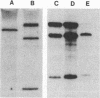
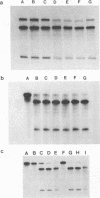
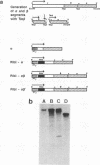
Abstract
The development of safe and effective antiviral agents has been a slow process, largely because of the difficulty in distinguishing between virus and host functions; materials toxic to the virus are frequently harmful also to the host in which the agent resides. Recently, techniques which target nucleic acid sequences as a means of reducing gene expression have emerged. This antisense armamentarium includes ribozymes, RNA enzymes which cleave other RNA molecules in a sequence-specific manner. We wish to assess the ability of ribozymes to control animal virus infection. Reasoning that the viruses most vulnerable to ribozyme intervention will be those whose complete life cycle is based on RNA (with no DNA stage), we have begun to develop ribozymes directed toward lymphocytic choriomeningitis virus (LCMV), the prototype of the arenavirus family. Using ribozymes of the hammerhead variety, we have identified several sites on the LCMV genome which can be efficiently cleaved in trans. The efficiency of cleavage is site dependent, and we demonstrate that secondary structure at the target site can abolish ribozyme cleavage. Computer-assisted analysis indicates that much of the LCMV genome may be involved in base pairing, which may render it similarly resistant to ribozyme attack. The few remaining open regions of LCMV lack a GUC target site, on which most studies to date have relied. Here we show that AUC, CUC, and AUU are alternative sites which can be cleaved by trans-acting ribozymes. This finding is important given the aforementioned restriction of available sites, imposed by secondary structure.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bishop D. H., Auperin D. D. Arenavirus gene structure and organization. Curr Top Microbiol Immunol. 1987;133:5–17. doi: 10.1007/978-3-642-71683-6_2. [DOI] [PubMed] [Google Scholar]
- Buchmeier M. J., Oldstone M. B. Protein structure of lymphocytic choriomeningitis virus: evidence for a cell-associated precursor of the virion glycopeptides. Virology. 1979 Nov;99(1):111–120. doi: 10.1016/0042-6822(79)90042-4. [DOI] [PubMed] [Google Scholar]
- Cameron F. H., Jennings P. A. Specific gene suppression by engineered ribozymes in monkey cells. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9139–9143. doi: 10.1073/pnas.86.23.9139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T. R., Zaug A. J., Grabowski P. J. In vitro splicing of the ribosomal RNA precursor of Tetrahymena: involvement of a guanosine nucleotide in the excision of the intervening sequence. Cell. 1981 Dec;27(3 Pt 2):487–496. doi: 10.1016/0092-8674(81)90390-1. [DOI] [PubMed] [Google Scholar]
- Celander D. W., Cech T. R. Visualizing the higher order folding of a catalytic RNA molecule. Science. 1991 Jan 25;251(4992):401–407. doi: 10.1126/science.1989074. [DOI] [PubMed] [Google Scholar]
- Cotten M., Birnstiel M. L. Ribozyme mediated destruction of RNA in vivo. EMBO J. 1989 Dec 1;8(12):3861–3866. doi: 10.1002/j.1460-2075.1989.tb08564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dange V., Van Atta R. B., Hecht S. M. A Mn2(+)-dependent ribozyme. Science. 1990 May 4;248(4955):585–588. doi: 10.1126/science.2185542. [DOI] [PubMed] [Google Scholar]
- Epstein L. M., Gall J. G. Self-cleaving transcripts of satellite DNA from the newt. Cell. 1987 Feb 13;48(3):535–543. doi: 10.1016/0092-8674(87)90204-2. [DOI] [PubMed] [Google Scholar]
- Forster A. C., Symons R. H. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell. 1987 Apr 24;49(2):211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- Goodchild J., Kohli V. Ribozymes that cleave an RNA sequence from human immunodeficiency virus: the effect of flanking sequence on rate. Arch Biochem Biophys. 1991 Feb 1;284(2):386–391. doi: 10.1016/0003-9861(91)90313-8. [DOI] [PubMed] [Google Scholar]
- Grosshans C. A., Cech T. R. Metal ion requirements for sequence-specific endoribonuclease activity of the Tetrahymena ribozyme. Biochemistry. 1989 Aug 22;28(17):6888–6894. doi: 10.1021/bi00443a017. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983 Dec;35(3 Pt 2):849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Haseloff J., Gerlach W. L. Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature. 1988 Aug 18;334(6183):585–591. doi: 10.1038/334585a0. [DOI] [PubMed] [Google Scholar]
- Hutchins C. J., Rathjen P. D., Forster A. C., Symons R. H. Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res. 1986 May 12;14(9):3627–3640. doi: 10.1093/nar/14.9.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi M., Hayase Y., Iwai S., Kamiya H., Inoue H., Ohtsuka E. Design of RNA enzymes distinguishing a single base mutation in RNA. Nucleic Acids Res. 1989 Sep 12;17(17):7059–7071. doi: 10.1093/nar/17.17.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi M., Ohtsuka E. Effects of phosphorothioate and 2-amino groups in hammerhead ribozymes on cleavage rates and Mg2+ binding. Biochemistry. 1991 May 28;30(21):5145–5150. doi: 10.1021/bi00235a005. [DOI] [PubMed] [Google Scholar]
- McSwiggen J. A., Cech T. R. Stereochemistry of RNA cleavage by the Tetrahymena ribozyme and evidence that the chemical step is not rate-limiting. Science. 1989 May 12;244(4905):679–683. doi: 10.1126/science.2470150. [DOI] [PubMed] [Google Scholar]
- Partono S., Lewin A. S. The rate and specificity of a group I ribozyme are inversely affected by choice of monovalent salt. Nucleic Acids Res. 1991 Feb 11;19(3):605–609. doi: 10.1093/nar/19.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prody G. A., Bakos J. T., Buzayan J. M., Schneider I. R., Bruening G. Autolytic processing of dimeric plant virus satellite RNA. Science. 1986 Mar 28;231(4745):1577–1580. doi: 10.1126/science.231.4745.1577. [DOI] [PubMed] [Google Scholar]
- Ruffner D. E., Dahm S. C., Uhlenbeck O. C. Studies on the hammerhead RNA self-cleaving domain. Gene. 1989 Oct 15;82(1):31–41. doi: 10.1016/0378-1119(89)90027-9. [DOI] [PubMed] [Google Scholar]
- Ruffner D. E., Stormo G. D., Uhlenbeck O. C. Sequence requirements of the hammerhead RNA self-cleavage reaction. Biochemistry. 1990 Nov 27;29(47):10695–10702. doi: 10.1021/bi00499a018. [DOI] [PubMed] [Google Scholar]
- Salvato M. S., Shimomaye E. M. The completed sequence of lymphocytic choriomeningitis virus reveals a unique RNA structure and a gene for a zinc finger protein. Virology. 1989 Nov;173(1):1–10. doi: 10.1016/0042-6822(89)90216-x. [DOI] [PubMed] [Google Scholar]
- Salvato M., Shimomaye E., Oldstone M. B. The primary structure of the lymphocytic choriomeningitis virus L gene encodes a putative RNA polymerase. Virology. 1989 Apr;169(2):377–384. doi: 10.1016/0042-6822(89)90163-3. [DOI] [PubMed] [Google Scholar]
- Salvato M., Shimomaye E., Southern P., Oldstone M. B. Virus-lymphocyte interactions. IV. Molecular characterization of LCMV Armstrong (CTL+) small genomic segment and that of its variant, Clone 13 (CTL-). Virology. 1988 Jun;164(2):517–522. doi: 10.1016/0042-6822(88)90566-1. [DOI] [PubMed] [Google Scholar]
- Sarver N., Cantin E. M., Chang P. S., Zaia J. A., Ladne P. A., Stephens D. A., Rossi J. J. Ribozymes as potential anti-HIV-1 therapeutic agents. Science. 1990 Mar 9;247(4947):1222–1225. doi: 10.1126/science.2107573. [DOI] [PubMed] [Google Scholar]
- Saxena S. K., Ackerman E. J. Ribozymes correctly cleave a model substrate and endogenous RNA in vivo. J Biol Chem. 1990 Oct 5;265(28):17106–17109. [PubMed] [Google Scholar]
- Toh H., Imamura A., Kanda K. Role of Mg2+ in the ribozyme system. FEBS Lett. 1987 Jul 27;219(2):279–282. doi: 10.1016/0014-5793(87)80235-1. [DOI] [PubMed] [Google Scholar]
- Uhlenbeck O. C. A small catalytic oligoribonucleotide. Nature. 1987 Aug 13;328(6131):596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- Wu H. N., Lin Y. J., Lin F. P., Makino S., Chang M. F., Lai M. M. Human hepatitis delta virus RNA subfragments contain an autocleavage activity. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1831–1835. doi: 10.1073/pnas.86.6.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M. Q., Kathe S. D., Goodrich-Blair H., Nierzwicki-Bauer S. A., Shub D. A. Bacterial origin of a chloroplast intron: conserved self-splicing group I introns in cyanobacteria. Science. 1990 Dec 14;250(4987):1566–1570. doi: 10.1126/science.2125747. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Grosshans C. A., Cech T. R. Sequence-specific endoribonuclease activity of the Tetrahymena ribozyme: enhanced cleavage of certain oligonucleotide substrates that form mismatched ribozyme-substrate complexes. Biochemistry. 1988 Dec 13;27(25):8924–8931. doi: 10.1021/bi00425a008. [DOI] [PubMed] [Google Scholar]