Abstract
Unicellular eukaryotes (protists) are key components of marine food webs, yet knowledge of their diversity, distributions and respective ecologies is limited. We investigated uncultured protists using 18S rRNA gene sequencing, phylogenetic analyses, specific fluorescence in situ hybridization (FISH) probes and other methods. Because few studies have been conducted in warm water systems, we focused on two Atlantic subtropical regions, the Sargasso Sea and the Florida Current. Cold temperate waters were also sampled. Gene sequences comprising a unique eukaryotic lineage, herein termed ‘biliphytes’, were identified in most samples, whether from high- (30°C) or from low- (5°C) temperature waters. Sequences within this uncultured group have previously been retrieved from high latitudes. Phylogenetic analyses suggest biliphytes are a sister group to the cryptophytes and katablepharids, although the relationship is not statistically supported. Bootstrap-supported subclades were delineated but coherence was not obvious with respect to geography or physicochemical parameters. Unlike results from the initial publication on these organisms (therein ‘picobiliphytes’), we could not detect a nucleomorph, either visually, or by targeted primers. Phycobilin-like fluorescence associated with biliphyte-specific FISH-probed cells supports the hypothesis that they are photosynthetic. Our data indicate the biliphytes are nanoplanktonic in size, averaging 4.1 ± 1.0 × 3.5 ± 0.8 μm (±SD) for one probed group, and 3.5 ± 0.9 × 3.0 ± 0.9 μm (±SD) for another. We estimate biliphytes contributed 28 (±6)% of the phytoplanktonic biomass in tropical eddy-influenced surface waters. Given their broad thermal and geographic distribution, understanding the role these protists play in biogeochemical cycling within different habitats is essential.
Introduction
Marine protists are vital components in the global carbon cycle. Knowledge of their diversity and respective ecologies is nascent yet rapidly growing as culture independent approaches are applied in a variety of natural settings. Application of polymerase chain reaction (PCR)-based approaches in marine systems has revealed a tremendous degree of eukaryotic diversity, using markers such as the plastid-encoded 16S ribosomal RNA (rRNA) gene (Rappéet al., 1998) and the RuBisCo large subunit (rbcL) gene (Paul et al., 2000) as well as the nuclear encoded 18S rRNA gene (Diez et al., 2001). Phylogenetic analyses of environmental sequence data have demonstrated the existence of many novel clades within the eukaryotic tree of life, with many sequences from ‘pico’-size (generally defined as < 2–3 μm) fractionated samples (Lopez-Garcia et al., 2001; Moon-van der Staay et al., 2001; Massana et al., 2004a; Groisillier et al., 2006; Not et al., 2007a). The majority of these studies have concentrated in ‘local’ coastal zones (Massana et al., 2004b; Romari and Vaulot, 2004; Countway et al., 2005; Worden, 2006), or extreme environments, such as deep or polar waters (Lopez-Garcia et al., 2001; Lovejoy et al., 2007). Consequently, there is a dearth of knowledge with respect to the diversity and abundance of subtropical and tropical microbial eukaryotes. Furthermore, little is known about the morphological or functional attributes of the organisms from which such sequences are derived. This is largely due to the fact that there are no cultured representatives for many of the newly identified clades.
Linked investigations of the diversity, abundance and function of uncultured protists are critical to understanding evolutionary and ecological aspects of these populations, as well as for efforts to improve biogeochemical models. For prokaryotes, tremendous advances have been made via ecological genomics, whereby, in some cases it has been possible to begin to assign functional roles to uncultured bacteria (Moran and Miller, 2007). However, to date, we lack insights on uncultured eukaryotes akin to those gained from prokaryotic metagenomic studies (DeLong et al., 2006; Worden et al., 2006; Rusch et al., 2007) and the genomic potential of protistan communities remains virtually unknown. While metagenomic studies provide a first glimpse at possible metabolic capabilities, linking them with rigorous environmental characterization and activity measurements allows basic understanding of ecosystem level processes to advance (Azam and Worden, 2004; Moran and Miller, 2007). For uncultured protists, such approaches promise a means to elucidate their physiologies and ecologies. At this stage identification of novel eukaryotic groups and data on their distributions and abundance are needed to facilitate selection of appropriate sampling sites and populations for targeted metagenomics.
Two studies have recently highlighted the presence of a protistan group in high-latitude marine surface waters with unknown affinities to other eukaryotes and termed ‘picobiliphytes’ (Not et al., 2007a,b). 18S rRNA gene sequences were recovered from the Arctic Ocean, the Norwegian Sea, and irregularly in fall and winter coastal European waters, suggesting that these organisms are widespread in cold and polar waters (Hearn, 2007; Not et al., 2007b). However, enumeration of these putatively photosynthetic cells showed they were extremely rare, composing less than 1% of the eukaryotic community (Not et al., 2007b).
Few studies have explored molecular phylogenetic diversity in subtropical and tropical environments or linked data on novel eukaryotic sequences with organism characteristics or abundance. We undertook a comprehensive set of research expeditions to explore subtropical marine protist communities. Here, we investigate the phylogeny of a unique set of sequences discovered in samples from subtropical regions as well as from cold waters. We show that this enigmatic lineage, here termed ‘biliphytes’, is composed of nanoplanktonic (2–20 μm) organisms falling within genetically distinct clades, which also harbour the previously published ‘picobiliphyte’ sequences (Not et al., 2007b). Based on results to date, we infer a functional role for the biliphytes and assess their ecological significance in the subtropics.
Results
Study sites
To attain broadly representative results we investigated molecular diversity in an array of marine samples, and population characteristics in a subset of those. Three regions were explored during cruises undertaken in 2001 and 2005, two regions being subtropical and one a cold water environment (Fig. 1, Table 1). The first region, the Sargasso Sea, is an open-ocean subtropical Gyre system. Two sites were sampled within this area, the Bermuda Atlantic Times-series Study (BATS) station and a location in the northern Sargasso Sea. At the time of sampling the water column at BATS was under summer-like conditions, with the mixed layer extending to 30 m. At the Northern Sargasso station, the mixed layer extended slightly deeper, to 40 m. Water samples were collected from the deep chlorophyll maximum (DCM), located well below the seasonal thermocline, and from close to the surface (Table 1). The second region, the Florida Straits, is located between south Florida, USA and the Bahamas. The Florida Current, which separates the Florida Keys, USA from Cuba and connects the Gulf of Mexico with the Atlantic Ocean, flows at high velocity (Fig. 1B and C) through the Straits, and composes a large fraction of the Gulf Stream forming waters. At the latitude sampled, this current is in many aspects representative of tropical waters: it is close to the tropics, which spans the equator from 23°27′N to 23°27′S, in latitude (our transect being at 25°30′) and maintains high water temperatures year round, with extended periods around 30°C, unlike subtropical systems such as BATS which undergo greater temperature variations. The physical properties of the Straits, atmospheric conditions and current velocity, contribute to retention of tropical water characteristics in particular zones of the Straits (see Fig. 1 and satellite data at http://imars.usf.edu). This location was sampled intensively, on a series of three cruises representing different seasonal and event-driven variations within the system including the remnants of a cyclonic mesoscale eddy on the western side during spring (Fig. 1B, D and F), the highly stratified summer waters (Fig. 1C, E and G) and the mixing winter waters (Fig. S1). Physical and fluorescence parameters were measured at 16 stations distributed from the western side (south Florida) to the eastern side (Bahamas) in order to comprehensively map characteristics of the water mass. Water samples were taken at the three stations which best represent different water zones in the Straits: the western side (Station 01), the more central Gulf Stream forming waters (Station 04) and the eastern side which has a deeper bottom depth (Station 14, Table 1). Samples were collected at the surface and DCM for the three stations on a March/April cruise, and a July/August cruise. Only one sample, from the DCM at Station 14, was utilized from the December cruise due to limited sampling resulting from rough seas. Overall, five of the Florida Straits samples were from high-temperature waters, ranging to 30°C (28.4 ± 2.2°C, average and SD, Table 1). The third region investigated was continental slope waters south-east of Cape Cod, USA. Two sites were sampled at the surface and DCM: over the continental slope (bottom depth of 2750 m) and over the continental slope close to the continental shelf boundary (bottom depth of 706 m). Temperatures for these water samples ranged from 5.3°C to 14.2°C.
Fig. 1.
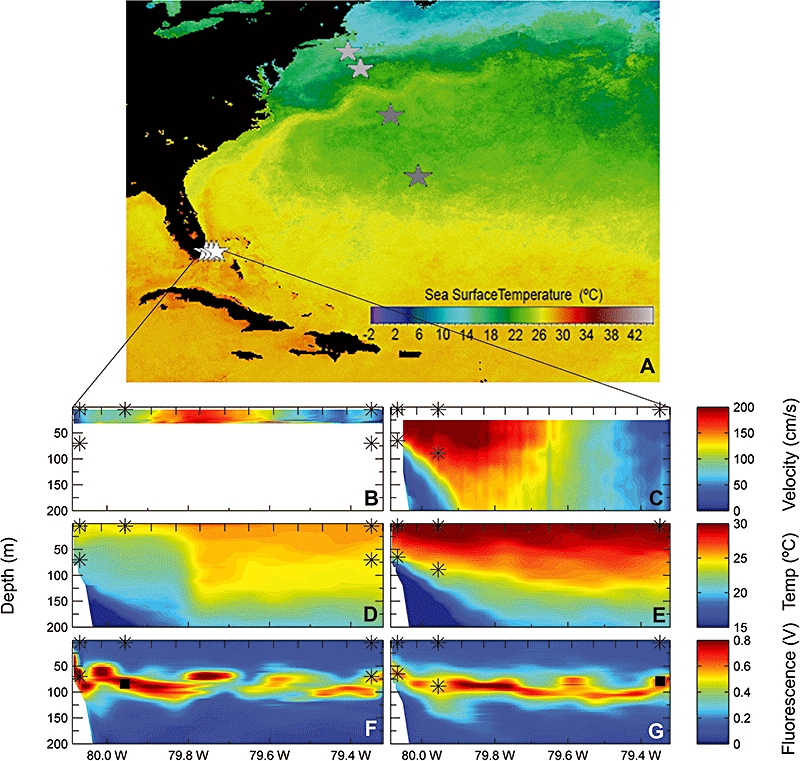
Geographic location, physical and biological parameters of the sample regions and sites. Three regions were investigated during the cruises. In the Sargasso Sea (dark grey stars), two sites were sampled: BATS (bottom star) and the Northern Sargasso (top star). In the continental shelf area (light grey stars), two sites were sampled: the continental edge (bottom star) and the continental slope (top star). In the Florida Straits (white stars), three sites were sampled for each of the three cruises (March 2005, July/August 2005, December 2005): Station 01 (left star), Station 04 (middle star) and Station 14 (right star) (A). Bottom panels show the cross-straits vertical profiles of the north component of current during March (B) and July (C); temperature during March (D) and July (E); and fluorescence during March (E) and July (F). Sampling stations and depths containing biliphytes are indicated with an asterisk. Florida Straits sampling depths where biliphyte sequences were not recovered are shown by black squares in (F) and (G). Tick marks on the upper x-axis indicate stations at which CTD casts were performed to measure environmental parameters. During the March cruise the ship was only equipped with a 600 kHz ADCP capable of measuring shallow currents, hence (B) only shows velocity in the upper 30 m of the water column. SeaWIFS data (A) are derived from http://oceancolor.gsfc.nasa.gov/cgi/browse.pl from the integrated June 2005 sea surface temperature data.
Table 1.
Coordinates, dates, environmental characteristics and associated 18S rDNA sequences for sample sites.
| Site | Latitude (N) | Longitude (W) | Date (d/m/y) | Sample depth (m) | Bottom depth (m) | Temperature (°C) | Salinity (ppt) | Sequence names |
|---|---|---|---|---|---|---|---|---|
| BATS | 31°39′20′′ | 64°37′21′′ | 05/29/05 | 75 | 4397 | 20.08a | 36.79a | OC413BATS_O043_75m |
| BATS | 31°39′20′′ | 64°37′21′′ | 06/01/05 | 15 | 4397 | 25.51a | 36.68a | OC413BATS_P006_15m OC413BATS_P082_15m |
| NSS | 35°09′24′′ | 66°33′46′′ | 06/05/05 | 15 | 4980 | 21.37–21.56a | 36.52–36.56a | OC413NSS_Q007_15m OC413NSS_Q040_15m OC413NSS_Q086_15m |
| CSS | 40°15′07′′ | 70°25′23′′ | 04/09/01 | 20 | 706 | 5.31 | 32.71 | EN351CTD040_20m |
| CSS | 40°15′07′′ | 70°25′23′′ | 04/09/01 | 4 | 706 | 5.60 | 32.70 | EN351CTD040_4m |
| CS | 39°10′55′′ | 69°31′08′′ | 04/09/01 | 30 | 2750 | 14.24 | 35.78 | EN351CTD039_30m |
| FS01 | 25°30′07′′ | 80°04′04′′ | 03/30/05 | 5 | 125 | 24.26 | 36.43 | FS01B026_30Mar05_5m FS01B029_30Mar05_5m FS01B033_30Mar05_5m FS01B048_30Mar05_5m |
| FS01 | 25°30′07′′ | 80°04′04′′ | 03/30/05 | 70 | 125 | 21.15 | 36.41 | FS01C040_30Mar05_70m |
| FS01 | 25°30′04′′ | 80°03′59′′ | 08/01/05 | 5 | 100 | 30.08 | 36.10 | FS01AA11_01Aug05_5m FS01AA94_01Aug05_5m |
| FS01 | 25°30′04′′ | 80°03′59′′ | 08/01/05 | 65 | 100 | 23.51 | 36.32 | FS01D014_01Aug05_65m FS01D022_01Aug05_65m FS01D031_01Aug05_65m FS01D054_01Aug05_65m FS01D057_01Aug05_65m FS01D065_01Aug05_65m |
| FS04 | 25°30′01′′ | 79°57′20′′ | 03/31/05 | 5 | 375 | 24.68 | 36.31 | FS04E037_31Mar05_5m FS04E081_31Mar05_5m |
| FS04 | 25°30′04′′ | 79°57′18′′ | 08/01/05 | 5 | 350 | 30.29 | 36.01 | FS04GA46_01Aug05_5m FS04GA95_01Aug05_5m FS04G188_01Aug05_5m FS04G190_01Aug05_5m |
| FS04 | 25°30′04′′ | 79°57′18′′ | 08/01/05 | 89 | 350 | 24.22 | 36.54 | FS04H169_01Aug05_89m FS04H153_01Aug05_89m |
| FS14 | 25°29′59′′ | 79°20′58′′ | 03/30/05 | 5 | 729 | 25.65 | 36.20 | FS14JA65_30Mar05_5m FS14JA72_30Mar05_5m |
| FS14 | 25°29′59′′ | 79°20′58′′ | 03/30/05 | 70 | 729 | 24.65 | 36.61 | FS14I06_30Mar05_70m |
| FS14 | 25°29′55′′ | 79°20′54′′ | 07/31/05 | 5 | 650 | 29.88 | 36.04 | FS14K017_31Jul05_5m FS14K025_31Jul05_5m |
| FS14 | 25°30′01′′ | 79°21′04′′ | 12/08/05 | 58 | N/A | 26.18 | 36.26 | FS14M008_08Dec05_58m FS14M021_08Dec05_58m |
Parameters measured with CTD detector prior to the sample collection (if a single measurement) or prior to and the following day (if a range of measurements), as samples were collected with a single GO-FLO bottle not equipped with a CTD detector.
BATS, Bermuda Atlantic Time-series Study Station; NSS, Northern Sargasso Sea; CSS, Continental Shelf-edge/Slope; CS, Continental Slope; FS, Florida Straits, Station 01, South Florida, USA side; Station 04, core Gulf Stream forming waters; Station 14, Bahamas side; N/A, data not available.
Gene sequences and phylogenetic analyses
The unique 18S rRNA gene sequences were retrieved from 17 out of 20 clone libraries (Fig. 1, Table 1). Phylogenetic analyses (Fig. 2) of the newly obtained sequences were performed in the context of publicly available data covering the known breadth of eukaryotic diversity. The results delineate a eukaryotic group at the first rank taxon level (Adl et al., 2005). Using an alignment of 126 taxa, a strongly supported clade was identified, containing 18S rRNA gene sequences derived from the subtropical regions, including remnants of a tropical eddy, and the continental slope sites. The ‘picobiliphyte’ sequences previously obtained from cold surface waters of the Arctic Ocean, the Norwegian Sea, as well as fall and winter coastal European waters (Not et al., 2007b) were scattered among these sequences (Fig. 2). Within this clade, no strong correlation between phylogenetic relatedness and geographic location was apparent, although several highly supported clusters of sequences are apparent (e.g. BP2; Fig. 2). As observed previously (Not et al., 2007b), our phylogenies resolve the biliphytes as a potential sister group to the cryptophytes and katablepharids. This relationship is, however, not statistically supported, similar to results obtained for most of the backbone of the 18S rRNA gene tree.
Fig. 2.
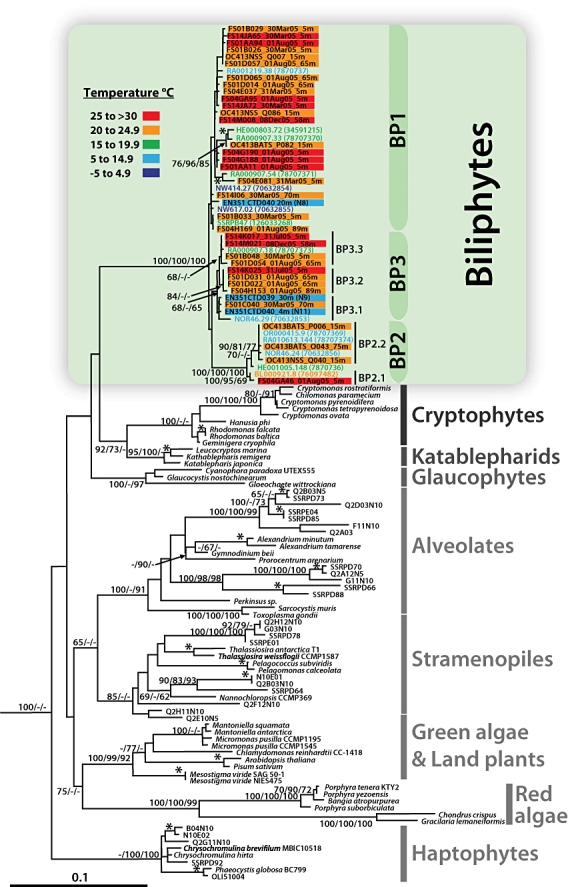
Phylogenetic analysis of 18S rRNA gene sequences from environmental clone libraries (all non-italic sequence names) and cultured representatives of eukaryotic first rank taxa. Sequences in coloured bars are reported for the first time in this study and are derived from 17 environmental clone libraries, three from the Sargasso Sea (prefixes: OC413BATS_P, Q; OC413NSS), 11 from three stations in the Florida Straits (prefixes: FS01, FS04, FS14) over multiple dates and depths (see Table 1), as well as three from continental shelf-edge and slope waters (prefixes: EN351). Biliphyte sequences from previous work are shown in coloured font. Bar and font colours correspond to five temperature ranges as indicated on the figure. Other Sargasso Sea and Pacific Ocean environmental sequences (non-italic, black font sequence names without coloured bars) are included to illustrate the distinct nature of biliphyte sequences relative to other uncultured eukaryotes retrieved from the same waters. The tree shown was inferred by maximum likelihood (ML) methods using the model TrN + I +Γ (I = 0.2332; Γ = 0.4748) with global rearrangements, randomized sequence input, 10 jumbles, Transition/Transversion = 1.9433 and six rate categories. Node values reflect bootstrap support in the order maximum likelihood/neighbour-joining distance/parsimony as percentages of 100/1000/100 replicates. Bootstrap support for terminal nodes (*) is indicated only if over 90% by all three methods. Two radiolarian sequences (126033309 and 126033226) served as an outgroup (not shown). The scale bar indicates the estimated number of nucleotide substitutions per site.
In sum, we isolated biliphyte 18S rRNA gene sequences from all five subtropical sites, BATS, the northern Sargasso Sea and the three Florida Straits stations. Populations were detected during all time periods investigated, including waters up to 30°C and in slope waters as cold as 5.3°C (Figs 1 and 2). Biliphyte sequences were retrieved from the surface and the DCM, close to the base of the euphotic zone. It should be noted that three other sequenced clone libraries, generated from samples taken on the same research expeditions, did not contain biliphytes among sequenced clones (data not shown). These samples were from the Florida Straits DCM at Station 04 in March and Station 14 in July as well as the DCM in the northern Sargasso Sea Station. No biliphyte-like 18S rRNA gene sequences were identified within the Global Ocean Sampling (GOS) data or CAMERA database.
Characteristics and abundance
Because cellular abundance cannot be inferred based on clone library numbers and function cannot be assigned purely based on 18S rRNA gene phylogeny, water samples were interrogated using two biliphyte-specific, rRNA-targeted fluorescence in situ hybridization (FISH) probes. Probes targeted a subset of sequences that fell within clades BP1 and BP2 (Fig. 2). These revealed a phycobilin-like autofluorescence (orange) within the fluorescein-labelled, probed cells, indicating that these organisms may be photosynthetic in nature, similar to the findings of Not and colleagues (2007b). Chlorophyll-like autofluorescence was not observed, nor expected, given that it is an alcohol soluble pigment and filters were treated with an alcohol dehydration series prior to FISH.
Cells identified using the specific FISH probes fell within the nanoplankton size class (2–20 μm). Those hybridized by the BP1 probe were slightly larger than those with the BP2 probe. The former averaged 4.1 ± 1.0 × 3.5 ± 0.8 μm (±SD, n = 60), while the latter averaged 3.5 ± 0.9 × 3.0 ± 0.9 μm (±SD, n = 45). Note that our FISH samples were not size fractionated, but rather represented ‘whole’ seawater. We also enumerated cells at the surface and DCM for three Florida Straits stations in March, for which the average water sample temperature was 25.0 ± 0.6°C. Biliphyte concentrations were 228 ± 42 and 312 ± 57 cells ml−1 at the surface for Stations 01 and 04 respectively. Cells were also detected at Station 01 and 04 in the DCM, but counts were too low to provide statistically reliable cell concentrations. The same was the case for surface waters at Station 14; however, biliphytes were relatively abundant at DCM (211 ± 39 cells ml−1). Note that FISH-based cell concentrations reflect a conservative estimate as these probes have mismatches to six of the 18S rDNA sequences reported herein (one from March), including all sequences within BP3.1, and mismatches to four sequences from the study by Not and colleagues.
Pigment signatures are a common means of analysing marine flow cytometry (FCM) samples. We returned to archived FCM data from the same water as collected for FISH and DNA analyses, where greater than 100 biliphytes ml−1 were detected (by FISH), to see if a biliphyte population could be identified. The FCM run volume for these samples was 250 μl, which in theory would yield a cluster of at least 25 cells. While particles were visible that had large (but dissimilar) light scatter, relative to 0.75 μm beads, and containing orange and red fluorescence signatures, a coherent population was not.
Biomass estimate
Based on the hypothesis that biliphytes are photosynthetic, we performed a simple calculation to estimate their contribution to phytoplankton biomass. First, cell volume was estimated using cell size measurements (n = 45) from samples for which we had corresponding HPLC Chl a data (reflecting total phytoplankton biomass; J.A. Hilton, F. Not, A.Z. Worden and others, unpublished). Given that epifluorescence microscopy only allows measurement of two dimensions, a conservative estimate was used for the third dimension by taking the smaller of the two measured dimensions. Therefore, cell biovolume, V, was estimated from the longest cell dimension, L, and the shortest cell dimension, S, both in units of μm using the equation
A conservative carbon conversion factor (2.37 × 10−7 μg C μm−3; Worden et al., 2004) was then combined with the cell abundance and biovolume information. The measured concentration of total Chl a was 0.048 and 0.061 μg Chl a l−1 in surface March 2005, Stations 01 and 04 respectively. These values were similar to previously reported Chl a concentrations in the Sargasso Sea (0.051 μg Chl a l−1) during a period of stratified oligotrophic waters and over a comparable temperature range (24–27°C), and much lower than concentrations (0.26–0.42 μg Chl a l−1) in higher nutrient waters (Goericke and Welschmeyer, 1998). Therefore, biomass of the total phytoplankton community was estimated by converting total Chl a concentration using a ratio of 90 C : Chl a, as appropriate for oligotrophic waters (Eppley, 1968). Based on this series of calculations biliphytes composed an estimated 28 (± 6)% of the total phytoplanktonic carbon in the eddy-influenced waters.
Search for a nucleomorph
Should the evolutionary processes leading to extant biliphytes be similar to endosymbiotic events in other protists (see Discussion), a nucleomorph could be present. Because of the previous report of a potential nucleomorph-like structure (Not et al., 2007b), we searched for evidence of a nucleomorph in FISH-labelled, DNA (DAPI) counter-stained biliphyte cells. However, DAPI fluorescence was not detected specifically associated with the region of phycobilin-like orange fluorescence (indicating the plastid) that might suggest the presence of a nucleomorph in the FISH-illuminated cells. We also assessed approximately 40 Rhodomonas salina cells but could not visualize this structure based on DAPI counter staining.
Because evidence for a nucleomorph was not found in our microscopy work, we tested for the presence of a nucleomorph-like body in biliphytes using a molecular approach. Primers designed to motifs unique to red algae and/or cryptophyte nucleomorph 18S sequences (for rational see Discussion), but biased against the amplification of biliphyte nuclear 18S rRNA genes, were used for PCR and clone library construction. No sequence showing similarity to nucleomorph sequences (or their red algal homologues) was obtained. Of 62 clones for which reliable sequence data were obtained, 56 showed significant similarity to alveolate and heterokont sequences, with the remaining sequences showing the highest similarity to green algae (2), fungi (2), katablepharids (1) and choanoflagellates (1) (data not shown).
Discussion
Evolutionary relationships
Our phylogenetic analyses revealed a unique eukaryotic lineage (herein named biliphytes) composed of multiple clades with significant statistical support by maximum-likelihood and other phylogenetic methods (Fig. 2). Clade structure and significance were enhanced by the significant expansion of the number of biliphyte sequences as well as the removal of a likely chimeric sequence (clone HE000427.214, accession DQ222872) used in the previously reported phylogeny (Not et al., 2007b). In that initial analysis, a clade composed of three sequences and considered to be one of three overall clades fell in a basal position. This basal clade is probably an artefact, potentially caused by the inclusion of the chimeric sequence, which falls within it, and potentially also destabilizes other clades. At any rate, our analysis did not resolve the basal clade as seen previously (Not et al., 2007b). Most inner nodes (backbone) did not retain support, as is typically the case for 18S rRNA gene trees, nor did the node which would more firmly establish the sister group to the biliphytes (where bifurcation of the biliphytes from the cryptophytes/katablepharids occurs). This latter node was not bootstrap supported in the analysis of Not and colleagues either, although a high Bayesian posterior probability value was provided (Not et al., 2007b). Bayesian posterior probability values are now known to be inflated and should thus be interpreted with caution (Simmons et al., 2004).
18S rRNA phylogenies place the cryptophytes and katablepharids as the closest eukaryotic lineages to the biliphytes, albeit without statistical support (Fig. 2; Not et al., 2007b). The cryptophytes (e.g. R. salina) acquired photosynthesis secondarily through the uptake and retention of a red algal endosymbiont and are one of only two lineages known to still retain the nucleus of the engulfed eukaryote, i.e. the nucleomorph (Douglas et al., 2001; Palmer, 2003; Archibald, 2007). In the cryptophytes, this DNA-containing structure is associated with the plastid, which also contains phycobiliproteins. Furthermore, 18S rDNA genes are a universal feature of cryptophyte nucleomorph genomes (Lane et al., 2006). In view of what is known about cryptophytes, the placement of the biliphytes as a potential sister clade is intriguing given that the FISH- and DAPI-counterstained biliphyte images of Not and colleagues (2007b) indicate the presence of a DAPI-fluorescent body in close association with the plastid. These results suggest the presence of a nucleomorph in the biliphytes. We did not observe such a body in association with biliphyte plastids. However, we did not use an identical counterstaining technique and the fairly high background observed under the DAPI channel could interfere with unambiguous detection of this small structure, which is similar in size to the many marine bacteria that are also stained by DAPI. Not only were we unable to visualize localized DAPI staining in the vicinity of pigment fluorescence in biliphyte cells, we could not in cultures of the cryptophyte R. salina (an organism known to have a nucleomorph) using our whole-cell staining methods. Nor could we in a R. salina image provided in supplementary materials of the former publication (Not et al., 2007b). In the case of cryptophytes, an earlier study using embedded and sectioned cells showed that DAPI-stained nucleomorph DNA can indeed be observed, but also revealed a punctate distribution of DAPI-stained plastid DNA (Ludwig and Gibbs, 1987). Thus, although the findings of Not and colleagues (2007b) are reminiscent of the situation in cryptophytes, their results do not exclude the possibility that the plastid-localized fluorescence attributed to nucleomorph DNA is in fact plastid DNA. Currently then, the presence of a nucleomorph in biliphytes remains unclear.
If a nucleomorph does exist in the biliphytes, there are (at least) two evolutionary explanations for its presence. First, the organelle could be homologous to the nucleomorph in cryptophytes, which is reasonable given a possible biliphyte-cryptophyte/katablepharid connection in molecular phylogenies. Under this scenario, the secondary endosymbiosis that gave rise to the plastid and nucleomorph in the two groups would have occurred in their common ancestor. These organelles would then have been completely lost secondarily in katablepharids and the cryptophyte Goniomonas (which unlike other cryptophytes does not harbour a nucleomorph). Nucleomorph loss has been proposed for haptophytes, a secondary plastid-containing lineage that large-scale nuclear gene phylogenies suggest is the sister group to cryptophytes (Burki et al., 2007; Patron et al., 2007). Another scenario would be that biliphytes acquired photosynthesis independently through the tertiary engulfment of a cryptophyte or a red alga, which would explain the presence of pigments with phycobiliprotein-like fluorescence. Our attempt to amplify a nucleomorph 18S ribosomal gene did not reveal any red algal or cryptophyte nucleomorph-like sequences. This suggests that the nucleomorph 18S ribosomal sequence of biliphytes may be quite divergent and, hence, their plastids probably did not arise through recent uptake of algal cells. However, it is also possible that sequencing a greater number of clones might have yielded such a sequence. Alternatively, the biliphytes may not have retained a nucleomorph. Understanding the origin of biliphyte plastids will likely require ultrastructural, molecular phylogenetic and genomic investigations.
Ecological range and significance in warm waters
Our data offer new insights on the ecological range and genetic diversity of biliphytes. Sequences falling within this lineage were first detected in high-latitude marine waters, as cold as −1°C (Not et al., 2007b). We show that their range extends through temperate and subtropical oceans, including tropically influenced features, and throughout the euphotic zone (Fig. 2). While the Sargasso Sea sites provide excellent representation of subtropical systems, the proximity of the Florida Straits sampling transect to the tropics makes this region more representative of tropical waters than common subtropical settings, such as BATS. Typical of tropical waters, such as those that form the Florida Current, high temperatures are maintained year round in the Straits (with extended periods around 30°C). Furthermore, tropical features can persist (see below) within the Straits. We detected sequences in waters ranging from 5°C to 30°C, with nearly one-third of the sequences recovered being from waters above 25°C and sampled over multiple dates (Fig. 2).
Fluorescence in situ hybridization results from the environmental samples support the hypothesis that these organisms are photosynthetic (Not et al., 2007b), given their phycobilin-like fluorescence. The abundance of biliphytes in warm water samples was greater than reported elsewhere to date. The only other published biliphyte cell concentrations show that the average (55 ± 23 biliphytes ml−1) for five dates at a cold water site in the English Channel was only a minor portion of the eukaryotic community (on average 1% of total count reported), not including data from five dates when none were detected (Not et al., 2007b). The average temperature at this site for five dates spread over the course of the year was 12.5 ± 2.0°C (D. Vaulot, pers. comm.)
In general, no clear relationship between water temperature and clade structure was visible, suggesting that the organisms within BP1, BP2 and BP3 are able to survive over an extremely broad thermal range (Fig. 2). Only two supported subclades of 18S rRNA gene sequences (BP2.1, BP3.2; Fig. 2) were composed of sequences from a more narrow temperature range (waters above 20°C), but this observation should be viewed with caution until more data are available. In support of our findings for warm waters, sequences falling within the biliphytes, although not identified as such when published, have also been obtained from an environmental clone library in 23°C summer-time surface coastal waters (Countway et al., 2005). The ‘picobiliphytes’ were initially reported only in cold surface waters of the Arctic Ocean, the Norwegian Sea, and in some fall and winter coastal European waters (Romari and Vaulot, 2004; Not et al., 2007b) as well as a single sequence from the permanent thermocline (500 m) near BATS (Not et al., 2007a). The fact that a sequence (SSRPB47, accession EF172850) was retrieved from 500 m could have several inferences, e.g. (i) that primary production by these organisms can be exported to the deep sea, serving to sequester atmospheric CO2, or (ii) that these organisms are mixotrophic or even heterotrophic (although heterotrophy seems unlikely).
Biliphyte sequences were not found within three clone libraries, all from the DCM at stations for which sequences were retrieved from surface libraries. The March Florida Straits Station 04 DCM library was unusual relative to most other Florida Straits libraries. It had a large number of prasinophyte sequences [Bathycoccus, Micromonas and Ostreococcus composed one-third of surface Station 04 sequences, with the majority of others belonging to the novel alveolates, as commonly found (Groisillier et al., 2006; Worden, 2006; Not et al., 2007a), data not shown] and no biliphytes sequences were recovered. Nevertheless, biliphytes were detected by FISH although at extremely low abundance (see Results). Another library from which we did not attain biliphyte sequences was the northern Sargasso Sea DCM library. Here again, and unlike other Sargasso Sea samples, a large number of prasinophyte sequences were retrieved (data not shown).
It is noteworthy that during the March 2005 cruise, conditions in the western half of the sampling transect were strongly influenced by remnants of a mesoscale cyclonic eddy. Continuous surface measurements of temperature, salinity and fluorescence recorded underway indicate a distinct front was present on 31 March 2005 at around 79.85°W. Colder, more saline (data not shown) euphotic zone waters with higher fluorescence occurred on the Florida side of this front (Fig. 1D). Satellite imagery from around this time (http://imars.usf.edu) indicates these characteristics were associated with upwelled waters of a mesoscale eddy translating downstream. Previous work has shown that these cyclonic features, termed Tortugas eddies, form as frontal eddies along the tropical Loop Current, translate into the southern portion of the Florida Straits, and remain resident there for a period of time before moving downstream where they elongate and shear apart off of the upper Florida Keys (Lee et al., 1994; Fratantoni et al., 1998). The shoaling of the thermocline in cyclonic eddies can increase nutrient supply to the euphotic zone and ultimately primary production (McGillicuddy et al., 2007). Enumeration of biliphytes by FISH for the March cruise reveals the highest biliphyte abundances at the surface of Station 01 and 04, both located on the Florida side of the front, where tropical eddy waters had an influential role on the water mass (Fig. 1).
Biliphytes were abundant in the surface waters of Station 01 and 04 (the eddy influences waters) and DCM of Station 14 in March. During the same expedition biliphytes were low in abundance in other samples (see Results). Although phylogenetic analysis showed no clear relationship between clade structure and water temperature, the fact that waters with higher abundances were isothermal (24°C; Fig. 1D, Table 1) suggests a potential optima related to temperature or associated environmental parameters. Further analysis and enumeration of biliphytes and other taxa in a range of conditions is clearly needed. The higher abundances were found in both low-light (DCM) and high-light (surface) isothermal waters. These waters (west of the front, influenced by the eddy; and DCM waters east of the front, influenced by higher nutrient concentrations found at the base of the euphotic zone) likely had relatively high nutrient concentrations. With respect to niche differentiation, it is then possible that conditions during which prasinophytes proliferated [particularly the significantly lower temperature DCM waters (Fig. 1D) at Florida Straits Stations 01 and 04 in March, data not shown] are less advantageous to biliphytes. The eddy waters clearly contributed to a shoaling of the thermocline at these stations, likely yielding higher nutrients in the warm surface waters where the biliphytes appeared to thrive.
Cell size, enumeration and biomass
The nanoplanktonic size of these organisms seems at odds with the initial designation ‘picobiliphytes’ (Not et al., 2007b). Whether the denomination ‘pico’ refers to cells < 2 μm in diameter (Sieburth et al., 1978; Stockner, 1988; Li et al., 2006), or, as defined by a subset of studies, to cells < 3 μm diameter (Romari and Vaulot, 2004; Not et al., 2007b), it is evident that a large portion of the probed cells are greater in diameter than indicated by either of these definitions. Images in the supplementary materials of the former publication (Not et al., 2007b) show several relatively spherical cells (akin to our findings) which, based on the scale bar provided, are approximately 5 × 5 μm, different from the shape and size estimate provided within the text (2 × 6 μm cells, n = 9). Together with our cell size data, this makes the denomination ‘pico-’ a confusing prefix. Competition processes, including those related to optimization of surface area to volume ratios, photosynthetic properties and grazing are all critical factors for population dynamics and are linked to organism size. The fact that sequences from larger cells are retrieved from our size-fractionated (< 2 μm) DNA samples is not necessarily surprising, as fragile cells and even appendages are known to break during the pre-filtration process, resulting in presence of some sequence data from larger organisms (see Worden, 2006; Not et al., 2007a). At the same time, no biliphtye sequences were identified in the GOS data (Rusch et al., 2007), which used a < 0.8 μm size fractionation step. Sequences from known picoplanktonic organisms, larger than this cut-off, in particular the prasinophyte Micromonas pusilla, which averages from 1.4 to 1.6 μm in diameter (R.M. Welsh, A.Z. Worden and others, unpublished), are present at several GOS sites, showing that at least small phytoplankton did pass through the pre-filter. The absence of biliphytes could be based on either the larger cell size of these organisms leading to complete exclusion, or its relative rareness, making the sequencing coverage inadequate for recovering an 18S gene fragment among genetic material from many more abundant organisms. Given the importance of ecological inferences associated with marine plankton size classes, and given that there are many plankton species falling exclusively within the < 2 μm size fraction, precise size classification is a fundamental ecological distinction. Hence, while lack of cultured isolates hinders further characterization, it seems prudent that this novel group be named ‘biliphytes’, in keeping with Not and colleagues (2007b), but removing the prefix referring to size classification.
Overall, re-evaluation of previous work on phytoplankton dynamics and biomass contributions will be needed in view of the widespread distribution of this new group. Cryptophytes havebeen reported in FCM and HPLC studies, but few cryptophyte-like sequences appear in open-ocean clone libraries (Countway et al., 2007; Not et al., 2007a). In the case of FCM, the similar orange fluorescence between cryptophytes and biliphytes may have caused biliphytes to mistakenly be identified as cryptophytes in earlier studies, if natural populations of these plankton are enumerated under current methodologies. This will depend on factors such as suitability of FCM fixation methods for preservation. Flow cytometry fixation and cryo-freezing procedures can be disruptive, particularly for eukaryotic nanoplankton. We quantified cell loss for mid-exponential growth cultures and found that while cyanobacterial numbers quantified in either glutaraldehyde (0.25%, final concentration) or paraformaldehyde (1%, final concentration) cryo-frozen samples were 100% of live counts, eukaryotes generally suffer some losses (R.M. Welsh, A.Z. Worden and others, unpublished). The prasinophytes Ostreococcus tauri and M. pusilla as well as the prymnesiophyte Isochrysis sp. CCMP1244 preserved relatively well, with losses ranging from 0% to 22%, depending on the cell type and fixative used (fixative always used in conjunction with flash freezing in liquid nitrogen). However, the cryptophyte R. salina, a cell wall-less nanoplankter, suffered 40 ± 7% loss in glutaraldehyde and 93 ± 3% loss in paraformaldehyde, both with cryo-freezing. Similar observations have previously been reported for R. salina, using a range of fixatives although without cryo-freezing (Klein Breteler, 1985). Because specific fixation effects cannot be unambiguously quantified without cultures, we can only infer, based on our FCM data and any potential similarities to species such as R. salina, that at least some fraction of biliphytes are likely lost via many of the common oceanographic FCM preservation methods. This could result in both a lack of an identifiable biliphyte population and an underestimation of the total eukaryotic phytoplankton community.
With respect to HPLC, these analyses do not utilize phycobiliproteins to quantify specific groups and therefore the presence of phycobilins in both cryptophytes and biliphytes would not confound interpretation of previous HPLC studies. However, currently it is unclear whether the signature pigments (Latasa et al., 2001; Latasa, 2007) used for cryptophytes (alloxanthin) might also be present in biliphytes; this would depend on the nature of the secondary endosymbiosis event and downstream processes.
Given their large size relative to the numerically dominant phytoplankton in oligotrophic environments (e.g. picoplankton such as Prochlorococcus), low numbers of these putative primary producers can contribute disproportionately to phytoplankton biomass. In the Florida Straits, the picocyanobacteria Prochlorococcus was always responsible for > 30% of the phytoplankton biomass, as might be expected for warm stratified oligotrophic environments (J.A. Hilton, F. Not, A.Z. Worden and others, unpublished). Nevertheless, biliphytes contributed an estimated 28% of the total phytoplanktonic carbon in the eddy-influenced, surface waters. This is a large fraction for a single taxon, and could be related to eddy dynamics, which often enhance primary production (McGillicuddy et al., 2007). Previous findings at high-latitude sites (Not et al., 2007b) influenced by Atlantic Ocean currents could potentially be a reflection of residual populations from warmer waters. Clearly, much remains to be learned about the abundance, dynamics and controls of biliphytes. Their widespread distribution is remarkable and presumably reflects a high capacity for acclimation to dramatic temperature variations.
Conclusions
The discovery of biliphytes at a well-studied time-series site (i.e. BATS) reflects the current paucity of our knowledge concerning eukaryotic plankton, unless these novel eukaryotes are indeed a recent addition to the community. While the relative importance of such novel microbes to primary production remains to be determined, their discovery necessitates reconsideration of phytoplankton dynamics. Our data from warm subtropical and cool temperate environments, combined with previously reported sequences from cold high-latitude waters (Not et al., 2007b), underscore the need to define and quantify the role of biliphytes in primary production and carbon cycling. Dynamics of phytoplankton with broad thermal ranges may be particularly important when considering future climatic scenarios.
Experimental procedures
Field sites and sample collection
Five cruises were conducted, two transects from coastal New England to the BATS site (R/V Endeavor cruise EN351 and R/V Oceanus cruise OC413) and three transects across the Florida Straits (R/V Walton Smith cruises WS0503, WS0518, WS0528) with characteristics as in Table 1. Samples were collected in surface and DCM waters using a Sea-Bird Niskin Rosette equipped with standard CTD and PAR detectors (see also below). For DNA extraction samples, 1 l of seawater was gravity filtered through 2 μm pore size filters (GE Osmonics, Minnetonka, MN, USA) and then onto a 0.45 μm pore size (2001) or 0.2 μm pore size (2005) Supor filter (Pall Gelman, Ann Arbor, MI, USA) using vacuum. For FISH samples whole seawater (no size fractionation) was preserved with 1% paraformaldahyde (final concentration) for at least 1 h in the dark at 4°C. For each replicate, volumes of 180 ml or more of seawater were gently filtered onto a 25 mm, 0.2 μm Anodisc filter (Whatman, Maidstone, England). The filters were then subjected to an ethanol dehydration series at 50%, 80%, 100% (diluted in sterile, 18.2 ΩO H2O) for 3 min each and stored at −80°C. Flow cytometry samples were preserved with 0.25% (final concentration) fresh electron microscopy grade glutaraldehyde (Tousimis, Rockville, MD, USA), flash frozen in liquid nitrogen and moved to −80°C for long-term storage.
DNA extraction and PCR amplification
DNA was extracted using the DNeasy kit (Qiagen, Germantown, MD, USA). 18S rRNA genes were amplified using primers (5′-ACCTGGTTGATCCTGCCAG-3′ and 5′-TGATCCTTCYGCAGGTTCAC-3′) complimentary to conserved regions proximal to the gene termini, designed as universal eukaryotic primer set, but likely with some biases (Moon-van der Staay et al., 2001). Polymerase chain reaction was performed with an initial ‘hot start’ for 15 min at 95°C, proceeded by 32 cycles at 94°C for 30 s, 55°C for 30 s and 72°C for 1 min; followed by a final extension at 72°C for 15 min. One microlitre of PCR product was ligated into the vector pCR2.1 (Invitrogen, Carlsbad, CA, USA) and transformed, after colony selection and growth plasmids were purified. Sequencing protocols were based on the di-deoxy sequencing method. Multi-well cycle-sequencing reaction plates were prepared with plasmid template DNA and sequencing reactions were completed using the Big Dye Terminator chemistry and sequenced with either two plasmid targeted primers (M13F and M13R), with the former two primers and primers internal to the PCR product, 1174R and 502F, or with just the latter (Worden, 2006). Reaction mixtures, thermal cycling profiles and electrophoresis conditions were optimized to reduce the volume of the Big Dye Terminator mix (Applied Biosystems) and to extend read lengths on the AB3730xl sequencers (Applied Biosystems). Ninety-six clones were sequenced for each of the 17 libraries (14 of which yielded biliphyte sequences) constructed from 2005 cruise samples. For 2001 samples 48 clones were sequenced for each of the three libraries, all of which yielded biliphyte sequences.
Primers were also designed to preferentially amplify red algal and/or cryptophyte nucleomorph 18S rRNA sequences (NMR-SSU-1102-F: 5′-GAAATCAAAGTSTHTGGGTTCT-3′; NMR-SSU-1607-R: 5′-TACAAAGGGCAGGGACGTAT-3′; NM-SSU-0465-F: 5′-AATCCTGAYTMAGGGAGGTAGC-3′; NMR-SSU-0881-R: 5′-TCACCTCTGRCAAYGRARTAC-3′; NMR-SSU-0150-F: 5′-CTACKTGGATAMCCGTAG-3′; NMR-SSU-0748-R: 5′-GCYWTGAACACTCTAHTTTNTTC-3′). Polymerase chain reaction and nested PCR was performed with these primers using 30 cycles of 95°C for 30 s/1 min, 45°C for 1 min and 72°C for 30 s/1 min. Polymerase chain reaction products were cloned using the TOPO-TA cloning kit (Invitrogen) as per manufacturer's instruction and sequenced on a Beckman Coulter CEQ8000 (Beckman Coulter, Fullerton, CA, USA).
Phylogenetic analyses
blastn and preliminary phylogenetic analysis were used to identify biliphyte-like sequences in the GenBank non-redundant (May 2006; March 2007) and CAMERA (April 2007) databases. It became apparent that several deposited clones (Not et al., 2007b) had been deposited previously (Romari and Vaulot, 2004) under other accession numbers. Here only one sequence version per clone is used. Check_Chimera (Maidak et al., 2001) and manual screening were used to identify and remove likely chimeras as necessary. The alignment used for final phylogenetic analyses was performed in clustalw, manually adjusted and masked to use only those bases within regions of unambiguous alignment and considered homologous. Prior to this, Chimera Check (RDP II) and manual screening of alignments were used to identify and remove likely chimeras. Several short originally unidentified sequences from a previous publication (Countway et al., 2005) were not used in the alignment, none of which fell in BP2. Model selection, number of rate categories, proportion of invariable sites, transition/transversion (TiTv) and the gamma distribution parameter were determined within Modeltest (Posada and Crandall, 1998). Phylogenetic analysis was performed using maximum likelihood, neighbour-joining distance methods and parsimony, all within the Phylip modules (Felsenstien, 2005). For maximum likelihood 100 data sets were used for bootstrapping, with global rearrangements, randomized input, 10 jumbles and six categories (fraction of invariable sites = 0.2332; TiTv = 1.9433; 1/α = 1.451258). The same alignment and parameters were used for neighbour-joining distance (1000 replicates, 10 jumbles) and parsimony analysis (100 replicates, 2 jumbles).
Cell enumeration, measurements and biomass determinations
Whole-cell 18S rRNA-targeted FISH was performed on replicate filter pieces in conjunction with tyramide signal amplification (TSA) using a modification of a previously published method (Not et al., 2002) and newly designed biliphyte-targeted probes targeting BP1 (5′-GCGTGATGCCAAAATCCG-3′) and BP2 (5′-ATATGCCCGTCAAACCGT-3′), as verified and used in Not and colleagues (2007b). For the hybridizations, the filters were partitioned into multiple pieces for use with different probes. One microlitre of probe stock solution (50 ng probe μl−1 stock solution) and 9 μl of 40% hybridization buffer were mixed and added to each filter piece, then incubated for 3 h in a humid chamber. In order to make the hybridization buffer, a solution of blocking reagent [150 mM NaCl, 100 mM maleic acid, 10% blocking reagent (w:v)] was first prepared. The final concentrations for the hybridization buffer were: 40% deionized formamide, 20 mM Tris-HCl, pH 7.5, 0.01% sodium dodecyl sulfate, 2% blocking reagent (w:v), 20 mM maleic acid, 930 mM NaCl (final concentration including the NaCl used for the blocking reagent). Following the incubation, the samples were washed twice for 20 min at 37°C (5 mM EDTA, 0.01% SDS, 20 mM Tris, 37 mM NaCl) and equilibrated in TNT buffer (100 mM Tris HCl, pH 7.5, 150 mM NaCl, 0.074% Tween 20) at room temperature (RT) for 15 min. The TSA Fluorescein Tyramide Reagent Pack (PerkinElmer) was used for amplification of the signal. Ten microlitres of kit reagent mix (1:1 amplification diluent and 40% dextran sulfate, 1:50 Fluorescein Tyramide) was added to the filter and incubated at RT in the dark for 30 min. The reaction was stopped by two consecutive baths (20 min each) in TNT buffer at 55°C. The filter pieces were equilibrated in nanopure water at RT for 7 min in the dark and counterstained with 4′, 6-diamidino-2-phenylindole (DAPI) 2.5 μg ml−1 for 5 min. Finally, the filters were rinsed for 5 min at RT in nanopure water and briefly dipped in 80% ethanol. Seven microlitres of mounting solution [1:5 antifading solution AF1 (Citifluor) and VECTASHIELD mounting medium (H-100, Vector Laboratories)] was added to each filter piece and a coverslip was fixed with nail polish. The filters were counted within 24 h. Probe specificity was initially verified using cultures of non-target species: R. salina (CCMP1319), Isochrysis cf sp. (CCMP1244), M. pusilla (NOUM17), O. tauri (OTH95) and Paraphysomonas imperforata to ensure that these cells were not illuminated by the biliphyte probes. The bacterial antisense NON338 probe was used as a negative control for all hybridizations. In addition to procedures used in Not and colleagues (2007b), two negative controls (BP1 and BP2 probes on cultured R. salina CCMP1319 cells; and NON338 on all field samples) were employed alongside hybridization of all field samples as well as a no-probe control. Fifty fields were enumerated per sample using a 100× oil-immersion objective on a Olympus BX61 epifluorescence microscope, probe signal in the FITC channel and associated DAPI fluorescence was enumerated. For cell sizing a total of 105 cells were measured using a calibrated sizing grid and reported as the average shortest dimension by the average longest dimension.
Florida Straits physical and fluorescence characteristics analyses
At each of the 16 Florida Straits stations physical measurements were made through the entire depth of the water column using a CTD equipped with Seabird sensors measuring temperature, salinity, fluorescence, oxygen, beam transmission and light levels. Continuous measurements of the currents were made using a 600 kHz (samples 2 m depth bins from 4 to 35 m) Acoustic Doppler Current Profiler (ADCP; Teledyne RD Instruments) in March and a 75 kHz (samples 16 m depth bins from 25 to 400 m) ADCP in July. All physical data were processed using the manufacturer-provided software, and were plotted using Matlab (Mathworks), with a cubic interpolation between stations. Some temporal aliasing is introduced into the plots due to the sampling of the eastern half of the transect (e.g. Stations 07–15 in March 2005) on the first night of the cruise, and the western half (e.g. Stations 06–00) of the transect on the second night of the cruise. In July, sampling of the eastern half of the transect (e.g. Stations 08, 10–15) occurred at the first night, and other stations (e.g. Stations 09, 07–00) at the second night of the cruise.
Nucleotide sequence accession numbers
Biliphyte sequences from this study have been deposited in GenBank under Accession No. EU368003–EU368039.
Acknowledgments
We thank R. Gausling, R. Welsh and K. Penn for assistance with clone libraries; R. Cowen and B.J. Binder for cruise space as well as the captain and crews of the R/V Oceanus, R/V Endeavor, R/V Walton Smith; C. Guigand for facilitating cruises and developing figures; A. Engman, J.A. Hilton, F. Not, R. Welsh and L. Zamora for cruise assistance; M. Latasa for HPLC analysis; M. Everett for Model Test results; we are indebted to S. McDonald and especially D. Hansell for helpful discussions and comments. Research in the lab of J.M.A. was supported by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada. This work was funded by a Young Investigator Award from the Gordon and Betty Moore Foundation and an NSF Grant OCE-0623928, both to A.Z.W.
Supplementary material
The following supplementary material is available for this article online:
Characteristics of the Florida Straits in December 2004 and 2005. Cross-straits vertical profiles of the north component of current (A) during December 2005; temperature (B) December 2004, with profiles for December 2005 superimposed on the December 2004 data for three stations (CTD data for December 2005 was not available for other stations due to rough weather conditions); and fluorescence during December 2004 (C), with profiles for December 2005 superimposed on the December 2004 data for three stations (data for December 2005 for reasons as above). Only a single library was processed, indicated by an asterisk, and this did contain biliphyte sequences. Tick marks on the upper x-axis indicate stations at which CTD casts were performed to measure environmental parameters. This material is available as part of the online article from http://www.blackwell-synergy.com
Please note: Blackwell Publishing is not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Adl SM, Simpson AG, Farmer MA, Andersen RA, Anderson OR, Barta JR, et al. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol. 2005;52:399–451. doi: 10.1111/j.1550-7408.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- Archibald JM. Nucleomorph genomes: structure, function, origin and evolution. Bioessays. 2007;29:392–402. doi: 10.1002/bies.20551. [DOI] [PubMed] [Google Scholar]
- Azam F, Worden A. Microbes, molecules, and marine ecosystems. Science. 2004;303:1622–1624. doi: 10.1126/science.1093892. [DOI] [PubMed] [Google Scholar]
- Burki F, Shalchian-Tabrizi K, Minge M, Skjaeveland A, Nikolaev SI, Jakobsen KS, Pawlowski J. Phylogenomics reshuffles the eukaryotic supergroups. PLoS ONE. 2007;2:e790. doi: 10.1371/journal.pone.0000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Countway PD, Gast RJ, Savai P, Caron DA. Protistan diversity estimates based on 18S rDNA from seawater incubations in the western North Atlantic. J Eukary Microbiol. 2005;52:95–106. doi: 10.1111/j.1550-7408.2005.05202006.x. [DOI] [PubMed] [Google Scholar]
- Countway PD, Gast RJ, Dennett MR, Savai P, Rose JM, Caron DA. Distinct protistan assemblages characterize the euphotic zone and deep sea (2500 m) of the western North Atlantic (Sargasso Sea and Gulf Stream) Environ Microbiol. 2007;9:1219–1232. doi: 10.1111/j.1462-2920.2007.01243.x. [DOI] [PubMed] [Google Scholar]
- DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard NU, et al. Community genomics among stratified microbial assemblages in the ocean's interior. Science. 2006;311:496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- Diez B, Pedros-Alio C, Massana R. Study of genetic diversity of eukaryotic picoplankton in different oceanic regions by small-subunit rRNA gene cloning and sequencing. Appl Environ Microbiol. 2001;67:2932–2941. doi: 10.1128/AEM.67.7.2932-2941.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas S, Zauner S, Fraunholz M, Beaton M, Penny S, Deng LT, et al. The highly reduced genome of an enslaved algal nucleus. Nature. 2001;410:1091–1096. doi: 10.1038/35074092. [DOI] [PubMed] [Google Scholar]
- Eppley RW. An incubation method for estimating the carbon content of phytoplankton in natural samples. Limnol Oceanogr. 1968;13:574–582. [Google Scholar]
- Felsenstien J. PHYLIP (Phylogeny Inference Package) Version 3.6. Seattle, WA, USA: Department of Genome Sciences, University of Washington; 2005. [Google Scholar]
- Fratantoni PS, Lee TN, Podesta GP, Muller-Karger F. The influence of the Loop Current perturbations on the formation and evolution of Tortugas eddies in the southern Straits of Florida. J Geophys Res-Oceans. 1998;103:24759–24779. [Google Scholar]
- Goericke R, Welschmeyer NA. Response of phytoplankton community structure and taxon-specific growth rates to seasonally varying physical forcing in the Sargasso Sea off Bermuda. Limnol Oceanogr. 1998;43:921–935. [Google Scholar]
- Groisillier A, Massana R, Valentin K, Vaulotl D, Guilloul L. Genetic diversity and habitats of two enigmatic marine alveolate lineages. Aquat Microb Ecol. 2006;42:277–291. [Google Scholar]
- Hearn K. Bizarre new form of life found in Arctic Ocean, scientists announce. National Geographic News. 2007. National Geographic.
- Klein Breteler WCM. Fixation artifacts of phytoplankton in zooplankton grazing experiments. Aquat Ecol. 1985;19:1386–2588. [Google Scholar]
- Lane CE, Khan H, MacKinnon M, Fong A, Theophilou S, Archibald JM. Insight into the diversity and evolution of the cryptomonad nucleomorph genome. Mol Biol Evol. 2006;23:856–865. doi: 10.1093/molbev/msj066. [DOI] [PubMed] [Google Scholar]
- Latasa M. Improving estimations of phytoplankton class abundances using Chemtax. Mar Ecol Prog Ser. 2007;329:13–21. [Google Scholar]
- Latasa M, vanLenning K, Garrido JL, Scharek R, Estrada M, Rodriguez F, Zapata M. Losses of chlorophylls and carotenoids in aqueous acetone and methanol extracts prepared for RPHPLC analysis of pigments. Chromatographia. 2001;53:385–391. [Google Scholar]
- Lee TN, Clark ME, Williams E, Szmant AF, Berger T. Evolution of the Tortugas Gyre and its influence on recruitment in the Florida Keys. Bull Mar Sci. 1994;54:621–646. [Google Scholar]
- Li WKW, Harrison WG, Head EJH. Coherent assembly of phytoplankton communities in diverse temperate ocean ecosystems. Proc R Soc B Biol Sci. 2006;273:1953–1960. doi: 10.1098/rspb.2006.3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Garcia P, Rodriguez-Valera F, Pedros-Alio C, Moreira D. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature. 2001;409:603–607. doi: 10.1038/35054537. [DOI] [PubMed] [Google Scholar]
- Lovejoy C, Vincent WF, Bonilla S, Roy S, Martineau MJ, Terrado R, et al. Distribution, phylogeny, and growth of cold-adapted picoprasinophytes in arctic seas. J Phycol. 2007;43:78–89. [Google Scholar]
- Ludwig M, Gibbs SP. Are the nucleomorphs of cryptomonads and Chlorarachnion the vestigial nuclei of eukaryotic endosymbionts? Ann NY Acad Sci. 1987;503:198–211. [Google Scholar]
- McGillicuddy DJ, Anderson LA, Bates NR, Bibby T, Buesseler KO, Carlson CA, et al. Eddy/wind interactions stimulate extraordinary mid-ocean plankton blooms. Science. 2007;316:1021–1026. doi: 10.1126/science.1136256. [DOI] [PubMed] [Google Scholar]
- Maidak BL, Cole JR, Lilburn TG, Parker CT, Saxman PR, Farris RJ, et al. The RDP-II (Ribosomal Database Project) Nucleic Acids Res. 2001;29:173–174. doi: 10.1093/nar/29.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massana R, Balague V, Guillou L, Pedros Alio C. Picoeukaryotic diversity in an oligotrophic coastal site studied by molecular and culturing approaches. FEMS Microl Ecol. 2004a;50:231–243. doi: 10.1016/j.femsec.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Massana R, Castresana J, Balague V, Guillou L, Romari K, Groisillier A, et al. Phylogenetic and ecological analysis of novel marine stramenopiles. Appl Environ Microbiol. 2004b;70:3528–3534. doi: 10.1128/AEM.70.6.3528-3534.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon-van der Staay SY, DeWachter R, Vaulot D. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature. 2001;409:607–610. doi: 10.1038/35054541. [DOI] [PubMed] [Google Scholar]
- Moran MA, Miller WL. Resourceful heterotrophs make the most of light in the coastal ocean. Nat Rev Microbiol. 2007;5:792–800. doi: 10.1038/nrmicro1746. [DOI] [PubMed] [Google Scholar]
- Not F, Simon N, Biegala IC, Vaulot D. Application of fluorescent in situ hybridization coupled with tyramide signal amplification (FISH-TSA) to assess eukaryotic picoplankton composition. Aquat Microb Ecol. 2002;28:157–166. [Google Scholar]
- Not F, Gausling R, Azam F, Heidelberg JF, Worden AZ. Vertical distribution of picoeukaryotic diversity in the open ocean. Environ Microbiol. 2007a;9:1233–1252. doi: 10.1111/j.1462-2920.2007.01247.x. [DOI] [PubMed] [Google Scholar]
- Not F, Valentin K, Romari K, Lovejoy C, Massana R, Töbe K, et al. Picobiliphytes: a marine picoplanktonic algal group with unknown affinities to other eukaryotes. Science. 2007b;315:253–255. doi: 10.1126/science.1136264. [DOI] [PubMed] [Google Scholar]
- Palmer JD. The symbiotic birth and spread of plastids: how many times and whodunit? J Phycol. 2003;39:4–11. [Google Scholar]
- Patron NJ, Inagaki Y, Keeling PJ. Multiple gene phylogenies support the monophyly of cryptomonad and haptophyte host lineages. Curr Biol. 2007;17:887–891. doi: 10.1016/j.cub.2007.03.069. [DOI] [PubMed] [Google Scholar]
- Paul JH, Alfreider A, Wawrik B. Micro and macrodiversity in rbcL sequences in ambient phytoplankton populations from the Southeastern Gulf of Mexico. Mar Ecol Prog Ser. 2000;198:9–17. [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Rappé MS, Suzuki MT, Vergin KL, Giovannoni SJ. Phylogenetic diversity of ultraplankton plastid small-subunit rRNA genes recovered in environmental nucleic acid samples from the Pacific and Atlantic Coasts of the United States. Appl Environ Microbiol. 1998;64:294–303. doi: 10.1128/aem.64.1.294-303.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romari K, Vaulot D. Composition and temporal variability of picoeukaryote communities at a coastal site of the English Channel from 18S rDNA sequences. Limnol Oceanogr. 2004;49:784–798. [Google Scholar]
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, et al. The Sorcerer II Global Ocean Sampling Expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth JM, Smetacek V, Lenz J. Pelagic ecosystem structure: heterotrophic compartments of plankton and their relationship to plankton size fractions. Limnol Oceanogr. 1978;33:1225–1227. [Google Scholar]
- Simmons MP, Pickett KM, Miya M. How meaningful are Bayesian support values? Mol Biol Evol. 2004;21:188–199. doi: 10.1093/molbev/msh014. [DOI] [PubMed] [Google Scholar]
- Stockner JG. Phototrophic picoplankton: an overview from marine and freshwater ecosystems. Limnol Oceanogr. 1988;33:765–775. [Google Scholar]
- Worden AZ. Picoeukaryote diversity in coastal waters of the Pacific Ocean. Aquat Microb Ecol. 2006;43:165–175. [Google Scholar]
- Worden AZ, Nolan JK, Palenik B. Assessing the dynamics and ecology of marine picophytoplankton: the importance of the eukaryotic component. Limnol Oceanogr. 2004;49:168–179. [Google Scholar]
- Worden AZ, Cuvelier ML, Bartlett DH. In-depth analyses of marine microbial community genomics. Trends Microbiol. 2006;14:331–336. doi: 10.1016/j.tim.2006.06.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characteristics of the Florida Straits in December 2004 and 2005. Cross-straits vertical profiles of the north component of current (A) during December 2005; temperature (B) December 2004, with profiles for December 2005 superimposed on the December 2004 data for three stations (CTD data for December 2005 was not available for other stations due to rough weather conditions); and fluorescence during December 2004 (C), with profiles for December 2005 superimposed on the December 2004 data for three stations (data for December 2005 for reasons as above). Only a single library was processed, indicated by an asterisk, and this did contain biliphyte sequences. Tick marks on the upper x-axis indicate stations at which CTD casts were performed to measure environmental parameters. This material is available as part of the online article from http://www.blackwell-synergy.com


