Abstract
The premature human aging Werner syndrome (WS) is caused by mutation of the RecQ-family WRN helicase, which is unique in possessing also 3′–5′ exonuclease activity. WS patients show significant genomic instability with elevated cancer incidence. WRN is implicated in restraining illegitimate recombination, especially during DNA replication. Here we identify a Drosophila ortholog of the WRN exonuclease encoded by the CG7670 locus. The predicted DmWRNexo protein shows conservation of structural motifs and key catalytic residues with human WRN exonuclease, but entirely lacks a helicase domain. Insertion of a piggyBac element into the 5′ UTR of CG7670 severely reduces gene expression. DmWRNexo mutant flies homozygous for this insertional allele of CG7670 are thus severely hypomorphic; although adults show no gross morphological abnormalities, females are sterile. Like human WS cells, we show that the DmWRNexo mutant flies are hypersensitive to the topoisomerase I inhibitor camptothecin. Furthermore, these mutant flies show highly elevated rates of mitotic DNA recombination resulting from excessive reciprocal exchange. This study identifies a novel WRN ortholog in flies and demonstrates an important role for WRN exonuclease in maintaining genome stability.
Keywords: exonuclease, Drosophila, genome stability, homologous recombination, Werner syndrome, WRN
Introduction
Werner syndrome (WS) provides a very useful model system for the study of human aging at the molecular level, with patients manifesting many signs of normal aging in an accelerated manner (reviewed in Martin, 1985; Goto, 2001; Cox & Faragher, 2007; Kudlow et al., 2007). The syndrome is caused by mutation of WRN (Yu et al., 1996), a member of the RecQ DNA helicase family. WS patient-derived cells undergo highly premature replicative senescence, with cellular defects including aberrant DNA replication (Pichierri et al., 2001; Rodriguez-Lopez et al., 2002) and hyper-recombination (Salk et al., 1981; Scappaticci et al., 1982; Fukuchi et al., 1985). Hypersensitivity to the DNA-damaging agent 4-nitroquinoline oxide, and the topoisomerase I inhibitor camptothecin (CPT) is characteristic of WS cells (Poot et al., 1999; Prince et al., 1999; Pichierri et al., 2000b). These agents lead to replication fork arrest or collapse, suggesting a function for WRN in DNA replication, which is further supported by its presence at replication foci coincident with RP-A and PCNA (Constantinou et al., 2000; Rodriguez-Lopez et al., 2003) and aberrant replication fork progression in WS fibroblasts (Rodriguez-Lopez et al., 2002). The hyper-recombinant phenotype of human WS cells, suppression of illegitimate recombination in yeast Sgs1 mutants by human WRN (Yamagata et al., 1998), interaction with MRN on replication fork stalling (Franchitto & Pichierri, 2004), the recovery of proliferative capacity after ectopic expression of a Holliday junction resolvase in WS cells (Rodriguez-Lopez et al., 2002), and excessive chromosome breakage at fragile sites in the absence of WRN (Pirzio et al., 2008) all suggest an important role for WRN in homologous recombination after replication fork arrest, either in preventing the formation of homologous recombination intermediates or in their rapid resolution (Dhillon et al., 2007; Rodriguez-Lopez et al., 2007). The importance of WRN in regulating genome stability is highlighted by the high cancer incidence in WS patients, while epigenetic inactivation of WRN is also associated with human cancer (Agrelo et al., 2006).
Identification of the action of WRN in homologous recombination is complicated by the presence of two enzymatic activities within the same protein: the 3′–5′ helicase characteristic of all RecQ family members (reviewed in Bachrati & Hickson, 2003), and a 3′–5′ exonuclease activity (Huang et al., 1998) unique within this family, but which is closely related structurally to the DnaQ exonuclease superfamily (Perry et al., 2006). X-ray crystallographic analysis of the exonuclease domain of human WRN suggests a role in DNA end processing (Perry et al., 2006), possibly at the stage of strand resection after double-strand breaks, such breaks that as occur at collapsed replication forks. This would be consistent with the importance of WRN in DNA recombination. However, the closely related BLM helicase can, at least in vitro, promote dissolution of double Holliday junctions without intrinsic nuclease activity (Wu & Hickson, 2003); determining the relative contributions to homologous recombination of the helicase and exonuclease activities of WRN is therefore important. Moreover, there appears to be a complex interplay between the helicase and exonuclease activities of WRN (Opresko et al., 2001); for example, it is possible that helicase activity may be required to generate a template suitable for cleavage by the nuclease. The distinct roles are difficult to dissect in vertebrate cells since ablation of one activity may affect the other; indeed, point mutation of the helicase is suggested to act in a dominant negative manner (Crabbe et al., 2004). RNAi depletion of WRN, although highly effective in recapitulating some WS-like phenotypes (Dhillon et al., 2007), eliminates both helicase and exonuclease activities.
To study WRN's role in recombination at the organismal level, we sought to develop a model in which WRN activity may be evaluated at different developmental stages. Although murine models have been described which show some WS-like features on mutation of the WRN helicase alone or with co-mutation of either telomerase or PARP (Lebel, 2002; Lebel et al., 2003; Chang et al., 2004; Massip et al., 2006), the relatively long lifespan and complexity of genetic intervention pose severe limitations on their exploitation. In order to develop a model system more amenable to genetic and biochemical analysis of WRN exonuclease function in vivo, we set out to identify and characterise WRN exonuclease from Drosophila melanogaster.
Results
Identification of Drosophila WRNexo
We conducted a BLASTP search (Altschul et al., 1997) of the Drosophila melanogaster genome sequence (Release 4.0), using as a probe the sequence of human WRN protein. The Drosophila candidate gene encoding a WRN-like exonuclease is CG7670, with an E-value of 1 × 10−25 (Cox et al., 2007, as also noted by Sekelsky et al., 2000). Upon cloning from mRNA and sequencing multiple CG7670 cDNA clones, we found two alleles of CG7670 differing solely by the presence or absence of an AAG codon (lysine) at nucleotide 235, amino acid 79 (GenBank accession numbers EF680279 and EF680280, respectively). Variant 2 (EF680280) lacking lysine 79 was the more commonly occurring clone, encoding a predicted protein of 352 amino acids.
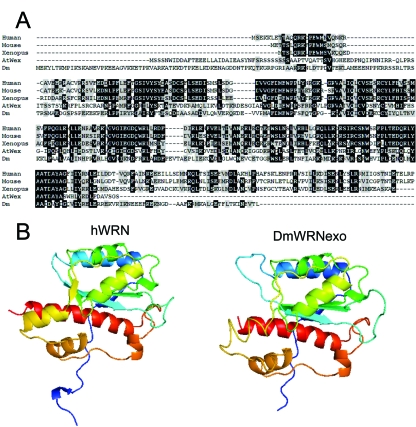
The predicted protein product of the CG7670 locus, which we call DmWRNexo, shares 35% identical and 59% similar amino acids with the exonuclease domain of human WRN over a region of 192 residues (Fig. 1A). Previous crystallographic studies of the human WRN exonuclease domain demonstrated that residues aspartate (D82) and glutamate (E84) within the nuclease catalytic site are essential for metal ion coordination (Perry et al., 2006). Importantly, these residues are conserved in DmWRNexo (Fig. 1A, asterisks). We have conducted SWISS-MODEL structural predictions (Peitsch, 1996; Schwede et al., 2003) of DmWRNexo from residues 118–312, which suggest that the protein might adopt a very similar configuration to human WRN exonuclease (Perry et al., 2006, PDB accession number 2fbyA, predicted similarity e-value 8.86 × 10−26), with conservation of key alpha helices and beta sheets comprising the nuclease active site (Fig. 1B).
Fig. 1.
Homology of DmWRNexo to human WRN. (A) Alignment of WRN protein sequences from human, mouse, Xenopus laevis and Arabidopsis thaliana (AtWEX) with full-length DmWRNexo (predicted from both our cloned CG7670 cDNA and genomic sequence (Flybase)). Black depicts residues identical in three or more species, with grey showing similarity. Note the highly conserved blocks of sequence flanking the catalytic core residues. D82 and E84 metal-ion co-coordinating residues (residues numbered for the human WRN protein) are marked with an asterisk. (B) SWISS-MODEL was used to predict the possible tertiary structure of DmWRNexo from residues 118–312, according to the known structure of human WRN exonuclease (2fbyA, Perry et al., 2006). Left panel shows the human WRN exonuclease domain while the right panel shows the predicted structure of Drosophila melanogaster DmWRNexo.
Hypomorphic allele of CG7670
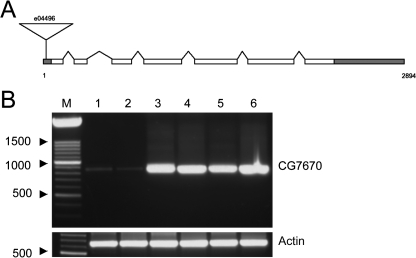
To assess the impact of DmWRNexo mutation on flies, we obtained an insertional mutant allele of CG7670, CG7670e04496, which contains a piggyBac{RB} element (Thibault et al., 2004) inserted within the 5′ UTR (Fig. 2A). Reverse transcription–polymerase chain reaction (RT-PCR) analysis shows that the CG7670e04496 allele is transcribed at an extremely low level in the homozygous mutant compared with CG7670 expression in heterozygous and wild-type flies (Fig. 2B); no band was detectable in negative controls (data not shown). Interestingly, CG7670e04496 homozygotes show no gross morphological abnormality, and while the females are sterile (eggs do not hatch), males are fertile. The location of CG7670 on chromosome 3R:14189966.14191859 (Flybase) and its identity with the gene encoding DmWRNexo is consistent with our mitotic recombination deficiency mapping studies (mwh1 CG7670e04496/Df(3R)Exel6178 flies display multiple wing hair clones; data not shown).
Fig. 2.
CG7670e04496 mutant is severely hypomorphic. (A) The predicted structure of the CG7670 transcript. The location of the piggyBac insertion e04496 is indicated. (B) Reverse transcription–polymerase chain reaction (RT-PCR) indicates that CG7670 is expressed at very low levels compared with heterozygotes or wild-type controls. cDNA was generated by one-step RT-PCR using primers specific to correctly spliced CG7670(upper panel) or actin 5C (lower panel), yielding products of 854 bp (CG7670) and 652 bp (actin). Lanes 1, 2 CG7670e04496 homozygotes; lanes 3, 4 CG7670e04496 heterozygotes; lanes 5, 6 wild type. Lanes 1, 3, 5 = male; lanes 2, 4, 6 = female.
DmWRNexo mutant flies are hypersensitive to CPT
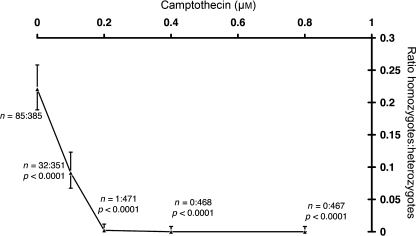
Werner syndrome cell lines are sensitive to the topoisomerase I poison CPT (Poot et al., 1999; Pichierri et al., 2000a) which causes replication fork collapse at the bound topoisomerase (Shao et al., 1999); such sensitivity can be partially complemented by expression of a bacterial Holliday junction nuclease (Rodriguez-Lopez et al., 2007), suggesting that WRN acts either to prevent accumulation of Holliday junctions at collapsed forks or to ensure rapid Holliday junction resolution. To test whether Drosophila mutant for DmWRNexo are similarly sensitive to CPT, larvae derived from crosses of CG7670e04496 heterozygotes were propagated on medium supplemented with varying concentrations of CPT, or vehicle-only control (0 µm CPT). Emerging flies were scored for heterozygosity or homozygosity of the CG7670e04496 allele. While the heterozygous flies appear to be fully viable at all concentrations of CPT used (Fig. 3), a significant loss of viability of flies homozygous for the CG7670e04496 allele (i.e. those with very low levels of expression of DmWRNexo) was observed even at 0.1 µm CPT, with almost total lethality from 0.2 µm (Fig. 3). Surviving homozygotes displayed roughened eyes, an indicator of cell death, and many died as pharate adults (data not shown), a typical lethal phase for flies exhibiting high levels of cell death. This is consistent with the high levels of apoptosis detected in human Werner syndrome cells exposed to CPT (Poot et al., 1999). Thus, loss of DmWRNexo results in hypersensitivity to CPT.
Fig. 3.
DmWRNexo mutant flies are sensitive to camptothecin (CPT). Flies heterozygous for the CG7670e04496 allele were crossed and progeny reared on medium supplemented with various concentrations of CPT. The proportion of all emerging adult flies that were homozygous for CG7670e04496 was plotted against CPT concentration; heterozygous flies were fully viable at all CPT concentrations tested. The lower frequency of homozygous flies observed at increasing drug doses demonstrates hypersensitivity to CPT. n represents the ratio of CG7670e04496 homozygous: heterozygous flies. p-values refer to the Fisher exact test for the probability that the proportion of homozygotes was less than that of the control value (0 µm CPT). The error bars show binomial 95% confidence intervals.
Genome instability in DmWRNexo mutant flies
Since hyper-recombination is a key phenotype of WS patient-derived cell lines (Salk et al., 1981; Scappaticci et al., 1982; Fukuchi et al., 1985), rates of chromosome breakage and/or mitotic recombination in DmWRNexo homozygous mutant flies were evaluated using the recessive multiple wing hairs marker (mwh, recombination map position 3-0.7); wing blade cells hemizygous or homozygous for mwh1 develop tufts of wing hairs instead of single hairs. Note that the adult wing consists of postmitotic cells arising from proliferating cells of the wing imaginal disc, so any recombination giving rise to clones of cells with the mwh phenotype must have occurred during cell proliferation in development.
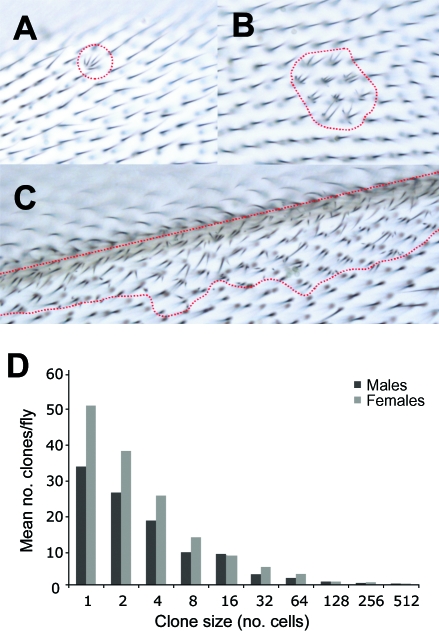
Wing blades were dissected from flies that were homozygous mutant for DmWRNexo but heterozygous for mwh (i.e. w1118; mwh1 CG7670e04496/CG7670e04496) and analysed microscopically (Fig. 4A–C). The frequency and size of clones showing multiple wing hairs was determined (Fig. 4D), demonstrating that mwh clones occur at a very high frequency in the DmWRNexo homozygous mutant flies, with an average of over 100 clones per fly. This is in sharp contrast to flies heterozygous for DmWRNexo which show a mean of 0.2 mwh clones per fly (data not shown). Furthermore, some mwh clones in DmWRNexo homozygous mutant wing blades were very large (> 500 cells) (Fig. 4D), while the rare clones observed in heterozygous flies were all single cells (data not shown). In addition to the very high rates of recombination detected in DmWRNexo mutant flies, these data also demonstrate that the CG7670e04496 allele is recessive, as reported for patient-derived human WRN mutations (Yu et al., 1996; Moser et al., 1999).
Fig. 4.
Hyper-recombination in DmWRNexo mutant flies. (A–C): Wing blades of flies homozygous for the hypomorphic allele of DmWRNexo (w; mwh1 CG7670e04496/CG7670e04496) were analysed microscopically; mwh clones are outlined with a broken line. (A) single-cell clone, (B) 10-cell clone, and (C) moderate-sized clone of mwh cells. (D) Frequency of mwh clones in wing blades of DmWRNexo homozygous mutant flies (w; mwh1 CG7670e04496/CG7670e04496) plotted against clone size, i.e. number of cells comprising the clone (number of clones counted: males n = 1278, females n = 1584). mwh clones were infrequent (0.2 per fly) in heterozygous controls, and all were single-cell clones (not shown).
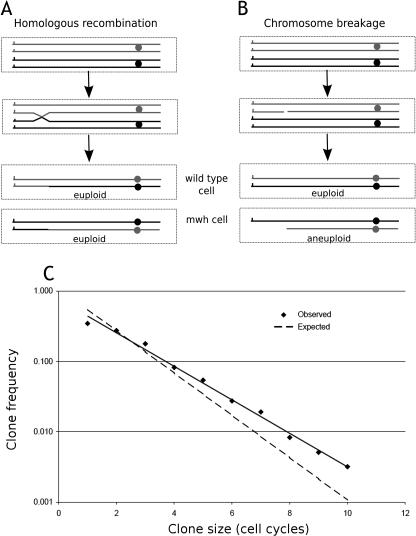
Cells in the wing blade showing the recessive multiple wing hairs phenotype could genetically be either homozygous or hemizygous for the mwh1 allele. Homozygous mwh1 cells would result from mitotic recombination via a reciprocal exchange event (Fig. 5A); daughter cells should be euploid without any loss of proliferative fitness. By contrast, chromosome loss or single chromosome/chromatid breakage events (Fig. 5B) would give rise to segmentally aneuploid hemizygous mwh cells, which would be predicted to proliferate more slowly than euploid cells. To distinguish between these possibilities, we have measured the size of each clone in terms of the number of cell cycles since its generation, and plotted the proportion of clones in each size class (on a logarithmic scale) against clone size (Fig. 5C). This type of analysis informs on the nature of the event leading to clone formation as the gradient of the plot reflects the growth rate of the clones (Baker et al., 1978). If clones proliferate at the same rate as surrounding normal cells, the gradient will closely parallel the expectation (that clones of size class n will be twice as numerous as clones of size class n + 1). Clones that proliferate more slowly, as would be expected for segmental aneuploids, would fit a line of steeper gradient. Our results (‘Observed’, Fig. 5C) indicate that cells comprising wing blade clones in DmWRNexo mutant flies proliferate essentially as expected for euploid cells. However, the gradient is slightly shallower than expected for two reasons. First, adjacent smaller clones may have been scored as a single larger clone, and second, the recombination events happening earlier in the lineage of the clone (yielding large clones) depletes the pool of cells from which later events (smaller clones) can occur. The actual frequency of recombination events occurring may also be higher than that observed, since sister chromatid exchange is not scored in this assay. We therefore propose that mitotic recombination is the predominant cause of mwh clones in these flies. Based on mathematical simulations (data not shown), we estimate the recombination frequency on chromosome arm 3L to be at least 0.01 event per cell division. Assuming a similar frequency throughout the genome, this corresponds to an overall frequency of at least 0.05 recombination events per cell division.
Fig. 5.
Reciprocal exchange is the major mechanism of mwh clone origin. (A, B) Recombinational origin of mwh clones. Flies are initially heterozygous for mwh1 and homozygous for CG7670e04496. The parental chromosomes (shown as two sister chromatids linked at the centromere) are depicted in grey (mwh+) and black (mwh1); the mwh locus is indicated by a tick mark. (A) Homologous recombination between mwh and the centromere gives rise to euploid mwh+/mwh+ and mwh1/mwh1 daughter cells. (B) Chromosome breakage between mwh+ and the centromere gives rise to one euploid mwh1/mwh+ daughter cell of wild-type phenotype (and so not scored) and one aneuploid daughter cell that is hemizygous for the mutant mwh1 allele, and which therefore shows the mwh phenotype. (C) The proliferation rate of mwh clones supports mitotic exchange as the principal cause of clone formation. Logarithmic plot of the frequency of clones of each clone size class against clone size, where clone size is expressed in numbers of cell cycles completed. If recombination results in euploid cells which proliferate at a normal rate, the expectation is that clones of size n cell cycles should be twice as frequent as clones of cell cycle n + 1. This results in a line indicated as ‘Expected’ (broken line) in the graph. Should the causative mechanism result in slowly growing cells (for example, if the marked clones are derived from aneuploid cells), the distribution of clone frequencies would tend towards a line of steeper gradient. The gradient of the observed distribution (diamonds) corresponds well to the gradient expected (broken line) for euploid cells (see text for further details).
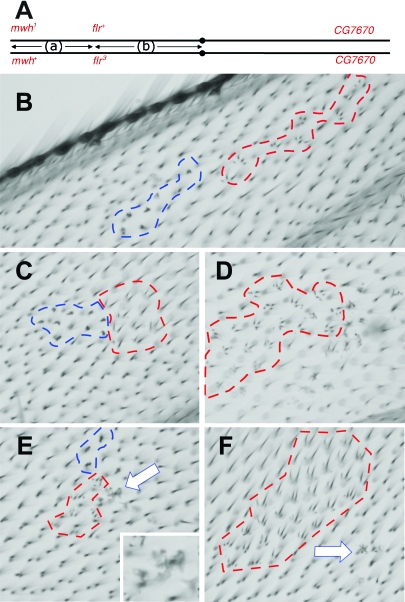
To further distinguish between chromosome breakage and homologous recombination as the principal mechanism for the elevated frequency of wing blade clones in CG7670e04496 homozygotes, we used a second cuticular marker, flare (flr, recombination map position 3–38.8), which causes wing blade hairs to be malformed. Wing blades of flies mutant for DmWRNexo and trans-heterozygous for mwh1 and flr3 (w1118; mwh1 CG7670e04496/flr3 CG7670e04496) were examined for mwh clones (arising as a consequence of recombination in the chromosomal interval between mwh and flr) and neighbouring mwh and flr clones (twin spots), resulting from recombination proximal to flr (Fig. 6). If the primary mechanism of genome instability in DmWRNexo mutants is chromosome breakage, mwh flr twin spots should be rare, while if mitotic recombination is the principal cause of wing blade clones, twin spots will be frequent. The observation of mwh flr twin spots at high frequency (Fig. 6E,F) strongly suggests that wing blade clones in the DmWRNexo mutants arise predominantly through mitotic recombination. Because of the high frequency of recombination in CG7670e04496 homozygotes, numerical analysis of the twin-spot frequency is difficult and further compounded by sequential repeated recombination events in clone lineages, as inferred from the observation of putative mwh flr double mutant cells (Fig. 6E,F). Nonetheless, it is clear that a significant proportion of wing blade clones are derived from homologous exchange. This is in marked contrast to the high rates of chromosomal breakage and loss observed in flies mutant for the RecQ homolog DmBLM (mus309) (Kusano et al., 2001). Additionally CG7670e04496 homozygote males are fully fertile, inconsistent with chromosomal breakage.
Fig. 6.
Twin-spot analysis confirms that hyper-recombination occurs through reciprocal exchange. (A) Position of mwh1 and flr3 markers relative to the centromere and CG7670 locus on chromosome 3 (not to scale). (B–F) Wing blade twin-spot mwh flr clones were analysed microscopically. mwh clones are outlined with red broken lines and flr clones by blue broken lines. (B, C) mwh-flr twin spots resulting from recombination proximal to flr (within interval (b) in diagram). (D–F) complex clones indicating sequential recombination events. Arrows in (E), (F) and the inset in (E) indicate a possible mwh flr double mutant cell.
Discussion
We have identified the locus CG7670 as the Drosophila ortholog of human WRN exonuclease. The encoded DmWRNexo protein shows significant structural and sequence similarities to human WRN exonuclease domain, and moreover, a severe hypomorphic mutation of the locus results in both hyper-recombination and CPT hypersensitivity in flies, features characteristic of human WS cells.
Our data demonstrate that loss of DmWRNexo function leads to very high levels of recombination in the developing Drosophila wing (and presumably also in other dividing tissues), consistent with hyper-recombination reported in cells from WS patients (Salk et al., 1981; Scappaticci et al., 1982; Fukuchi et al., 1985). The high frequency of twin spots in wing blade clones seen here strongly supports the assertion that the majority of marked clones in the mutant flies arise as a result of homologous recombination rather than chromosomal breakage. We cannot at this stage rule out the possibility that nonhomologous end joining is also aberrant, as is the case in human cells lacking functional WRN (Chen et al., 2003; Otsuki et al., 2007), since such end joining is unlikely to yield scoreable clones in the assays used here.
Mechanistically, it has been difficult to differentiate between the impact of the exonuclease and helicase activities of WRN in vivo since RNAi ablates all activities, while point mutants may act as dominant negatives (Crabbe et al., 2004). By studying a model organism in which the WRN exonuclease activity is encoded on a genomic locus distinct from any putative partner helicase, we can readily ablate the exonuclease activity without the possibility of creating dominant negative complexes. Our data presented here clearly demonstrate the importance of WRN exonuclease in restraining mitotic recombination. Furthermore, it is likely that at least some of the recombination detected in the DmWRNexo mutants occurs as a result of deficiencies in resolving aberrant DNA structures arising during DNA replication. The observed hypersensitivity of CG7670e04496 homozygotes to CPT is indicative of a role for DmWRNexo at collapsed replication forks, as predicted from human studies (Pichierri et al., 2001; Rodriguez-Lopez et al., 2002, 2007; Pirzio et al., 2008). Human WRN can regress replication forks in vitro (Machwe et al., 2006); how it supports re-establishment of collapsed forks in vivo is less clear, but our data suggest that the exonuclease activity of WRN may be important in preventing hyper-recombination at this stage. We speculate that DmWRNexo, like human WRN (Saintigny et al., 2002; Dhillon et al., 2007; Rodriguez-Lopez et al., 2007), may act to prevent Holliday junction accumulation at stalled or collapsed replication forks, and that DNA end processing activity of the exonuclease (Perry et al., 2006) may be critical to direct fork re-establishment. Such end processing could result in removal of DNA strands that would otherwise be used in the strand invasion step in homologous recombination. Thus, in cells lacking DmWRNexo, collapsed replication forks (such as at CPT-induced breaks) would persist, and promote Holliday junction formation and homologous recombination.
This study demonstrates the strength of using a genetically amenable model system for analysis of genes associated with genomic instability and human aging, even though the adult is largely postmitotic; absence of DmWRNexo function through fly development manifests as a hyper-recombinant phenotype in the mature adult. This raises the exciting possibility of using the short-lived fruit fly as model system for analysis and experimental modulation of WRN function in vivo.
Experimental procedures
Bioinformatics
BLAST searches (Altschul et al., 1997) were conducted against Release 4.0 of the Drosophila melanogaster genome sequence, and reciprocally against the human protein RefSeq database. BioEdit was used to generate the alignments following processing with Clustal W. Structural predictions were carried out using SWISS-MODEL (Peitsch, 1996; Schwede et al., 2003) based on the structure of human WRN exonuclease domain (Perry et al., 2006).
DNA and RNA analysis
Total RNA was extracted from flies using RNeasy spin columns (Qiagen, Crawley, West Sussex, UK) and quantitated using a Qubit fluorometer (Invitrogen, Paisley, UK). For analysis of transcript levels, one-step RT-PCR (Qiagen) was carried out with gene-specific primers (CG7670 Exon1–2 F: 5′-ATGAAGTTCCCAAGGAAGAGG-3′; CG7670 Exon1–2 R: 5′-GATGGCGGCGTACATTAGTT-3′; actin 5C F: 5′-CACCGGTATCGTTCTGGACT-3′, actin 5C R: 5′-GGACTCGTCGTACTCCTGCT-3′) using 0.5 µg total RNA. Products were analysed on 0.9% agarose gels stained with ethidium bromide, against a 100-bp ladder (Roche, Burgess Hill, West Sussex, UK).
To clone CG7670, cDNA was prepared from freshly isolated RNA from female flies (TM6B/TM3, wild type for CG7670) using Omniscript reverse transcriptase (Qiagen) and random hexamer primers (Operon, Cologne, Germany) or oligo-dt(16) primers (Applied Biosystems, Warrington, UK). The cDNA was PCR-amplified using pfx50 proofreading DNA polymerase (Invitrogen) and primers Forward F1A (CGGGTTATGGAAAAATATTTAACAAAAATGCCC) and Reverse R-2 A (AGCTTACAGAGTCACCTCGTTGATCTTGG), to yield a blunt-end PCR product which was cloned into TOPO vector (Zero Blunt®TOPO® PCR Cloning Kit, Invitrogen). DNA sequencing was performed in-house by Geneservice on an ABI 3730xl DNA Analyser.
Fly stocks
Fly stocks were obtained from the Bloomington Drosophila Stock Center (http://flystocks.bio.indiana.edu/), and were maintained on a standard oatmeal, yeast, molasses and agar medium. Wing blades were dissected from flies stored in 70% ethanol, mounted in Gary's Magic Mountant (Lawrence et al., 1986) and analysed by brightfield microscopy.
Camptothecin sensitivity studies
Fly medium containing 60 g L−1 each of dextrose and yeast, 3% w/v nipagin and 3% v/v propionic acid was supplemented with CPT in a 5% ethanol/5% Tween-20 solution to achieve final CPT concentrations of 0–0.8 µm in vials containing 10 mL fly food (Cunhe et al., 2002). Heterozygous CG7670e04496/TM6B flies were crossed and eggs were seeded into 4–5 vials per dose at ~200 eggs/vial, according to Clancy & Kennington (2001) and allowed to develop at 18 °C. Surviving heterozygous and homozygous adult flies were scored.
Acknowledgments
We thank Christine Borer for technical support. We are grateful to the BBSRC for funding this research. I.B. is supported by grant no. BB/E000924/1 to L.S.C.; D.J.C. is supported by grant no. BB/E002072/1 to R.D.C.S.
References
- Agrelo R, Cheng WH, Setien F, Ropero S, Espada J, Fraga MF, Herranz M, Paz MF, Sanchez-Cespedes M, Artiga MJ, Guerrero D, Castells A, von Kobbe C, Bohr VA, Esteller M. Epigenetic inactivation of the premature aging Werner syndrome gene in human cancer. Proc. Natl Acad. Sci. USA. 2006;103:8822–8827. doi: 10.1073/pnas.0600645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachrati CZ, Hickson ID. RecQ helicases: suppressors of tumorigenesis and premature aging. Biochem. J. 2003;374:577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BS, Carpenter AT, Ripoll P. The utilization during mitotic cell division of loci controlling meiotic recombination and disjunction in Drosophila melanogaster. Genetics. 1978;90:531–578. doi: 10.1093/genetics/90.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Multani AS, Cabrera NG, Naylor ML, Laud P, Lombard D, Pathak S, Guarente L, DePinho RA. Essential role of limiting telomeres in the pathogenesis of Werner syndrome. Nat. Genet. 2004;36:877–882. doi: 10.1038/ng1389. [DOI] [PubMed] [Google Scholar]
- Chen L, Huang S, Lee L, Davalos A, Schiestl RH, Campisi J, Oshima J. WRN, the protein deficient in Werner syndrome, plays a critical structural role in optimizing DNA repair. Aging Cell. 2003;2:191–199. doi: 10.1046/j.1474-9728.2003.00052.x. [DOI] [PubMed] [Google Scholar]
- Clancy DJ, Kennington WJ. A simple method to achieve consistent larval density in bottle cultures. Drosoph. Inf. Serv. 2001;84:168–169. [Google Scholar]
- Constantinou A, Tarsounas M, Karow JK, Brosh RM, Bohr VA, Hickson ID, West SC. Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 2000;1:80–84. doi: 10.1093/embo-reports/kvd004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox LS, Faragher RGA. From old organisms to new molecules: integrative biology and therapeutic targets in accelerated human ageing. Cell Mol. Life Sci. 2007;64:2620–2641. doi: 10.1007/s00018-007-7123-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox LS, Clancy DJ, Boubriak I, Saunders RD. Modeling Werner syndrome in Drosophila melanogaster: hyper-recombination in flies lacking WRN-like exonuclease. Ann. N Y Acad. Sci. 2007;1119:274–288. doi: 10.1196/annals.1404.009. [DOI] [PubMed] [Google Scholar]
- Crabbe L, Verdun RE, Haggblom CI, Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- Cunha KS, Reguly ML, Graf U, Rodrigues de Andrade HH. Comparison of camptothecin derivatives presently in clinical trials: genotoxic potency and mitotic recombination. Mutagenesis. 2002;17:141–147. doi: 10.1093/mutage/17.2.141. [DOI] [PubMed] [Google Scholar]
- Dhillon KK, Sidorova J, Saintigny Y, Poot M, Gollahon K, Rabinovitch PS, Monnat RJ., Jr Functional role of the Werner syndrome RecQ helicase in human fibroblasts. Aging Cell. 2007;6:53–61. doi: 10.1111/j.1474-9726.2006.00260.x. [DOI] [PubMed] [Google Scholar]
- Franchitto A, Pichierri P. Werner syndrome protein and the MRE11 complex are involved in a common pathway of replication fork recovery. Cell Cycle. 2004;3:1331–1339. doi: 10.4161/cc.3.10.1185. [DOI] [PubMed] [Google Scholar]
- Fukuchi K, Tanaka K, Nakura J, Kumahara Y, Uchida T, Okada Y. Elevated spontaneous mutation rate in SV40–transformed Werner syndrome fibroblast cell lines. Somat Cell Mol. Genet. 1985;11:303–308. doi: 10.1007/BF01534688. [DOI] [PubMed] [Google Scholar]
- Goto M. Clinical characteristics of Werner syndrome and other premature aging syndromes: pattern of aging in progeroid syndromes. In: Goto M, Miller RW, editors. From Premature Gray Hair to Helicase – Werner Syndrome: Implications for Aging and Cancer. Tokyo, Japan: Japan Scientific Societies Press; 2001. pp. 27–39. [Google Scholar]
- Huang S, Li B, Gray MD, Oshima J, Mian IS, Campisi J. The premature ageing syndrome protein, WRN, is a-3′–5′ exonuclease. Nat. Genet. 1998;20:114–116. doi: 10.1038/2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudlow BA, Kennedy BK, Monnat RJ., Jr Werner and Hutchinson–Gilford progeria syndromes: mechanistic basis of human progeroid diseases. Nat. Rev. Mol. Cell Biol. 2007;8:394–404. doi: 10.1038/nrm2161. [DOI] [PubMed] [Google Scholar]
- Kusano K, Johnson-Schlitz DM, Engels WR. Sterility of Drosophila with mutations in the Bloom syndrome gene – complementation by Ku70. Science. 2001;291:2600–2602. doi: 10.1126/science.291.5513.2600. [DOI] [PubMed] [Google Scholar]
- Lawrence PA, Johnston P, Morata G. Methods of marking cells. In: Roberts DB, editor. Drosophila: A Practical Approach. Oxford, UK: IRL Press; 1986. pp. 229–242. [Google Scholar]
- Lebel M. Increased frequency of DNA deletions in pink-eyed unstable mice carrying a mutation in the Werner syndrome gene homologue. Carcinogenesis. 2002;23:213–216. doi: 10.1093/carcin/23.1.213. [DOI] [PubMed] [Google Scholar]
- Lebel M, Lavoie J, Gaudreault I, Bronsard M, Drouin R. Genetic cooperation between the Werner syndrome protein and poly(ADP-ribose) polymerase-1 in preventing chromatid breaks, complex chromosomal rearrangements, and cancer in mice. Am. J. Pathol. 2003;162:1559–1569. doi: 10.1016/S0002-9440(10)64290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machwe A, Xiao L, Groden J, Orren DK. The Werner and Bloom syndrome proteins catalyze regression of a model replication fork. Biochemistry. 2006;45:13939–13946. doi: 10.1021/bi0615487. [DOI] [PubMed] [Google Scholar]
- Martin GM. Genetics and aging: the Werner syndrome as a segmental progeroid syndrome. Adv. Exp. Med. Biol. 1985;190:161–170. [Google Scholar]
- Massip L, Garand C, Turaga RV, Deschenes F, Thorin E, Lebel M. Increased insulin, triglycerides, reactive oxygen species, and cardiac fibrosis in mice with a mutation in the helicase domain of the Werner syndrome gene homologue. Exp. Gerontol. 2006;41:157–168. doi: 10.1016/j.exger.2005.10.011. [DOI] [PubMed] [Google Scholar]
- Moser MJ, Oshima J, Monnat RJ., Jr WRN mutations in Werner syndrome. Hum. Mutat. 1999;13:271–279. doi: 10.1002/(SICI)1098-1004(1999)13:4<271::AID-HUMU2>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Opresko PL, Laine JP, Brosh RM, Jr, Seidman MM, Bohr VA. Coordinate action of the helicase and 3′ to 5′ exonuclease of Werner syndrome protein. J. Biol. Chem. 2001;276:44677–44687. doi: 10.1074/jbc.M107548200. [DOI] [PubMed] [Google Scholar]
- Otsuki M, Seki M, Kawabe Y, Inoue E, Dong YP, Abe T, Kato G, Yoshimura A, Tada S, Enomoto T. WRN counteracts the NHEJ pathway upon camptothecin exposure. Biochem. Biophys. Res. Commun. 2007;355:477–482. doi: 10.1016/j.bbrc.2007.01.175. [DOI] [PubMed] [Google Scholar]
- Peitsch MC. ProMod and Swiss-Model: Internet-based tools for automated comparative protein modelling. Biochem. Soc. Trans. 1996;24:274–279. doi: 10.1042/bst0240274. [DOI] [PubMed] [Google Scholar]
- Perry JJ, Yannone SM, Holden LG, Hitomi C, Asaithamby A, Han S, Cooper PK, Chen DJ, Tainer JA. WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing. Nat. Struct. Mol. Biol. 2006;13:414–422. doi: 10.1038/nsmb1088. [DOI] [PubMed] [Google Scholar]
- Pichierri P, Franchitto A, Mosesso P, Palitti F. Werner's syndrome cell lines are hypersensitive to camptothecin-induced chromosomal damage. Mutat. Res. 2000a;456:45–57. doi: 10.1016/s0027-5107(00)00109-3. [DOI] [PubMed] [Google Scholar]
- Pichierri P, Franchitto A, Mosesso P, Proietti de Santis L, Balajee AS, Palitti F. Werner's syndrome lymphoblastoid cells are hypersensitive to topoisomerase II inhibitors in the G2 phase of the cell cycle. Mutat. Res. 2000b;459:123–133. doi: 10.1016/s0921-8777(99)00065-8. [DOI] [PubMed] [Google Scholar]
- Pichierri P, Franchitto A, Mosesso P, Palitti F. Werner's syndrome protein is required for correct recovery after replication arrest and DNA damage induced in S-phase of cell cycle. Mol. Biol. Cell. 2001;12:2412–2421. doi: 10.1091/mbc.12.8.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirzio LM, Pichierri P, Bignami M, Franchitto A. Werner syndrome helicase activity is essential in maintaining fragile site stability. J. Cell Biol. 2008;180:305–314. doi: 10.1083/jcb.200705126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poot M, Gollahon KA, Rabinovitch PS. Werner syndrome lymphoblastoid cells are sensitive to camptothecin-induced apoptosis in S-phase. Hum. Genet. 1999;104:10–14. doi: 10.1007/s004390050903. [DOI] [PubMed] [Google Scholar]
- Prince PR, Ogburn CE, Moser MJ, Emond MJ, Martin GM, Monnat RJ., Jr Cell fusion corrects the 4-nitroquinoline 1-oxide sensitivity of Werner syndrome fibroblast cell lines. Hum. Genet. 1999;105:132–138. doi: 10.1007/s004399900078. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Lopez AM, Jackson DA, Iborra F, Cox LS. Asymmetry of DNA replication fork progression in Werner's syndrome. Aging Cell. 2002;1:30–39. doi: 10.1046/j.1474-9728.2002.00002.x. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Lopez AM, Jackson DA, Nehlin JO, Iborra F, Warren AV, Cox LS. Characterisation of the interaction between WRN, the helicase/exonuclease defective in progeroid Werner's syndrome, and an essential replication factor, PCNA. Mech. Ageing Dev. 2003;124:167–174. doi: 10.1016/s0047-6374(02)00131-8. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Lopez AM, Whitby MC, Borer CM, Bachler MA, Cox LS. Correction of proliferation and drug sensitivity defects in the progeroid Werner's syndrome by Holliday junction resolution. Rejuvenation Res. 2007;10:27–40. doi: 10.1089/rej.2006.0503. [DOI] [PubMed] [Google Scholar]
- Saintigny Y, Makienko K, Swanson C, Emond MJ, Monnat RJ., Jr Homologous recombination resolution defect in werner syndrome. Mol. Cell Biol. 2002;22:6971–6978. doi: 10.1128/MCB.22.20.6971-6978.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salk D, Au K, Hoehn H, Martin GM. Cytogenetics of Werner's syndrome cultured skin fibroblasts: variegated translocation mosaicism. Cytogenet Cell Genet. 1981;30:92–107. doi: 10.1159/000131596. [DOI] [PubMed] [Google Scholar]
- Scappaticci S, Cerimele D, Fraccaro M. Clonal structural chromosomal rearrangements in primary fibroblast cultures and in lymphocytes of patients with Werner's Syndrome. Hum. Genet. 1982;62:16–24. doi: 10.1007/BF00295599. [DOI] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekelsky JJ, Brodsky MH, Burtis KC. DNA repair in Drosophila: insights from the Drosophila genome sequence. J. Cell Biol. 2000;150:F31–F36. doi: 10.1083/jcb.150.2.f31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao RG, Cao CX, Zhang H, Kohn KW, Wold MS, Pommier Y. Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA:DNA-PK complexes. EMBO J. 1999;18:1397–1406. doi: 10.1093/emboj/18.5.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, Buchholz R, Demsky M, Fawcett R, Francis-Lang HL, Ryner L, Cheung LM, Chong A, Erickson C, Fisher WW, Greer K, Hartouni SR, Howie E, Jakkula L, Joo D, Killpack K, Laufer A, Mazzotta J, Smith RD, Stevens LM, Stuber C, Tan LR, Ventura R, Woo A, Zakrajsek I, Zhao L, Chen F, Swimmer C, Kopczynski C, Duyk G, Winberg ML, Margolis J. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 2004;36:283–287. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- Wu L, Hickson ID. The bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- Yamagata K, Kato J, Shimamoto A, Goto M, Furuichi Y, Ikeda H. Bloom's and Werner's syndrome genes suppress hyperrecombination in yeast sgs1 mutant: implication for genomic instability in human diseases. Proc. Natl Acad. Sci. USA. 1998;95:8733–8738. doi: 10.1073/pnas.95.15.8733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, Martin GM, Mulligan J, Schellenberg GD. Positional cloning of the Werner's syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]