Abstract
Background
The determination of genetic variation in sperm competitive ability is fundamental to distinguish between post-copulatory sexual selection models based on good-genes vs compatible genes. The sexy-sperm and the good-sperm hypotheses for the evolution of polyandry require additive (intrinsic) effects of genes influencing sperm competitiveness, whereas the genetic incompatibility hypothesis invokes non-additive genetic effects. A male's sperm competitive ability is typically estimated from his fertilization success, a measure that is dependent on the ability of rival sperm competitors to fertilize the ova. It is well known that fertilization success may be conditional to genotypic interactions among males as well as between males and females. However, the consequences of effects arising from the random sampling of sperm competitors upon the estimation of genetic variance in sperm competitiveness have been overlooked. Here I perform simulations of mating trials performed in the context of sibling analysis to investigate whether the ability to detect additive genetic variance underlying the sperm competitiveness phenotype is hindered by the relative nature of fertilization success measurements.
Results
Fertilization success values render biased sperm competitive ability values. Furthermore, asymmetries among males in the errors committed when estimating sperm competitive abilities are likely to exist as long as males exhibit variation in sperm competitiveness. Critically, random effects arising from the relative nature of fertilization success lead to an underestimation of underlying additive genetic variance in sperm competitive ability.
Conclusion
The results show that, regardless of the existence of genotypic interactions affecting the output of sperm competition, fertilization success is not a perfect predictor of sperm competitive ability because of the stochasticity of the background used to obtain fertilization success measures. Random effects need to be considered in the debate over the maintenance of genetic variation in sperm competitiveness, and when testing good-genes and compatible-genes processes as explanations of polyandrous behaviour using repeatability/heritability data in sperm competitive ability. These findings support the notion that the genetic incompatibility hypothesis needs to be treated as an alternative hypothesis, rather than a null hypothesis, in studies that fail to detect intrinsic sire effects on the sperm competitiveness phenotype.
Background
A wealth of research inspired by Parker's [1] seminal study on sperm competition has provided overwhelming evidence that mating success is not always equivalent to reproductive success [1-4]. Sperm competition, a direct consequence of polyandry (females mating with different partners within a single reproductive episode), can contribute to the maintenance of promiscuity if females accrue benefits from encouraging the overlap of ejaculates at the site of fertilization. The sexy-sperm hypothesis, a post-copulatory analogue of the sexy-sons model for the evolution of female preferences and male attractiveness [5-7], suggests that polyandrous females could enhance their fitness if fertilization success is genetically correlated to female mating frequency. The good-sperm hypothesis, an analogue of the pre-copulatory good-genes model for mate choice, also suggests that females will accrue genetic quality for their offspring through facilitation of sperm competition, in this case if males with higher sperm competitive ability sire offspring with higher viability [8,9]. Importantly, both the sexy-sperm and the good-sperm model are based on additive (intrinsic) effects of genes influencing a male's sperm competitiveness [10,11]. On the contrary, polyandrous females have also been suggested to benefit from processes based on non-additive genetic effects if multiple mating served as a defence against genetic incompatibilities between them and their partners. The genetic incompatibility hypothesis [12-15] was originally proposed to account for benefits of polyandry in the form of enhanced viability of offspring (i.e., through processes occurring after fertilization). However, avoidance of genetic incompatibility at the pre-fertilization stage, through sperm selection mechanisms based on, for instance, gamete recognition, can also play a role in determining fertilization output [16-18].
In the context of selection based on good-genes, sperm competitive ability is defined as the investment in traits that influence a male's ability to win fertilizations in conditions of sperm competition, under the assumption that males of higher genetic quality can allocate more resources to traits involved in sperm competition [9]. An increasing body of research focuses on investigations of the transitivity or repeatability of fertilization success, or the genetic architecture of traits influencing fertilization success [see reviews in [11,19]]. Support for the notion that sperm competitive ability can be an intrinsic trait comes from different sources. These include studies looking at the repeatability or heritability of fertilization success or paternity success scores [20-27], or examining phenotypic traits thought to be important in sperm competition [11,19,28], but also selection experiments [29] and analysis of the condition-dependence of these traits [28,30-32]. However, current views agree that some factors limit the evolution of sperm competition traits via good-genes processes and contribute to the maintenance of genetic variation among males in traits determining the outcome of sperm competition. These factors include male × female interactions [e. g., [26,30,33-36]], antagonistic pleiotropy [37], sex-biased inheritance [38-41], and ejaculate × ejaculate interactions [34,42,43].
Accurate estimates of sperm competitiveness values for individual males are thus critical for the study of post-copulatory sexual selection, and in particular, to (1) investigate the genetic basis of sperm competitive ability, (2) distinguish between competing hypotheses based on additive or non-additive genetic effects, and (3) further test predictions from these hypotheses. For instance, testing the good-sperm model in phenotypic studies relies on the correlation between a male's sperm competitive ability and his offspring viability [9,44]. However, a pervasive problem that has received little attention arises from the fact that a male's sperm competitiveness is an absolute measure that has to be estimated from a relative measure: fertilization success. Sperm competitive ability of a given male can only be holistically assessed by looking at the output of fertilization trials involving other males in the population acting as sperm competitors. In other words, a male's fertilization success depends not only on his sperm competitive ability but also on the ability of the rival males to win fertilizations.
This study asks for the first time whether fertilization success measures are good estimators of individual sperm competitiveness values. By simulating sperm competition experiments involving pairs of males extracted from a large population characterized by a given distribution of sperm competitiveness I first show that fertilization success is not a perfect predictor of sperm competitiveness. The study further examines the consequences of random effects arising from the relative nature of fertilization success for studies of post-copulatory sexual selection. Simulations of mating trials performed in the context of sibling analysis indicate that the heritability of sperm competitive ability is dramatically underestimated when fertilization success values are used. I discuss the implications of these results for investigations on the genetic nature and the maintenance of genetic variance in sperm competitiveness, and for the study of the evolution of polyandry.
Methods
General methods
The analyses consist of three steps. First, simulations of sperm competition experiments involving pairs of males extracted from a large population characterized by a given distribution of sperm competitiveness. Second, the assessment of the efficacy of fertilization success as a predictor of sperm competitive ability. Third, the assessment of the consequences of the deviations between individual fertilization success values and sperm competittive ability values.
Sperm competitiveness is influenced by a number of traits that may include ejaculate volume [45], sperm quality [46,47], sperm morphology [30,48], seminal fluid products [49-51] and genital morphology [24,52], among others. For simplicity, here I will refer to sperm competitive ability as a male's investment in ejaculate volume (numbers of sperm inseminated), which will be assumed to be the primary determinant of fertilization success in a hypothetical species. Nevertheless, this simplification imposes no limitation to the interpretation of the results. The analyses involve the calculation of fertilization success for the second male to mate a doubly mated female. Fertilization success for this arbitrarily chosen male is defined as the proportion of ova that he fertilizes and is denoted F2. In practice, fertilization success is generally estimated from P2, the observed proportion of offspring sired by the second male calculated at birth or hatching [53]. Here the denomination F2 is preferred over P2 to emphasize that the analyses focus on "true" fertilization success, i.e., estimated just at conception. The congruence between F2 and P2 will depend to a large extent on the subsequent viability of offspring between fertilization and paternity assessment [54-56].
Two examples of distributions for sperm competitive ability are used: S and Snormal, both containing 60000 values of sperm competitiveness in a scale from 0 to 1. Mean sperm competitiveness for the bell-shaped S distribution is 0.495 (SD = 0.19, range 0.192–0.999; further details of this distribution can be found elsewhere [56] and in the Additional file 1). Although the distribution S exhibits to some extent a shape similar to that of a normal distribution, it deviates significantly from normality (K-S d = 0.08, p < 0.01 ; Lilliefors p < 0.01). A second distribution of values for sperm competitive ability (Snormal) uses a normal distribution (x = 0.5, SD = 0.12, Min. = 0.013, Max. = 0.994).
Fertilization success as a predictor of sperm competitive ability
I have simulated sperm competition trials involving pairing of males from the hypothetical distribution of S, or Snormal, to calculate fertilization success (F2) values assuming a sperm mixing mechanism of sperm competition following the fair raffle principle of Parker [57]. Under the fair raffle, the fertilization success of a male is a function of his investment in sperm competition relative to the investment of the competitor male, and thus F2 = s2/(s1+s2), where s1 and s2 are sperm competitiveness for the first and second male, respectively, on a scale from 0–1 (without including zero). I have calculated F2 values using values of s1 taken at random from the distribution of sperm competitive abilities and fixed s2 values of 0.2, 0.3, 0.4, ..., 1 (0.192 is the minimum sperm competitiveness value for the distribution S), or 0.02, 0.3, 0.4, ..., 1 (0.013 is the minimum sperm competitiveness value for the distribution Snormal). I have simulated 5000 double matings for each category of s2. Subsequently, from the array of F2 values obtained for each fixed category of s2 I have calculated the probability that F2 deviates from s2 in such a way that the absolute difference between F2 and s2 is higher than 0.1, 0.2 or 0.3.
Differences in the variance between a set of F2 values and the s2 values from which they originate were explored in a second set of simulations. Here I simulated 10000 sperm competition experiments (n = 50 double matings each experiment) by random resampling of s1 and s2 values in the distribution of sperm competitiveness S or Snormal. Subsequently, for each simulated experiment, the coefficients of variation for the set of s2 values, and for the set of F2 values, were calculated. The coefficients of variation were compared using a T-test for dependent samples.
Consequences of the relative nature of fertilization success on the estimation of genetic variance in sperm competitiveness
Results arising from the analyses above suggested that the relative nature of F2 calculations may have important consequences for investigations of the genetic variance in sperm competitive ability. These consequences have been explored with an approach that calculates the intraclass correlation coefficient following the simulation of hypothetical sibling analyses. The genetic design simulated involves screening of the fertilization success of 8 offspring for each of 50 sires. Sperm competitive ability for each sire (s2-Sire) has been assigned by random extraction of sperm competitive ability values from the distribution S. The values of sperm competitive ability for each individual offspring of each sire have been calculated as s2-Offspring = (s2-Sire + R)/2, where R is a random number between 0 and 1 from a uniform distribution. This protocol simulates heritable variation for sperm competitive ability, and results in moderate random deviations of s2-Offspring from s2-Sire. For each experiment (sibling analysis involving 50 sires), a one-way Anova is carried out to obtain the variance components (among sires, and within sire), which allows the calculation of the intraclass correlation coefficient. The intraclass correlation coefficient, t, is defined as the phenotypic correlation between sibs [58], which provides an estimate of the fraction of the phenotypic variance attributable to differences among sires. The simulated set up can be taken as a paternal half-sib design in which a single offspring from each of 8 unrelated females mated to a male is assayed [58], or a full sib design in which each sire is mated to only one female, and 8 offspring per female are assayed. In the case of the paternal half-sib analysis, heritability, h2 = 4t, under the assumption of negligible epistasis and common environmental effects, whereas in the case of the full sib analysis, h2 = 2t, provided that there were no dominance effects and no common environmental effects [58].
The key point of the analysis performed is that the protocol allows the calculation of the "true" intraclass correlation coefficient for the trait "sperm competitive ability" (i.e., s2). To investigate the influence of the relative nature of F2 measures on estimates of the additive genetic variance of sperm competitive ability this "true" t is compared to the t that would be obtained in empirical studies; i. e., inferring genetic additive variance in sperm competitive ability from fertilization success values (F2). Therefore, the values of sperm competitiveness for the offspring in the hypothetical design have been put in the context of fertilization success values following double matings. I have calculated fertilization success values for the offspring (F2-Offspring) using each offspring's s2-Offspring value (generated from the s2-Sire) and random-extracted s1 values from the distribution S. Subsequently, in the same way that t was calculated using "real" sperm competitiveness values, I have calculated the intraclass correlation coefficient using the F2-Offspring values derived from the s2-Offspring. The difference between the intraclass correlation coefficient for s2-Offspring and for F2-Offspring would therefore inform on how the estimations of additive genetic variance in sperm competitive ability are affected by the relative nature of fertilization success values. The whole protocol has been replicated 10000 times, each time following a new random extraction of sperm competitive ability values for the sires. The intraclass correlation coefficients using s2 data and their associated intraclass correlation coefficients using F2 data have been compared using a T-test for dependent samples. The same methodology has been used using sperm competitive ability values from the Snormal distribution. Simulations have been carried out using PopTools 2.7.5 [59], while Statistica 6.0 [60] has been used for data analyses.
Results
Fertilization success as a predictor of sperm competitive ability
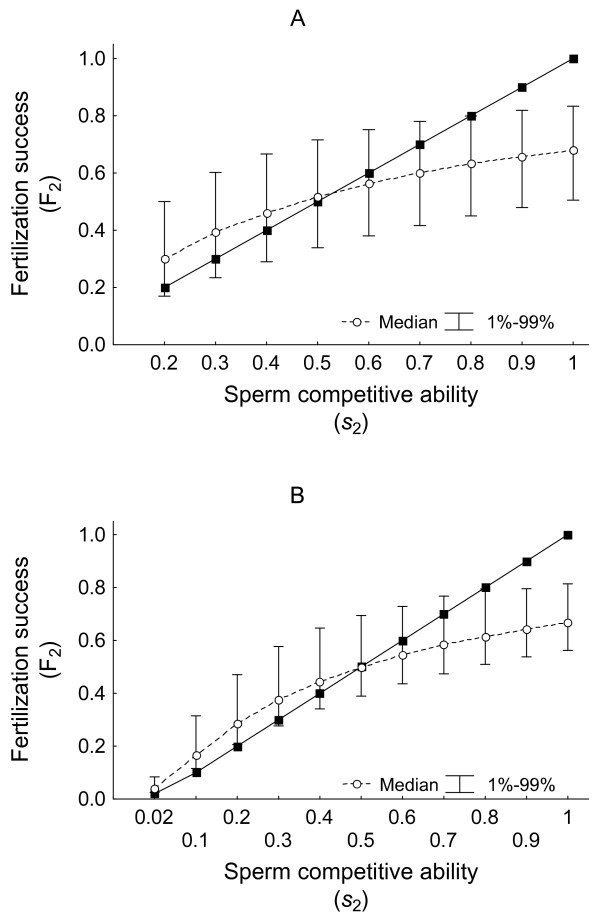
Every single value of sperm competitive ability can generate a wide range of fertilization success values as a result of variation in the sperm competitive ability of rival males (Figs. 1A and 1B). The probabilities of mistakenly inferring sperm competitive ability values can be high (see Additional file 2). The probability curves reflect assymetries in the degree and direction of biases, which depend on the sperm competitive ability value being tested and the influence of values that are overrepresented in the population (e.g., average sperm competitiveness values in distributions suggestive of stabilizing selection on sperm competitive ability)(see Additional file 2).
Figure 1.
Variation in fertilization success values. The figure represents the variation in fertilization success values for the second male (open circles and whiskers) that can be originated from the same sperm competitive ability value for the second male. The example uses fixed values of s2, and s1values extracted at random from the distribution of sperm competitiveness S (A) or S normal (B). If F2 were a perfect estimator of sperm competitive ability the solid line should be obtained.
An important consequence of the patterns of bias is that the variance for the observed ability of males to win fertilizations is reduced. This is confirmed in the analysis in which the coefficients of variation for s2 values and the associated F2 values are calculated following the simulation of sperm competition experiments. When sperm competitive ability values are randomly extracted from the distribution S, the mean (± SD) coefficient of variation for the set of s2 values is 38.1% ± 3.5 (n = 10000 experiments of sample size 50 each) compared to 25.7% ± 2.4 for the set of associated F2 values (T-test for dependent samples t9999 = 391.6, p <<0.0001). A lower but still highly significant decrease in the coefficient of variation is found when sperm competitive ability values are extracted from the Snormal distribution: 23.8% ± 2.5 compared to 17.7% ± 2.0, for s2 values and associated F2 values respectively (T-test for dependent samples t9999 = 286.9, p <<0.0001).
The relative nature of fertilization success and the estimation of genetic variance in sperm competitiveness
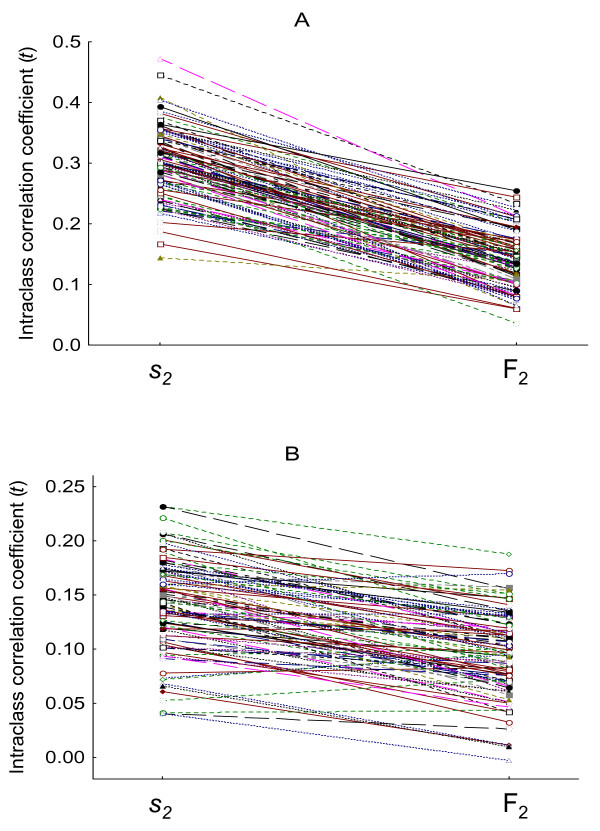
The consequences of the relative nature of F2 values for investigations of genetic variance in sperm competitive ability have been examined with analyses of the intraclass correlation coefficient (t), recreating hypothetical studies using fertilization success values as proxy of sperm competitive ability (see Methods), such as it is done in real experiments. Results from these analyses show that the heritability of sperm competitive ability, as inferred from fertilization success, can be seriously underestimated. The comparison between t generated from "true" sperm competitive ability values and that generated from the F2 values arising from those sperm competitive ability values shows that t is decreased to a great extent using F2 values. When sperm competitive ability values are randomly extracted from the distribution S, the mean (± SD) t for sperm competitive ability (s2) calculated from 10000 experimental genetic designs (50 sires and 8 offspring per sire in each experiment) is 0.30 ± 0.06 compared to 0.14 ± 0.04 when using the set of associated F2 values (T-test for dependent samples t9999 = 366.4, p <<0.0001) (See Fig. 2A). This means an average reduction for t of around 50%. For example, under the conditions described in the methods, heritabilities of sperm competitiveness calculated in full-sib experiments of around 0.6 will be on average estimated as being 0.3 because of the deviations that fertilization success values impose on the real sperm competitive ability values. Similarly, a significant decrease in the intraclass correlation coefficient is obtained when sperm competitive values are extracted from the distribution Snormal: 0.15 ± 0.05 compared to 0.10 ± 0.04, for s2 values and associated F2 values respectively (T-test for dependent samples t9999 = 162.5, p <<0.0001) (Fig. 2B).
Figure 2.
Reduction in the intraclass correlation coefficient due to variance arising from the relative nature of fertilization success. The figure shows the intraclass correlation coefficient for sperm competitive ability (s2) and the associated coefficient when sperm competitive ability is estimated using fertilization success values (F2). Each line pertains to a single sib analysis (involving 50 sires and 8 offspring per sire). Only a random subsample of 100 sib analyses is shown. Sperm competitive ability values were extracted from the distribution S (A) or Snormal (B).
Discussion
Research on sperm competition has increased exponentially since the first formal account of its implications [1] and it is now recognized as a powerful force shaping numerous traits in males and females [2], including polyandrous behaviour [5]. In many cases the understanding of post-copulatory processes relies on the measurement of one critical parameter: sperm competitive ability of individual males. However, accurately determining sperm competitive ability presents some problems, partially because it involves third parties. This study cautions that fertilization success is not a perfect predictor of sperm competitive ability, because of the relative nature of the calculation of fertilization success measures. Fertilization success values render biased sperm competitive ability values. Importantly, this bias is not constant along all the range of sperm competitiveness values and a consequence of the patterns of bias is that variance for the observed ability of males to win fertilizations is significantly reduced. Critically, as shown in simulations of sibling analyses, the heritability of sperm competitive ability is seriously underestimated when fertilization success (or paternity success) values are used. Thus, the ability to detect additive genetic variance underlying the sperm competitiveness phenotype is hindered.
A number of factors, including interactions between male and female genotypes [36,43], have been suggested to contribute towards the maintenance of genetic variation in sperm competitive ability or in traits determining sperm competitive ability [reviewed in [11,19,61]]. The existence of male × male interactions may further complicate the determination of the unit of selection for sperm competition [34,43]. However, the role of male × male interactions in maintaining genetic variation is still uncertain. Recently, Bjork et al [42] have carried out a thorough examination of ejaculate × female and ejaculate × ejaculate interactions on sperm offense and defence in Drosophila melanogaster using a complex experimental set up that allowed them to measure the repeatabilities of sperm precedence (estimated from paternity success values) across multiple sperm competition trials. Bjork et al [42] found that both sperm offense and defense were highly repeatable in matings involving the same pair of males and the same female. These repeatabilities declined when the same pair of males was mated to different females, indicating that the outcome of sperm competition responds to interactions between male × female genotypes. Finally, the repeatabilities of paternity scores disappeared when males competed each time against different rival males within different females. It has been suggested that this kind of result supports the notion that the maintenance of non-heritable variation in sperm competitive ability is due to interactions between competing males [34,42]. It is important to know, however, whether scores showing low consistency when rival males are changed are due to non-additive variance arising from genotypic male by male interactions or to random, purely environmental, effects due to the relative nature of fertilization success (or paternity success) measures. Ejaculate × ejaculate interactions can be due to genotypic interactions sensu stricto between competing males (i.e., due to polymorphism in genes responsible for sperm competitiveness), implying non-transitivity in sperm competitive ability. Prout and Bundgaard [62] provide an example of this scenario, in which genotype AA outcompetes genotype Aa, and Aa is superior that aa, but aa is superior than AA. In this case, it would not be possible to rank males on the basis of their sperm competitiveness, and as Clark [34] points out, the success in sperm competition of a particular genotype will depend on the presence and frequencies of other male genotypes in the population. It seems clear that this type of ejaculate × ejaculate interaction would indeed contribute to the maintenance of polymorphism in genes determining sperm competitive ability [34,43]. Alternatively, ejaculate × ejaculate interactions can be observed as the result of the relative nature of fertilization/paternity success calculations, even if sperm competitive ability is an intrinsic trait allowing ranking of males: males will score better or worse depending on the sperm competitive abilities of rival males. In this case, ejaculate × ejaculate interactions would explain variance in the outcome of sperm competition, but it is less clear that these interactions contributed towards the maintenance of genetic variation in sperm competitive ability. The evolutionary consequences of these two types of ejaculate × ejaculate interactions differs, and it seems important to investigate the existence of genetic variation in sperm competitive ability controlling, if possible, for random effects arising from the relative nature of fertilization success. Accounting for these effects would inform on whether sperm competitive ability can be treated as an intrinsic trait or whether it should be treated as interacting phenotype [19,63-65].
As it has been recently pointed out by Dowling et al. [65], studies examining genetic variance in the outcome of sperm competition have generally found lower levels of additive genetic variance than studies focusing on specific traits with a role in sperm competition. Results in this paper highlight that confounding random effects arising from the relative nature of fertilization success could explain this discrepancy. While these effects do not affect estimations of genetic variance in specific sperm competition traits, because in these cases absolute measures are taken (e.g., testis size of individuals is measured), they are likely to confound estimations of genetic variance underlying holistic measures of sperm competitiveness. This, however, does not mean that the study of absolute specific traits should be preferred over the study of fertilization success, or vice versa. Fertilization success is the only integrative measure for the outcome of sperm competition and the foremost predictor of absolute sperm competitive ability in general terms. The investigation of the sources of variation in the observable output of sperm competition is warranted [20,23,24,27,34,42,66], and both types of studies (those focusing on a few traits and those focusing on the sperm competitiveness phenotype) are important if we are to advance the understanding of post-copulatory sexual selection.
Interpretations on the plausibility of good-genes or compatible-genes underlying the genetic benefits of polyandrous behaviour often rely on the detection of additive (intrinsic) effects of genes influencing a male's sperm competitiveness [10,11]. However, obtaining evidence for intrinsic sire effects on the sperm competitiveness phenotype is impeded to some degree due to random effects arising from the background in which sperm competitiveness is estimated. This study, therefore, supports the notion that a lack of evidence for sire effects on fertilization success should be taken with caution, and that the genetic incompatibility hypothesis needs to be treated as an alternative hypothesis, rather than a null hypothesis, when testing for post-copulatory processes based on good genes.
The simulations of sperm competition mating trials demonstrate that there can be significant additive genetic variation in sperm competitive ability that researchers measuring fertilization success may fail to detect. Given that it is fertilization success that ultimately determines the patterns of paternity, it could be argued that females facilitating sperm competition will encounter the same problem as researchers staging random mating trials and that they will not be able to obtain genetic benefits. However, fertilization success is a multiple trait that is mainly determined by the action and interaction of absolute traits contributing to sperm competitiveness, which are the real targets of selection (a useful analogy could be, for instance, mating success, which may be determined by a number of traits such as body size, vigour, size of ornaments, etc.). Importantly, the genetic benefits that a polyandrous female can obtain will be determined by the existence of additive genetic variance in traits conferring sperm competitive ability, because these traits will be inherited by the offspring and they will determine to a great extent their subsequent fertilization success. We can imagine a scenario in which there is one male, M, with high absolute sperm competitive ability (0.9 on a scale from 0 to 1), mating with two females: female A, previously mated with another male of high absolute sperm competitive ability (0.85), and female B, previously mated with a male of low sperm competitive ability (0.01). Fertilization success for the male M is clearly not repeatable across these matings (fertilization success will be 0.51 when mated to female A, and 0.99 when mating with female B). However, if sperm competitive ability is heritable, females A and B will benefit through the offspring sired by male M because of the inheritance of high absolute sperm competitive ability that will confer, on average and despite stochastic influences (contingent on the distribution of sperm competitiveness in the population of males), high fertilization success. It is, therefore, the existence of additive genetic variance in traits contributing to sperm competitive ability that determines the potential for the acquisition of good genes through post-copulatory processes in polyandrous females.
It is important to bear in mind that the confounding effects imposed by the relative nature of fertilization success are particularly severe when sperm competitive ability for the individual is the parameter with relevance for inferring post-copulatory processes. In addition, it is worth mentioning that the magnitude of the random effects would depend on the distribution of sperm competitive ability and on the mechanism of sperm competition. However, these effects are likely to occur whenever there is variation among males in sperm competitive ability and some degree of sperm mixing (including the loaded raffle, [67]), or whenever the outcome of sperm competition is determined by interactions between the determinants of paternity of competing males. Finally, it is important to take into account that fertilization success is generally estimated from paternity success at hatching or birth. In this case, differential embryo viability across competing males may imply another source of variation that would further obscure examinations of additive genetic variance in sperm competitive ability [56].
The fact that individual fertilization success values are not always a reflection of sperm competitive ability would generate confounding effects in analyses based on the ranking of males, in particular those investigating the association between sperm competitiveness and phenotypic or life-history traits. Putting the measures of fertilization success in context with those from rival males can alleviate the problem imposed by its relative nature in these studies. Some other methodological measures could ameliorate the problems imposed by the relative nature of fertilization success. In general terms, random extraction of males from the population together with the use of large sample sizes to calculate fertilization success values are advisable to minimize random effects influencing the degree to which fertilization success does not reflect sperm competitiveness. In studies looking at the genetic variance of sperm competitiveness, several approaches could be adopted, although none of them is probably a perfect solution. Sperm competitive ability could be tested against standardised sperm competitors. The use of either the same male or the same set of tester rival males would minimize the problems imposed by the relative nature of fertilization success. This approach will analyse genetic variance in sperm competitive ability in absence of not only variation due to random effects, but also due to genotypic ejaculate × ejaculate interactions. This approach is, however, difficult to implement in most species with internal fertilization due to confounding age effects or mating history effects. In D. melanogaster, a promising line for the study of additive genetic variation is hemiclonal analysis (members of a single hemiclone share a random genomic haplotype) [27]. This methodology presents some important strengths including that it virtually screens the entire genome, and that measures of genetic variation are devoid of maternal effects, dominance, and practically epistatic variation [27]. Using this technique, Friberg et al. [27] recently found low but significant heritable variation in offensive and defensive sperm displacement in D. melanogaster. Hemiclones could be tested against standardized competitor males; for instance, members of a single hemiclone could be used as competitors for screening variation in sperm competitive ability among an array of hemiclones. In this way, differences among hemiclones without the influence of random effects due to differences in the background in which sperm competitiveness is assayed could be determined. Several species of external fertilizers or species for which artificial insemination/fertilization techniques are available could also be models to examine sources of variance in fertilization success using a standard background with which to assess sperm competitiveness.
Conclusion
Sperm competitive ability is typically estimated following fertilization trials involving rival males. Thus, a given value of sperm competitive ability, which represents the absolute investment in traits that convey advantage under conditions of sperm competition, has the potential to generate a wide range of different fertilization success values, depending on the sperm competitive ability of rival males. Here I have shown that the relative nature of the calculation of fertilization success means that the inference of sperm competitive ability is biased. Random effects arising from the way that fertilization success is calculated may confound investigations on the genetic nature and the maintenance of genetic variance in sperm competitiveness, an area that is generating considerable debate [24,25,34,37,38,42,49]. Indeed, results in this study demonstrate that the detection/estimation of additive genetic variance in sperm competitiveness is hampered when using fertilization success. Given that observed low additive genetic variance in fertilization success is often taken as support for post-copulatory processes based on genetic incompatibilities driving polyandrous behaviour [10,11,61], these results have important implications for studies of the evolution of polyandry. The effects shown in this study suggest that taking the genetic incompatibility hypothesis as the null-hypothesis when tests for repeatability/heritability in fertilization success fail to support good-genes processes is not advisable.
Authors' contributions
FG-G conceived the study, performed the simulations, analyzed the data, and wrote the manuscript.
Supplementary Material
S, the frequency distribution of sperm competitive ability used as an example.
Probabilities for the deviations between fertilization success and sperm competitive ability.
Acknowledgements
I thank Leigh Simmons and Jon Evans for useful comments on earlier drafts of the manuscript, and two anonymous reviewers for constructive suggestions that improved the final version. This work was supported by a Discovery Project fellowship from the Australian Research Council.
References
- Parker GA. Sperm competition and its evolutionary consequences in the insects. Biol Rev. 1970;45:525–567. [Google Scholar]
- Birkhead TR, Møller AP. Sperm Competition and Sexual Selection. San Diego, California, Academic Press; 1998. [Google Scholar]
- Smith RL. Sperm competition and the evolution of animal mating systems. New York, Academic Press; 1984. [Google Scholar]
- Simmons LW. Sperm competition and its evolutionary consequences in the insects. Princeton, Princeton University Press; 2001. [Google Scholar]
- Curtsinger JW. Sperm competition and the evolution of multiple mating. Am Nat. 1991;138:93–102. [Google Scholar]
- Keller L, Reeve HK. Why do females mate with multiple males? The sexually selected sperm hypothesis. Adv Stud Behav. 1995;24:291–315. [Google Scholar]
- Harvey PH, May RM. Out for the sperm count. Nature. 1989;337:508–509. doi: 10.1038/337508a0. [DOI] [PubMed] [Google Scholar]
- Sivinski J. In: Sperm Competition and the Evolution of Animal Mating Systems. Smith RL, editor. London, Academic Press; 1984. Sperm in competition; pp. 86–115. [Google Scholar]
- Yasui Y. A "good sperm" model can explain the evolution of costly multiple mating by females. Am Nat. 1997;149:573–584. [Google Scholar]
- Neff BD, Pitcher TE. Genetic quality and sexual selection: an integrated framework for good genes and compatible genes. Mol Ecol. 2005;14:19–38. doi: 10.1111/j.1365-294X.2004.02395.x. [DOI] [PubMed] [Google Scholar]
- Evans JP, Simmons LW. The genetic basis of traits regulating sperm competition and polyandry: can selection favour the evolution of good- and sexy-sperm? Genetica. 2008. in press . [DOI] [PubMed]
- Zeh JA, Zeh DW. The evolution of polyandry I: intragenomic conflict and genetic incompatibility. Proc R Soc Lond B. 1996;263:1711–1717. [Google Scholar]
- Zeh JA, Zeh DW. The evolution of polyandry II: post-copulatory defences against genetic incompatibility. Proc R Soc Lond B. 1997;264:69–75. [Google Scholar]
- Tregenza T, Wedell N. Genetic compatibility, mate choice and patterns of parentage. Mol Ecol. 2000;9(8):1013–1027. doi: 10.1046/j.1365-294x.2000.00964.x. [DOI] [PubMed] [Google Scholar]
- Jennions MD, Petrie M. Why do females mate multiply? A review of the genetic benefits. Biol Rev. 2000;75(1):21–64. doi: 10.1017/s0006323199005423. [DOI] [PubMed] [Google Scholar]
- Zeh JA, Zeh DW. Reproductive mode and the genetic benefits of polyandry. Anim Behav. 2001;61(6):1051–1063. [Google Scholar]
- Simmons LW. The evolution of polyandry: Sperm competition, sperm selection, and offspring viability. Annu Rev Ecol Evol Syst. 2005;36(1):125–146. [Google Scholar]
- Wedekind C. Mate choice and maternal selection for specific parasite resistances before, during and after fertilization. Philos Trans R Soc Lond B Biol Sci. 1994;346(1317):303–311. doi: 10.1098/rstb.1994.0147. [DOI] [PubMed] [Google Scholar]
- Simmons LW, Moore AJ. In: Sperm Evolution. Birkhead TR, Pitnick S, Hosken DJ, editor. London, Elsevier; 2008. Evolutionary quantitative genetics of sperm. in press . [Google Scholar]
- Konior M, Keller L, Radwan J. Effect of inbreeding and heritability of sperm competition success in the bulb mite Rhizoglyphus robini. Heredity. 2005;94(6):577–581. doi: 10.1038/sj.hdy.6800649. [DOI] [PubMed] [Google Scholar]
- House CM, Simmons LW. Relative influence of male and female genital morphology on paternity in the dung beetle Onthophagus taurus. Behav Ecol. 2005;16(5):889–897. [Google Scholar]
- Lewis SM, Austad SN. Sources of intraspecific variation in sperm precedence in red flour beetles. Am Nat. 1990;135:351–359. [Google Scholar]
- Radwan J. Heritability of sperm competition success in the bulb mite, Rhizoglyphus robini. J Evol Biol. 1998;11(3):321–327. doi: 10.1038/sj.hdy.6800649. [DOI] [PubMed] [Google Scholar]
- Arnqvist G, Danielsson I. Copulatory behavior, genital morphology, and male fertilization success in water striders. Evolution. 1999;53:147–156. doi: 10.1111/j.1558-5646.1999.tb05340.x. [DOI] [PubMed] [Google Scholar]
- Konior M, Radwan J, Kolodziejczyk M, Keller L. Strong association between a single gene and fertilization efficiency of males and fecundity of their mates in the bulb mite. Proc Biol Sci. 2006;273(1584):309–314. doi: 10.1098/rspb.2005.3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson N, Tubman SC, Eady PE, Robertson GW. Female genotype affects male success in sperm competition. Proc R Soc Lond B. 1997;264:1491–1495. [Google Scholar]
- Friberg U, Lew TA, Byrne PG, Rice WR. Assessing the potential for an ongoing arms race within and between the sexes: Selection and heritable variation. Evolution. 2005;59(7):1540–1551. [PubMed] [Google Scholar]
- Simmons LW, Kotiaho JS. Evolution of ejaculates: patterns of phenotypic and genotypic variation and condition dependence in sperm competition traits. Evolution. 2002;56:1622–1631. doi: 10.1111/j.0014-3820.2002.tb01474.x. [DOI] [PubMed] [Google Scholar]
- Hosken DJ, Garner TW, Ward PI. Sexual conflict selects for male and female reproductive characters. Curr Biol. 2001;11(7):489–493. doi: 10.1016/s0960-9822(01)00146-4. [DOI] [PubMed] [Google Scholar]
- García-González F, Simmons LW. Shorter sperm confer higher competitive fertilization success. Evolution. 2007;61:816–824. doi: 10.1111/j.1558-5646.2007.00084.x. [DOI] [PubMed] [Google Scholar]
- Schulte-Hostedde AI, Millar JS, Hickling GJ. Condition dependence of testis size in small mammals. Evol Ecol Res. 2005;7:143–149. [Google Scholar]
- Engqvist L, Sauer KP. Determinants of sperm transfer in the scorpionfly Panorpa cognata: male variation, female condition and copulation duration. J Evol Biol. 2003;16(6):1196–1204. doi: 10.1046/j.1420-9101.2003.00613.x. [DOI] [PubMed] [Google Scholar]
- Civetta A, Clark AG. Chromosomal effects on male and female components of sperm precedence in Drosophila. Genet Res. 2000;75(2):143–151. doi: 10.1017/s0016672399004292. [DOI] [PubMed] [Google Scholar]
- Clark AG. Sperm competition and the maintenance of polymorphism. Heredity. 2002;88:148–153. doi: 10.1038/sj.hdy.6800019. [DOI] [PubMed] [Google Scholar]
- Miller GT, Pitnick S. Sperm-female coevolution in Drosophila. Science. 2002;298:1230–1233. doi: 10.1126/science.1076968. [DOI] [PubMed] [Google Scholar]
- Clark AG, Begun DJ, Prout T. Female x male interactions in Drosophila sperm competition. Science. 1999;283(5399):217–220. doi: 10.1126/science.283.5399.217. [DOI] [PubMed] [Google Scholar]
- Fiumera AC, Dumont BL, Clark AG. Sperm competitive ability in Drosophila melanogaster associated with variation in male reproductive proteins. Genetics. 2005;169:243–257. doi: 10.1534/genetics.104.032870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzari T, Birkhead TR. The sexually-selected sperm hypothesis: sex-biased inheritance and sexual antagonism. Biol Rev. 2002;77(2):183–209. doi: 10.1017/s1464793101005863. [DOI] [PubMed] [Google Scholar]
- Zeh JA. Sexy sons: a dead end for cytoplasmic genes. Proc Biol Sci. 2004;271(Suppl 5):S306–S309. doi: 10.1098/rsbl.2004.0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froman DP, Kirby JD. Sperm mobility: phenotype in roosters (Gallus domesticus) determined by mitochondrial function. Biol Reprod. 2005;72:562–567. doi: 10.1095/biolreprod.104.035113. [DOI] [PubMed] [Google Scholar]
- Gemmell NJ, Metcalf VJ, Allendorf FW. Mother's curse: the effect of mtDNA on individual fitness and population viability. Trends Ecol Evol. 2004;19(5):238–244. doi: 10.1016/j.tree.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Bjork A, Starmer WT, Higginson DM, Rhodes CJ, Pitnick S. Complex interactions with females and rival males limit the evolution of sperm offence and defence. Proc Biol Sci. 2007;274(1619):1779–1788. doi: 10.1098/rspb.2007.0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Dermitzakis ET, Civetta A. Nontransitivity of sperm precedence in Drosophila. Evolution. 2000;54(3):1030–1035. doi: 10.1111/j.0014-3820.2000.tb00102.x. [DOI] [PubMed] [Google Scholar]
- Fisher DO, Double MC, Blomberg SP, Jennions MD, Cockburn A. Post-mating sexual selection increases lifetime fitness of polyandrous females in the wild. Nature. 2006;444(7115):89–92. doi: 10.1038/nature05206. [DOI] [PubMed] [Google Scholar]
- Gage MJG, Morrow EH. Experimental evidence for the evolution of numerous, tiny sperm via sperm competition. Curr Biol. 2003;13(9):754–757. doi: 10.1016/s0960-9822(03)00282-3. [DOI] [PubMed] [Google Scholar]
- García-González F, Simmons LW. Sperm viability matters in insect sperm competition. Curr Biol. 2005;15(3):271–275. doi: 10.1016/j.cub.2005.01.032. [DOI] [PubMed] [Google Scholar]
- Gage MJG, Macfarlane CP, Yeates S, Ward RG, Searle JB, Parker GA. Spermatozoal traits and sperm competition in Atlantic salmon: relative sperm velocity is the primary determinant of fertilization success. Curr Biol. 2004;14(1):44–47. [PubMed] [Google Scholar]
- Radwan J. Intraspecific variation in sperm competition success in the bulb mite: A role for sperm size. Proc R Soc Lond B. 1996;263(1372):855–859. [Google Scholar]
- Fiumera AC, Dumont BL, Clark AG. Associations between sperm competition and natural variation in male reproductive genes on the third chromosome of Drosophila melanogaster. Genetics. 2007;176(2):1245–1260. doi: 10.1534/genetics.106.064915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poiani A. Complexity of seminal fluid: a review. Behav Ecol Sociobiol. 2006;60(3):289–310. [Google Scholar]
- Wolfner MF. Tokens of love: Functions and regulation of Drosophila male accessory gland products. Insect Biochem Mol Biol. 1997;27(3):179–192. doi: 10.1016/s0965-1748(96)00084-7. [DOI] [PubMed] [Google Scholar]
- House CM, Simmons LW. Genital morphology and fertilization success in the dung beetle Onthophagus taurus: an example of sexually selected male genitalia. Proc Biol Sci. 2003;270(1514):447–455. doi: 10.1098/rspb.2002.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorman E, Parker GA. Sperm (ejaculate) competition in Drosophila melanogaster, and the reproductive value of females to males in relation to female age and mating status. Ecol Entomol. 1976;1:145–155. [Google Scholar]
- Olsson M, Pagel M, Shine R, Madsen T, Doums C, Gullberg A, Tegelstrom H. Sperm choice and sperm competition: suggestions for field and laboratory studies. Oikos. 1999;84(1):172–175. [Google Scholar]
- García-González F, Simmons LW. Paternal indirect genetic effects on offspring viability and the benefits of polyandry. Curr Biol. 2007;17:32–36. doi: 10.1016/j.cub.2006.10.054. [DOI] [PubMed] [Google Scholar]
- García-González F. Male genetic quality and the inequality between paternity success and fertilization success: consequences for studies of sperm competition and the evolution of polyandry. Evolution. 2008. in press . [DOI] [PubMed]
- Parker GA. Sperm competition games: raffles and roles. Proc Biol Sci. 1990;242:120–126. [Google Scholar]
- Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sunderland, MA, Sinauer; 1998. [Google Scholar]
- Hood GM. PopTools version 2.7.5. 2006. http://www.cse.csiro.au/poptools
- Statsoft. STATISTICA for Windows (Computer program manual) Tulsa; 1996. [Google Scholar]
- Zeh JA, Zeh DW. Maternal inheritance, epigenetics and the evolution of polyandry. Genetica. 2008. in press . [DOI] [PubMed]
- Prout T, Bundgaard J. The population genetics of sperm displacement. Genetics. 1977;85(1):95–124. doi: 10.1093/genetics/85.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AJ, Brodie ED, Wolf JB. Interacting phenotypes and the evolutionary process .1. Direct and indirect genetic effects of social interactions. Evolution. 1997;51(5):1352–1362. doi: 10.1111/j.1558-5646.1997.tb01458.x. [DOI] [PubMed] [Google Scholar]
- Moore AJ, Pizzari T. Quantitative genetic models of sexual conflict based on interacting phenotypes. Am Nat. 2005;165(5):S88–S97. doi: 10.1086/429354. [DOI] [PubMed] [Google Scholar]
- Dowling DK, Friberg U, Arnqvist G. A comparison of nuclear and cytoplasmic genetic effects on sperm competitiveness and female remating in a seed beetle. J Evol Biol. 2007;20(6):2113–2125. doi: 10.1111/j.1420-9101.2007.01433.x. [DOI] [PubMed] [Google Scholar]
- Birkhead TR, Chaline N, Biggins JD, Burke T, Pizzari T. Nontransitivity of paternity in a bird. Evolution. 2004;58(2):416–420. [PubMed] [Google Scholar]
- Parker GA. In: Sperm competition and sexual selection. Birkhead TR, Møller AP, editor. San Diego, California, Academic Press; 1998. Sperm competition and the evolution of ejaculates: towards a theory base; pp. 3–54. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S, the frequency distribution of sperm competitive ability used as an example.
Probabilities for the deviations between fertilization success and sperm competitive ability.