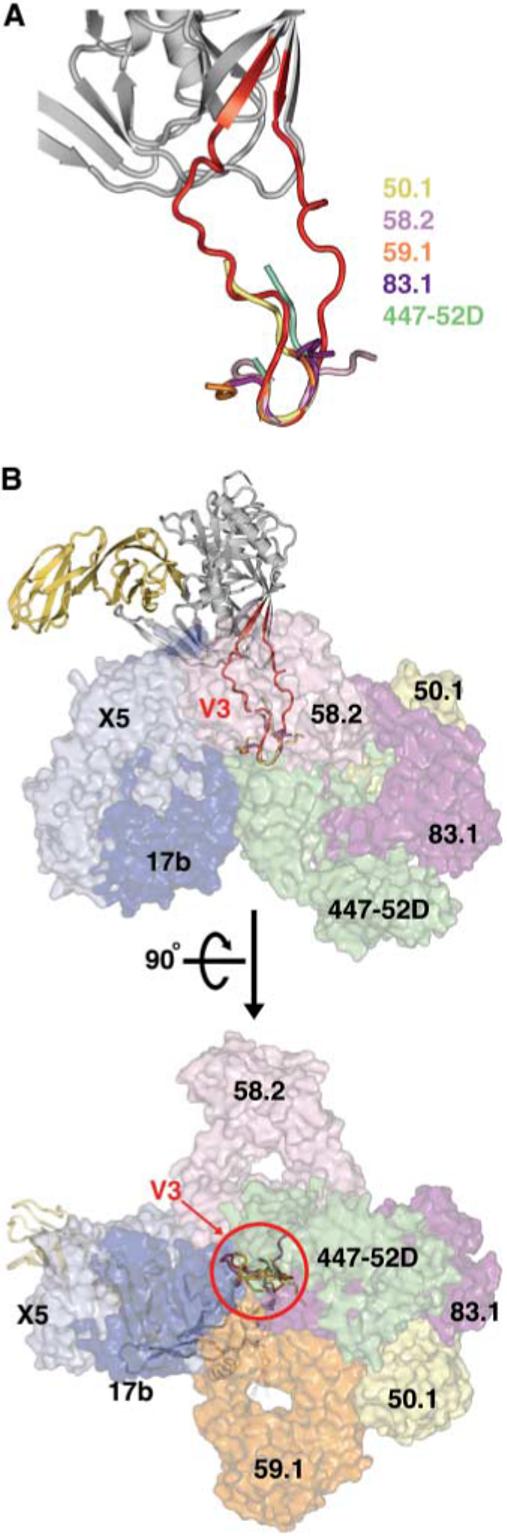
Accessibility of V3 to neutralizing antibodies. The molecular surfaces of neutralizing antibodies that block coreceptor binding are shown superimposed onto gp120 in the context of V3; antibodies 17b and ×5 bind to the conserved coreceptor binding site on the core, whereas monoclonal antibodies 50.1, 58.2, 59.1, 83.1, and 447−52D bind to V3. (A) Superposition of V3 structures. Core with V3 is shown with V3 peptides as extracted from peptide–anti-V3 neutralizing antibody complexes after superposition of the conserved V3 tip. (B) Antibody accessibility of V3. Core gp120 with V3 (ribbon representation) is shown in two perpendicular views with Fab fragments (molecular surface representation) of antibodies that bind at the coreceptor binding site on either core or V3. V3 is completely surrounded by neutralizing antibodies, suggesting a high degree of accessibility for generating an immune response.
The attributes that we observe for V3 (i.e., high relative surface area, chemically reactive backbone, conformational flexibility, and overall extended nature) may allow V3 to serve as a general molecular hook. Before CD4 binding, these attributes would enhance the ability of V3 to grasp neighboring protomers on the viral spike. Such quaternary interactions would explain V3's influence on overall neutralization sensitivity—for example, its ability to transfer neutralization resistance from YU2 to HXBc2 (11). After CD4 binding, the coreceptor binding site forms and V3 would jut prominently toward the target cell membrane. In this context, binding at the V3 tip may act as a “ripcord” to initiate gp41-mediated fusion. Our results provide a context for coreceptor interactions and suggest how V3, by altering quaternary interactions, can influence HIV evasion of the immune system and also trigger HIV entry into cells. The structure itself represents an elegant evolutionarily malleable solution that balances competing requirements of functional conservation and antigenic variation.
Supplementary Material
References and Notes
- 1.Wyatt R, Sodroski J. Science. 1998;280:1884. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 2.Dalgleish AG, et al. Nature. 1984;312:763. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 3.Sattentau QJ, Moore JP. J. Exp. Med. 1991;174:407. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng Y, Broder CC, Kennedy PE, Berger EA. Science. 1996;272:872. [Google Scholar]
- 5.Wu L, et al. Nature. 1996;384:179. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 6.Trkola A, et al. Nature. 1996;384:184. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 7.Chen B, et al. Nature. 2005;433:834. doi: 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- 8.Kwong PD, et al. Nature. 1998;393:648. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartley O, Klasse PJ, Sattentau QJ, Moore JP. AIDS Res. Hum. Retroviruses. 2005;21:171. doi: 10.1089/aid.2005.21.171. [DOI] [PubMed] [Google Scholar]
- 10.Hwang SS, Boyle TJ, Lyerly HK, Cullen BR. Science. 1991;253:71. doi: 10.1126/science.1905842. [DOI] [PubMed] [Google Scholar]
- 11.Sullivan N, et al. J. Virol. 1998;72:6332. doi: 10.1128/jvi.72.8.6332-6338.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Javaherian K, et al. Science. 1990;250:1590. doi: 10.1126/science.1703322. [DOI] [PubMed] [Google Scholar]
- 13.Gao F, et al. J. Virol. 2005;79:1154. doi: 10.1128/JVI.79.2.1154-1163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwong PD, et al. J. Biol. Chem. 1999;274:4115. doi: 10.1074/jbc.274.7.4115. [DOI] [PubMed] [Google Scholar]
- 15.Stevens RC, Wilson IA. Science. 2001;293:519. doi: 10.1126/science.293.5529.519. [DOI] [PubMed] [Google Scholar]
- 16.See supporting data on Science Online
- 17.The crystallized JR-FL was derived from a JR-FL variant with two point mutants, Asn301 → Gln and Thr388 → Ala. These mutations removed two N-linked glycans, and the resultant virus was more sensitive to neutralization but was otherwise functional (18)
- 18.Koch M, et al. Virology. 2003;313:387. doi: 10.1016/s0042-6822(03)00294-0. [DOI] [PubMed] [Google Scholar]
- 19.Moulard M, et al. Proc. Natl. Acad. Sci. U.S.A. 2002;99:6913. doi: 10.1073/pnas.102562599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryu SE, et al. Nature. 1990;348:419. doi: 10.1038/348419a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang JH, et al. Nature. 1990;348:411. doi: 10.1038/348411a0. [DOI] [PubMed] [Google Scholar]
- 22.Darbha R, et al. Biochemistry. 2004;43:1410. doi: 10.1021/bi035323x. [DOI] [PubMed] [Google Scholar]
- 23.Kwong PD, et al. Structure. 2000;8:1329. doi: 10.1016/s0969-2126(00)00547-5. [DOI] [PubMed] [Google Scholar]
- 24.Huang C, et al. Structure. 2005;13:755. [Google Scholar]
- 25.Rizzuto CD, et al. Science. 1998;280:1949. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 26.Rizzuto C, Sodroski J. AIDS Res. Hum. Retroviruses. 2000;16:741. doi: 10.1089/088922200308747. [DOI] [PubMed] [Google Scholar]
- 27.Cormier EG, Tran DNH, Yukhayeva L, Olson WC, Dragic T. J. Virol. 2001;75:5541. doi: 10.1128/JVI.75.12.5541-5549.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cormier EG, Dragic T. J. Virol. 2002;76:8953. doi: 10.1128/JVI.76.17.8953-8957.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stanfield RL, Ghiara JB, Ollman E, Saphire AT, Profy, Wilson IA. Virology. 2003;315:159. doi: 10.1016/s0042-6822(03)00525-7. [DOI] [PubMed] [Google Scholar]
- 30.Klco JM, Wiegand CB, Narzinski K, Baranski TJ. Nat. Struct. Mol. Biol. 2005;12:320. doi: 10.1038/nsmb913. [DOI] [PubMed] [Google Scholar]
- 31.Lin G, et al. paper presented at the 12th Conference on Retroviruses and Opportunistic Infections; Boston. 21 to 26 February 2005. [Google Scholar]
- 32.Kwong PD, Wyatt R, Sattentau QJ, Sodroski J, Hendrickson WA. J. Virol. 2000;74:1961. doi: 10.1128/jvi.74.4.1961-1972.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Resch W, Hoffman N, Swanstrom R. Virology. 2001;288:51. doi: 10.1006/viro.2001.1087. [DOI] [PubMed] [Google Scholar]
- 34.Werner A, Levy JA. J. Virol. 1993;67:2566. doi: 10.1128/jvi.67.5.2566-2574.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wyatt R, et al. Nature. 1998;393:705. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 36.Wei X, et al. Nature. 2003;422:307. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 37.Kuiken CL, et al. Sequence Compendium, H.I.V. 2002. Los Alamos National Laboratory; Los Alamos, NM: 2002. [Google Scholar]
- 38.The electron density corresponding to the V3 tip was strongly defined. For Pro313, however, carbonyl “bumps” corresponding to both type I and type II β-turns could be seen, suggesting that the tip was a mixture of conformations. Both type I and type II turns have been observed in V3 antibody-peptide complexes (29). We have modeled the V3 tip as an equal mixture of type I and type II β-turns. For visual simplicity, however, figures display only the type II conformation
- 39.Wu H, Kwong PD, Hendrickson WA. Nature. 1997;387:527. doi: 10.1038/387527a0. [DOI] [PubMed] [Google Scholar]
- 40.We thank J. Robinson for CD4i antibodies 17b, 48d, and 412d; J. Hoxie and G. Lin for discussions regarding the effects of V3 deletion on HIV entry; E. Berger, G. Nabel, and L. Shapiro for comments on the manuscript; and Syrrx for assistance with robotic screening. Supported by the Intramural Research Program of the NIH, Vaccine Research Center (R.W. and P.D.K.), and National Cancer Institute (D.S.D.) and by the International AIDS Vaccine Initiative (J.S. and I.A.W.), NIH grants AI24755, AI31783, AI39429, AI40895 (J.S.) and GM46192 (I.A.W.), and a grant from the Bill and Melinda Gates Foundation Grand Challenges in Global Health Initiative (D.S.D., R.W., and P.D.K.). Use of SER-CAT at the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract W-31−109-Eng-38. Coordinates and structure factors have been deposited with the Protein Data Bank under accession number 2B4C and are also available from the authors
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


