Abstract
The amino terminal sequence of the Candida albicans cell wall protein Int1 exhibited partial identity with the major histocompatibility complex (MHC) class II binding site of the Mycoplasma arthritidis superantigen MAM. Int1-positive C. albicans blastospores activated human T lymphocytes and expanded Vβ subsets 2, 3, and/or 14; Int1-negative strains were inactive. Release of interferon-γ (IFN-γ) but not of tumor necrosis factor–α or interleukin-6 was Int1 dependent; interleukin-4 and interleukin-10 were not detected. T lymphocyte activation, Vβ expansion, and IFN-γ release were associated with a soluble polypeptide that encompassed the first 263 amino acids of Int1 (Pep263). Monoclonal antibody 163.5, which recognizes an Int1 epitope that overlaps the region of identity with MAM, significantly inhibited these activities when triggered by Int1-positive blastospores or Pep263 but not by staphylococcal enterotoxin B. Histidine263 was required. Pep263 bound to T lymphocytes and MHC class II and was detected in the urine of a patient with C. albicans fungemia. These studies identify a candidal protein that displays superantigen-like activities.
Candida species have emerged as the fourth most common cause of nosocomial bloodstream infections, and Candida albicans typically predominates [1–4]. Although mucosal colonization is usually not a direct cause of mortality, entry of C. albicans into the bloodstream is associated with fatality rates of 10%−49% among patients with thermal burns, cardiopulmonary bypass or abdominal surgery, neutropenia or neutrophil dysfunction, or extreme prematurity (birth weight, <1000 g) [5]. Although most patients with candidemia have a prolonged course with an incremental cost of treatment ranging from $40,000 to $90,000 per patient [6, 7], a few rapidly progress to hypotension and death accompanied by concentrations of proinflammatory cytokines (i.e., interferon-γ [IFN-γ], tumor necrosis factor–α [TNF-α], and interleukin-6 [IL-6]) that exceed those found in bacterial infections [8–10].
With other opportunistic pathogens, such as Aspergillus fumigatus, a fulminantly fatal course may be facilitated by toxins, including aflatoxin or gliotoxin. Neither toxin has been identified in C. albicans [11], although provocative evidence for a superantigen-like toxin derives from the observation that human peripheral blood mononuclear cells (PBMCs) incubated with C. albicans skin test antigen increased gene transcripts for Vβ subsets 5.1 and 5.2 [12]. The amino terminus of the C. albicans cell wall protein Int1 (relative molecular mass, 220 kDa), named for its integrin-like sequence and putative roles in adhesion, morphogenesis, and virulence [13, 14], displays 56% identity with the major histocompatibility complex (MHC) class II binding site of the well-characterized superantigen MAM from Mycoplasma arthritidis [15] (figure 1). We therefore investigated the possibility of Int1-dependent superantigen activity in C. albicans.
Figure 1.
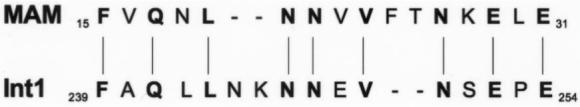
Identity between the amino terminus of the Candida albicans cell wall protein Int1 and the major histocompatibility complex class II binding site of Mycoplasma arthritidis superantigen MAM.
MATERIALS AND METHODS
Strains
C. albicans strains included CAF2 and its isogenic Int1-negative mutant CAG3 [14], as well as a second Int1-negative mutant (VBIDM2) constructed with polymerase chain reaction (PCR)–mediated mutagenesis [16] and expressing only the first 21 amino acids of Int1 (table 1).
Table 1.
Characteristics of Saccharomyces cerevisiae and Candida albicans strains used in this study.
| Organism, strain | Genotype | Int1 status | Reference |
|---|---|---|---|
| S. cerevisiae | |||
| BJ3501 | Matα pep4::HIS3 prb1 his3− ura3 can1 gal2 | Not available | ... |
| C. albicans | |||
| CAF2 | As SC5314 (wild-type), except URA3/ura3::λimm434 | Positive | [17] |
| CAI-4a | As CAF2, except ura3::λimm434/ura3::λimm434 | Positive | [17] |
| CAG3 | As CAI-4, except int1::hisG/int1::hisG-URA3-hisG | Negative | [14] |
| Efg1 null | As CAI-4, except efg1::hisG/efg1::hisG-URA3-hisG | Positive | [18] |
| BWP17a | ura3::λimm434/ura3::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG | Positive | [16] |
| BWP17wt | As BWP17, except arg4::hisG::ARG4::URA3/his1::hisG::HIS1 | Positive | [16] |
| DAY286 | As BWP17, except ARG4::URA3::arg4::hisG/arg4::hisG | Positive | [19] |
| VBIDM2 | As BWP17, except int1::ARG4/int1::URA3 | Negative | Present study |
| VBIDM6−2 | As VBIDM2, except int1::ARG4/int1::URA3/his1::hisG/HIS1 | Negative | Present study |
| VBIDM6−2R | As VBIDM6−2, except his1::hisG::HIS1-INT1 | Positive | Present study |
Used for strain construction but for experiments.
PCR-mediated disruption of INT1
Materials and methods are specified in the appendix, which is only available in the electronic edition of the journal.
Antibodies
The following antibodies were obtained: anti-human CD3, CD4, CD8, CD25, CD69, HLA-DR/IgG2a, and secondary conjugates (Becton Dickinson); anti-human TCR Vβ subsets (Beckman Coulter); and IgG1 isotype control for monoclonal antibody (mAb) 163.5 (Becton Dickinson). Typically, 10 μL were used to stain 106 cells. The immunizing peptide for mAb 163.5 (Inhibitex) was C-VNSEPEALTDMKLKRENFSNLSLDEKVNLY coupled to ovalbumin.
Activation of human PBMCs by C. albicans
Organisms were grown to saturation (optical density at 600 nm [OD600] = 0.5) in yeast extract/peptone/dextrose (YPD) medium with shaking at 30°C overnight; subcultures grown to early exponential phase in fresh YPD at 30°C (OD600 = 0.25) were washed twice in phosphate buffered saline (PBS) containing 0.2 μg/mL of amphotericin B (Invitrogen) to eliminate filamentous growth of C. albicans [23, 24]. Preliminary experiments showed that this concentration of amphotericin B did not inhibit the response of PBMCs to staphylococcal enterotoxin B (SEB) or Int1-positive C. albicans.
Whole blood was collected from adult donors by venipuncture, using heparinized vacutainer tubes (Becton Dickinson). PBMCs were isolated by standard methods, using Histopaque (Sigma). PBMCs washed with Hanks balanced salt solution were resuspended at 2.5 × 106 cells/mL in medium containing RPMI1640 supplemented with 2 mmol/L of l-glutamine, 100 U/mL of penicillin, 100 μg/mL of streptomycin, 1 mmol/L of sodium pyruvate, 0.1 mmol/L of nonessential amino acids, 20 mmol/L of Hepes, 10% certified fetal bovine serum, and 0.2 μg/mL of amphotericin B. In experiments with anti–HLA-DR antibodies, 10 μg/mL of azide free anti-HLA-DR or mouse IgG2a isotype control was added to PBMCs prior to plating.
A total of 5 × 105 blastospores in PBS were inoculated into each well of a 96-well plate and incubated at 37°C for 30 min. PBS manually aspirated from the culture wells containing adherent blastospores was replaced with 200 μL of complete medium containing 5 × 105 PBMCs, after ascertaining microscopically that wells contained approximately equal numbers of Int1-positive or Int1-negative yeast cells. SEB (Sigma; concentration, 10−100 ng/mL) served as a control. Plates were incubated at 37°C in 5% CO2.
Flow cytometry
For all experiments except those with data reported in figure 2B, PBMCs were harvested for antibody staining on the fourth day of coculture and were analyzed by flow cytometry for expression of the specified marker. Fluorochrome-labeled antibodies and conjugates were used in 2-color, 3-color, or 4-color flow cytometry with a FACSCalibur (BD Biosciences). Data were analyzed with Win-MDI software, version 2.8 (available at: http://facs.scripps.edu/software.html). Each experiment was performed in duplicate or triplicate wells; 30,000 cells from each well were analyzed.
Figure 2.
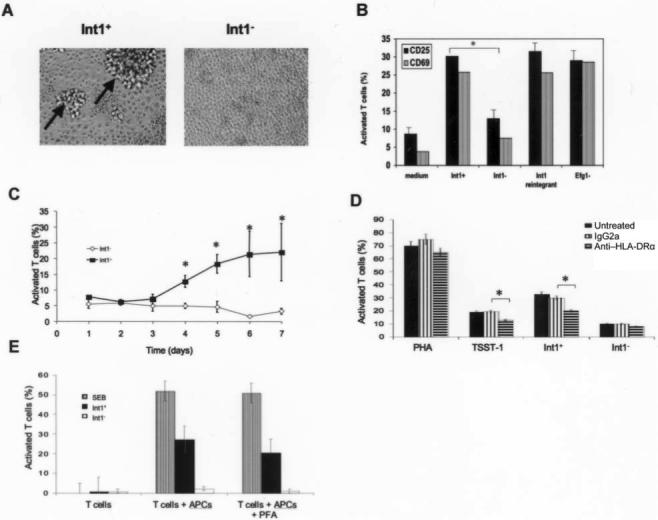
A, Foci of peripheral blood mononuclear cells (arrows) on the fourth day of coculture with Int1-positive (Int1+) Candida albicans blastospores or Int1-negative (Int1−) C. albicans blastospores. B, Peripheral blood mononuclear cell positivity for CD25 or CD69 after coculture with Int1+ C. albicans strain (DAY286), Int1− C. albicans strain (VBIDM2), Int1+ reintegrant (VBIDM6−2R), or the Efg1 null mutant. *P < .002 (n = 6 experiments). C, Activation of T cells from 5 healthy donors in response to Int1+ (CAF2) or Int1-negative (CAG3) C. albicans blastospores. Data are mean values ± 2 SDs. *P < .02. D, Effects of no treatment, IgG2a control antibody, or anti–HLA-DRα antibody on T cell activation evoked by phytohemagglutinin (PHA), toxic shock toxin (TSST-1), Int1+ C. albicans blastospores (CAF2), or Int1-negative C. albicans blastospores (CAG3). Data are mean values ±2 SDs. *P < .04 (n = 3 experiments). E, T cell activation induced by stimulation of T lymphocytes alone, T lymphocytes plus antigen-presenting cells, or T lymphocytes plus paraformaldehyde-fixed antigen-presenting cells after stimulation with staphylococcal enterotoxin B (SEB), Int1+ C. albicans, or Int1-negative C. albicans (n = 2 experiments).
Paraformaldehyde fixation of antigen-presenting cells and purification of T cells
PBMCs were plated in petri dishes at 37°C for 1 h. Adherent antigen-presenting cells were harvested by incubating cells at 4°C for 1 h in Ca2+-free and Mg2+-free PBS that contained 1 mmol/L EDTA and 0.5% glucose. T cells were purified from the nonadherent population to ≥95% purity, using a human T cell enrichment column kit (R&D Systems). PBS-washed antigen-presenting cells were resuspended in either complete medium or 0.06% paraformaldehyde (Sigma). After 5 min in paraformaldehyde at 37°C, ice cold 0.06% glycyl glycine was added (Sigma) [25]. After 2 PBS washes, paraformaldehyde-treated antigen-presenting cells were incubated in complete medium (CM; 0.17% Difco yeast nitrogen base, 0.5% NH4SO4, 2% glucose, and 1.5% Bacto-agar) at 37°C for an additional 30 min to remove residual paraformaldehyde before resuspension in fresh medium. Purified T cells and antigen-presenting cells were combined at a ratio of 2.3:1.
Cytokine measurement
Supernatants from T cell activations were analyzed for TNF-α, IFN-γ, IL-6, IL-4, and IL-10 by use of OptEIA ELISA II kits (BD Biosciences), according to the manufacturer's instructions.
Intracellular staining for IFN-γ
PBMCs cocultured with Int1-positive C. albicans, Int1-negative C. albicans, SEB, or medium alone were treated with brefeldin A (Golgi-Plug [BD Biosciences]) for 5 h before staining. Washed cells were stained for CD25, followed by intracellular staining for IFN-γ, according to the manufacturer's protocol (Cytofix/Cytoperm kit [BD Biosciences]).
Blockade of function with mAb 163.5
mAb 163.5 or an irrelevant mouse IgG1 was added to tissue culture wells daily at concentrations of 25 or 50 μg/mL.
Construction of 6XHis-Pep263 expression vector, mutagenesis of Pep263 codon 251, expression and purification of Pep263 and truncation mutants, and biotinylation of Pep263
Materials and methods are specified in the appendix.
Binding of Pep263 to T lymphocytes and MHC class II molecules
A total of 5 × 105 freshly isolated human PBMCs were incubated with 10 μg of biotinylated Pep263 and 5 × 105 Int1-negative blastospores (VBIDM2) for 4 days at 37°C. In control incubations, Pep263 or Int1-negative blastospores were omitted. All cells in each experimental mixture were washed with 1× PBS/3% BSA/0.02% sodium azide (FACS buffer), pelleted by centrifugation at 500 g, and stained for 25 min at room temperature with 10 μL of CD3-FITC (BD Pharmingen) and 10 μL of a 1:10 dilution of streptavidin–phyco-erythrin (PE) (BD Pharmingen). In the absence of biotinylated Pep263, no binding of streptavidin-PE to T lymphocytes was observed.
Experiments to measure the binding of biotinylated Pep263 to antigen-presenting cells were performed in the conditions described above, save that adherent antigen-presenting cells were detached from the wells by use of Versene (0.2 g/L EDTA-4Na in PBS [Gibco BRL]). Staining reagents included FITC-conjugated HLA-DR (BD; 10 μL for 106 cells) for class II molecules; mannosylated BSA-FITC (Sigma; 10 μL of 1 mg/mL) for mannose receptors on antigen-presenting cells, and streptavidin-PE for biotinylated Pep263 (10 μL of a 1:10 dilution). In the absence of biotinylated Pep263, no binding of streptavidin-PE to antigen-presenting cells was observed.
Western immunoblotting
Five milliliters of urine from a 43-day old premature male of 24 weeks' gestation with C. albicans fungemia was centrifuged at 2500 g at 4°C for 10 min to remove cellular debris. Clarified urine was concentrated 50-fold on a Centricon Plus-20 5000-molecular weight-cutoff filter (Millipore) at 4000 g at 4°C for 30 min. Urine from 4 healthy adults was treated identically. Retentates from each sample were denatured under reducing conditions and electrophoresed on a 7.5% SDS-PAGE gel. As an additional control, recombinant Pep263 was added to an aliquot from the retentate of 1 healthy donor. Proteins were transferred to a nitrocellulose membrane (Bio-Rad), blocked with TBST (50 mmol/L of Tris-HCl, 150 mmol/L of NaCl [pH 7.5], and 0.1% of Tween 20) that contained 5% skimmed milk powder (TBS-MT), and incubated at 4°C for 16 h with mAb 163.5 (stock concentration, 8.3 mg/mL) at a dilution of 1:500 in TBS-MT. The blot was washed with TBST and incubated at 25°C for 2 h with HRP-conjugated sheep anti-mouse IgG (GE Healthcare Bio-Science) at a dilution of 1:5000 in TBS-MT. After another wash with TBST, the membrane was developed with SuperSignal West Femto (Pierce) and exposed to Kodax Blue XB-1 film for 1 min.
Human subjects
Isolation of PBMCs from adults was approved by the Yale Human Investigations Committee (HIC #1047). Collection of urine from the infant received an exemption because the specimen would otherwise have been discarded.
Statistical analysis
Statistical comparisons were performed using an unpaired Student t test; a P value of <.05 was considered statistically significant.
RESULTS
Int1-dependent T cell activation, Vβ expansion, and IFN-γ secretion induced by C. albicans blastospores
Coculture of human PBMCs with Int1-positive C. albicans blastospores for 3.5 days induced an average of 15 foci of T cell blasts, each of which had a diameter of ≥5 μm (figure 2A); Int1-negative mutants missing amino acids 434−1664 [14, 17] or 21−1664 induced on average 1 foci of T cell blasts per well, which was equivalent to the induction rate in medium alone (figure 2A).
Unless otherwise stated, cells were harvested on the fourth day of coculture for analysis by flow cytometry. Both Int1-positive strains and their respective reintegrants activated T lymphocytes, as measured by up-regulation of the IL-2 receptor CD25; Int1-negative isogenic strains were significantly less active (P < .002). A mutant with 2 disrupted copies of EFG1 (which encodes a transcription factor linked to enhanced filamentous growth [18]) was not impaired (figure 2B). The pattern was identical with a second activation marker, CD69 (figure 2B).
Populations of CD3+ cells (T lymphocytes) that expressed CD25 (30.1% of cells) or CD69 (25.8% of cells) in response to Int1-positive C. albicans contained both CD4+ cells and CD8+ cells, in addition to CD4+CD8+ cells (table 2); a similar pattern was noted with SEB (table 2).
Table 2.
Rates of T cell responses to Int1-positive Candida albicans and staphylococcal enterotoxin B (SEB).
| Percentage of CD25+ T cells, by protein(s) expressed |
Percentage of CD69+ T cells, by protein(s) expressed |
|||||
|---|---|---|---|---|---|---|
| Stimulus | CD4+ | CD8+ | CD4+CD8+ | CD4+ | CD8+ | CD4+CD8+ |
| C. albicans | 42.4 | 27.9 | 26.3 | 36.9 | 35.0 | 12.5 |
| SEBa | 47.4 | 27.7 | 24.5 | 49.0 | 31.0 | 15.1 |
Responses were assayed on the fourth day of coculture.
In coculture with PBMCs from 5 adult donors, the Int1-positive strain was associated with up-regulation of the IL-2 receptor in all donors; isogenic Int1-negative mutants were inactive. These effects were statistically significant on days 4−7 (P < .02) (figure 2C).
A monoclonal antibody against the HLA-DR α-subunit had no effect on T lymphocyte activation in response to phytohemagglutinin but partially inhibited T cell activation induced by toxic shock toxin (TSST-1) or by Int1-positive C. albicans blastospores (P < .04). Blockade of HLA-DR did not affect the negligible T cell response induced by isogenic Int1-negative mutants (figure 2D).
In the absence of antigen-presenting cells, neither SEB, the Int1-positive strain, or the isogenic Int1-negative mutant induced T cell activation (figure 2E). In the presence of adherent antigen-presenting cells, T lymphocytes were activated by SEB and the Int1-positive strain. Paraformaldehyde fixation of antigen-presenting cells did not inhibit T cell activation induced by SEB or by Int1-positive blastospores. Int1-negative blastospores were inactive under all conditions.
The expansion of T lymphocyte populations bearing specific Vβ subsets is a hallmark of superantigens, although there is considerable donor-to-donor variation in the combination of subsets expanded [26]. For example, SEB expands T lymphocytes expressing Vβ subsets 3, 12, 14, 15, 17, or 20 [26, 27]. We defined “expansion” as a response to Int1-positive cells that was 5 times the response to Int1-negative cells. All donors expanded Vβ subsets 2, 3, or 14 in Int1-dependent fashion (figure 3). One donor also expanded Vβ7.1 (figure 3). In no case did we observe expansion of Vβ subsets by Int1-negative cells. In 3 experiments involving a donor with expansion of Vβ2 and Vβ14 in response to Int1-positive C. albicans, a mean (±SD) of 80.0% ± 6.6% cells expressing Vβ2 and 77.8% ± 6.7% expressing Vβ14 also expressed CD25; in response to SEB, 79.3% ± 6.4% of cells expressing Vβ14 also expressed CD25. Thus, >75% of Vβ2 and Vβ14 T cells were activated in response to Int1-positive C. albicans.
Figure 3.
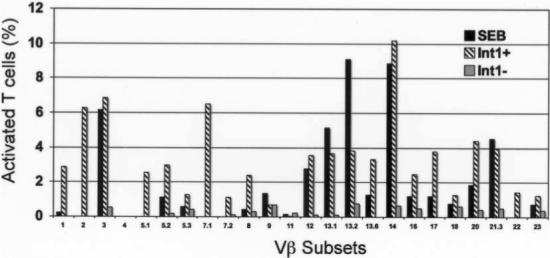
Expansion of Vβ subsets from a representative donor in response to staphylococcal enterotoxin B (SEB), Int1-positive (Int1+) Candida albicans blastospores (CAF2), or Int1-negative (Int1−) C. albicans blastospores (CAG3).
Expansion of Vβ subsets is accompanied by release of proinflammatory cytokines [28, 29]. C. albicans blastospores elicited TNF-α, IL-6, and IFN-γ, with the latter cytokine elicited in an Int1-dependent fashion (table 3); neither IL-4 nor IL-10 was detected, which was consistent with observations reported elsewhere [30]. Staining for intracellular IFN-γ showed that 97.1% of IFN-γ was produced by CD25+ cells in response to Int1-positive C. albicans, a result equivalent to what was observed with SEB.
Table 3.
Cytokine responses to staphylococcal enterotoxin B (SEB) and to Candida albicans blastospores with or without Int1.
| Cytokine, mean level ± SD, ng/mL |
|||||
|---|---|---|---|---|---|
| Stimulus | IFN-γ | TNF-α | IL-6 | IL-4 | IL-10 |
| C. albicans | |||||
| Int1 positive | 15.7 ± 5.3a | 1.8 ± 0.5b | 30.7 ± 21.7b | 0 | 0 |
| Int1 negative | 4.0 ± 0.5 | 2.8 ± 1.9 | 29.9 ± 17.7 | 0 | 0 |
| SEB | 66.4 ± 7.7 | 1.8 ± 0.2 | 5.3 ± 1.4 | 0 | 0 |
NOTE. Responses were assayed on the fourth day of coculture. INF-γ, interferon-γ; IL-4, interleukin-4; IL-6, interleukin-6; IL-10, interleukin-10; TNF-α, tumor necrosis factor—α.
P < .01, compared with Int1-negative strains.
P = not significant, compared with Int1-negative strains.
Characterization of a monoclonal antibody to the amino terminus of Int1
Linear peptides spanning amino acids 239−278 of Int1 were used to generate murine mAb. Only mAb 163.5 recognized Pep263, a construct encompassing the first 263 amino acids of Int1 (figure 4).
Figure 4.
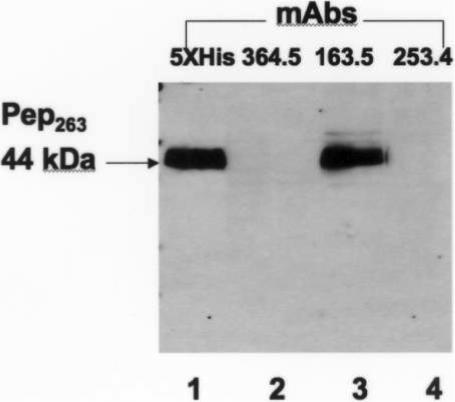
Recognition of purified Pep263 by monoclonal antibodies (mAbs). His-tagged Pep263 was identified by a penta-His mAb.
mAb 163.5 significantly inhibited activation of T lymphocytes (P < .035) (figure 5A) and expansion of Vβ subsets 2 (P < .02) and 14 (P < .02) (figure 5B) induced by Int1-positive C. albicans, compared with the activity of an irrelevant IgG1 control. Neither mAb 163.5 nor the isotype control inhibited T cell activation or expansion of the Vβ14 subset triggered by SEB (figure 5A and 5B). Therefore, mAb 163.5 was specific for Int1.
Figure 5.
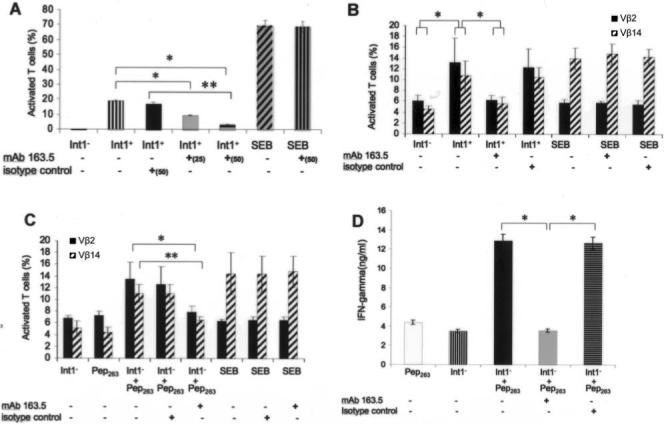
A and B, Inhibitory effects of monoclonal antibody (mAb) 163.5 or IgG1 isotype control mAb on T lymphocyte activation (A; *P < .002 and **P < .035) or expansion of Vβ subsets 2 and 14 (B; *P < .02) induced by Int1-negative (Int1−) C. albicans blastospores (VBIDM2) or Int1-positive (Int1+) C. albicans blastospores (DAY286). Staphylococcal enterotoxin B (SEB) served as control. Data are mean values ±2 SDs (n = 5 experiments). C and D, Inhibitory effects of mAb 163.5 or IgG1 isotype control mAb on expansion of Vβ subsets 2 and 14 (C; *P = .02 for Vβ2 and **P = .01 for Vβ14) and secretion of interferon-γ (IFN-γ) (D; *P < .01) induced by soluble Pep263 in the absence or presence of Int1− C. albicans blastospores (VBIDM2). Data are mean values ±2 SDs (n = 6 experiments).
mAb 163.5 also inhibited IFN-γ production by Int1-positive C. albicans by 50% (mean IFN-γ level [±SD], 22.89 ± 12.59 ng/mL vs. 11.26 ± 6.01 ng/mL in the presence of mAb 163.5; P = .002). Control mAb also showed an inhibitory effect of 18% (mean IFN-γ level [±SD], 22.89 ± 12.59 ng/mL vs. 18.75 ± 11.63 ng/mL in the presence of control mAb), but this effect was not statistically significantly different from the level of IFN-γ produced by Int1-positive C. albicans alone.
Localization of superantigen-like activity to the amino terminus of Int1p
Because of the inhibitory effects of mAb 163.5, a construct encompassing the first 263 amino acids of Int1 (Pep263) was expressed and purified. In the presence of Int1-negative blastospores, soluble Pep263 was associated with blastogenic foci of T lymphocytes (data not shown), expansion of Vβ subsets 2 and 14, and release of IFN-γ (figure 5C and 5D). mAb 163.5 significantly inhibited the expansion of Vβ subsets 2 (P = .02) and 14 (P = .01) and the release of IFN-γ (P < .01) in response to Pep263 (figure 5C and 5D), compared with the effects of an irrelevant IgG1 isotype control.
Structural requirements for the function of soluble Pep263
Mutation of key histidine residues in classical superantigens aborts their potency [31–33]. Although expression of native Pep263 ending in 260KLKH263 activated T lymphocytes and expanded Vβ subsets 2 and 14 as previously shown, truncation constructs ending in K262, L261, or K260 were no more active than medium alone. Substitution of A, D, or R for H263 did not restore the construct's ability to activate T lymphocytes or to expand Vβ subsets 2 and 14.
Requirement for Int1-negative blastospores in the binding of Pep263 to T lymphocytes and MHC class II molecules
Int1-negative strains were unable to activate T lymphocytes; expand Vβ subsets 2, 3, or 14; or elicit significant amounts of IFN-γ (figures 2, 3, and 5); nevertheless, the combination of soluble Pep263 and Int1-negative blastospores mediated these effects (figure 5C and 5D). Control experiments determined that heat-killed Int1-negative blastospores or live Saccharomyces cerevisiae could not substitute for replicating Int1-negative blastospores (figure 6). We hypothesized that Int1-negative blastospores were required for the interaction of Pep263 with T lymphocytes or antigen-presenting cells.
Figure 6.
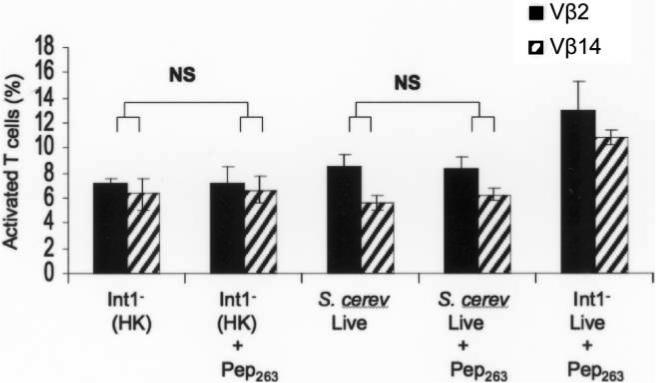
Expansion of T lymphocytes bearing Vβ subset 2 or 14 by replicating Int1-negative Candida albicans or a heat-killed Int1-negative C. albicans mutant (VBIDM2) and by replicating Saccharomyces cerevisiae, in the absence or presence of Pep263.
By FACS analysis, biotinylated Pep263 bound to a mean (±SD) of 5.8% ± 0.3% of CD3+ T lymphocytes in the presence of Int1-negative blastospores but only to 1.2% ± 0.5% of T lymphocytes in their absence. Pep263 bound equivalently to CD4+ and CD8+ T lymphocytes but preferentially to CD4+CD8+ cells in a ratio of 1:1:6.
Approximately 10%−11% of each PBMC preparation was positive for mannosylated bovine serum albumin as a marker for the mannose receptor (MR) on antigen-presenting cells; 90.3% of MR-positive cells were also positive for HLA-DR (figure 7). When biotinylated Pep263 was added, 96.9% of MR-positive cells also stained with streptavidin-PE, but the detection of HLA-DR was reduced more than 5-fold (from 90.3% of cells to 15.9% of cells). Thus, in the presence of Int1-negative blastospores, binding of Pep263 to antigen-presenting cells interfered with the detection of a class II antigen.
Figure 7.
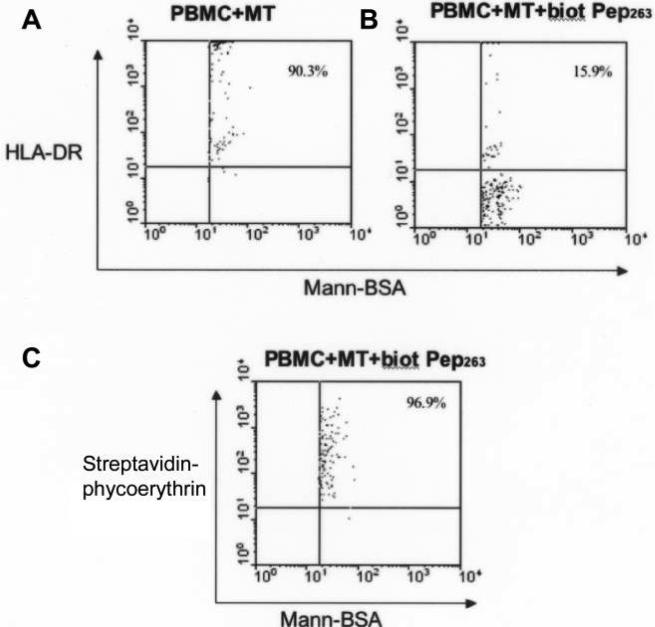
A, Binding of HLA-DR antibody to antigen-presenting cells and Int1-negative blastospores (MT) in the absence of biotinylated (biot) Pep263. B, Binding of HLA-DR antibody to antigen-presenting cells and MT in the presence of biot Pep263. C, Binding of Pep263 to antigen-presenting cells. Mann-BSA, mannosylated bovine serum albumin; PBMC, peripheral blood mononuclear cell.
Detection of Pep263 in a clinical specimen
Western immunoblotting with mAb 163.5 identified bands at 44 and 22 kDa in urine from a 43-day-old premature infant with catheter-associated C. albicans fungemia (figure 8); these bands corresponded to 2 fragments of purified Pep263; similar bands were not detected in uninfected urine. These results show that Pep263 was generated in vivo.
Figure 8.
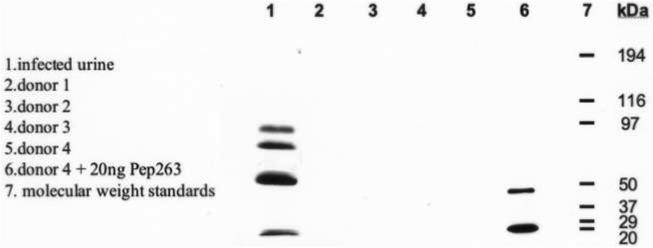
Western blotting with monoclonal antibody 163.5 for detection of Pep263. Lane 1, Urine specimen from a patient with Candida albicans fungemia. Lanes 2−5, Urine specimens from uninfected control donors. Lane 6, Urine specimen from an uninfected control donor plus 20 ng of purified Pep263. Lane 7, Molecular weight standards.
DISCUSSION
Superantigens elicit their effects by bridging the α or β subunit of the MHC class II locus and T lymphocytes bearing certain Vβ sequences at the T cell receptor [26, 34]. Although superantigens are not processed within the antigen-presenting cell or presented in the peptide groove, participation of MHC class II and costimulatory molecules such as B7−1 is required [35]. By obviating the mechanisms that control the well-modulated production of proinflammatory cytokines from antigen-specific T lymphocytes upon presentation of microbial antigens in the peptide groove, most bacterial superantigens recruit up to 10,000-fold more T lymphocytes of both CD4+ and CD8+ phenotypes, thereby triggering excessive release of cytokines such as IFN-γ, TNF-α, or IL-6.
Results reported here indicate that several effects associated with superantigens—activation of T lymphocytes independently of antigen processing and presentation, expansion of T cell populations expressing specific Vβ subsets, and elicitation of IFN-γ—are associated with a polypeptide (Pep263) derived from the amino terminus of the C. albicans cell wall protein Int1. As with classic superantigens, such as TSST-1 or staphylococcal enterotoxin A, the C-terminal histidine residue of Pep263 is essential for T lymphocyte activation and Vβ expansion [32, 33, 36]. Soluble Pep263 binds to T lymphocytes and to MHC class II molecules; however, because Pep263 requires the presence of Int1-negative blastospores for full activity, its effects might best be called “superantigen like.”
These activities amplify the putative roles of Int1 in adhesion, morphogenesis, and virulence derived from earlier mutational analyses [13, 14]. Localization of Int1 to the septin ring at the junction of the mother-daughter dyad in budding yeast cells, a key marker for polarity [37], may also provide a critical orientation that enables Pep263 to enhance virulence. Although elegant experiments have indicated the importance of mannosylation as a signal for Toll-like receptors and mannose receptors [38–40], Int1 appears not to be extensively modified by mannnosylation, despite several N-glycosylation sites in the sequence.
mAb 163.5, directed against a linear epitope of Pep263, combats these effects, whether induced by Int1-positive C. albicans blastospores or by soluble Pep263. Inhibitory mechanisms may include interference with the exposure of Pep263, its degradation, or its ability to link α or β subunits of MHC class II molecules to T lymphocytes expressing the relevant Vβ subsets. As proof of specificity, mAb 163.5 does not inhibit T cell activation, Vβ expansion, or IFN-γ release in response to SEB.
Two separate lines of evidence formed the critical underpinnings for these studies. First, a surprising identity between the extensively characterized superantigen MAM and the amino terminus of Int1 (figure 1) is much stronger than the identity of MAM with other superantigens, such as SEB, SEC-1, mouse mammary tumor virus, and HIV-1 gp160 [15].
Second, in vitro stimulation of human PBMCs with an extract prepared from boiled C. albicans blastospores led to a selective increase in gene transcripts for Vβ 5.1 and 5.2 [12]. Forty-eight hours after injection of a C. albicans filtrate into the skin, infiltrating T lymphocytes bearing Vβ 5.1 and 8.1 subsets were detected. As noted by Walsh et al. [12], these results raised the tantalizing prospect of a C. albicans–derived superantigen-like moiety; however, the reactive species was not further characterized.
There are at least 2 possible explanations for the differences between our results and those of Walsh et al. [12]. First, they used PCR for detection of Vβ transcripts, whereas we used flow cytometry to measure the binding of specific antibodies to expressed Vβ sequences. Second, they boiled their reagents, which may have significantly altered the predominant cell surface proteins of C. albicans. Expression of Pep263 in S. cerevisiae and purification by affinity chromatography should be less damaging to glycosylation and native conformation.
Although Pep263 induces T cell activation, specific Vβ expansion, and production of IFN-γ, the requirement for the Int1-negative mutant (figure 5C and 5D) does not fit the paradigm of a bacterial superantigen. Because heat-killed Int1-negative blastospores and live S. cerevisiae are ineffective (figure 6), some C. albicans–dependent, active process must be involved. Our experiments have not ruled out a role for Int1-negative blastospores in modifying the amino terminus of Pep263 or in contributing non–Int1-derived peptides to optimize its presentation, much as SEA126−131 peptides are required for optimal presentation of TSST-1 [41]. Mutant-induced proteolysis of the KLKH sequence at the carboxyl terminus of Pep263 seems unlikely, because removal of H263 nullifies activity. Interestingly, SEC-1, SEC-2, and SEC-3 have a KLKN sequence within the region that determines Vβ subset specificity [42]. Further experiments are underway to differentiate among these possibilities.
Although fulminant death from candidemia occurs in but a minority of patients, at least 2 potential clinical interventions are suggested by these studies. In addition to candidal mannans and β-glucans, which serve as potent elicitors of TNF-α [43, 44], the production of IFN-γ in response to Int1 and the possible contributions of Toll-like receptors, costimulatory molecules, and HLA alleles may permit modulation of the host response [38, 45]. In this regard, human immunodeficiency virus–infected subjects receiving recombinant IFN-γ showed reduced candidal infections and improved 3-year survival in a phase III trial [46]. As a first step to therapeutic neutralization or augmentation, a case-control study correlating HLA-DR haplotype, IFN-γ production, and outcome of candidemia could help to identify patients with overabundant or insufficient IFN-γ. Such approaches have proven extraordinarily informative in identifying genetic risk for superantigen-mediated morbidity during group A streptococcal infection [28, 47].
A humanized form of mAb 163.5 may also confer therapeutic benefits through its ability to modulate Int1-dependent T cell activation, Vβ expansion, and IFN-γ production. The identification of a C. albicans protein responsible for superantigen-like effects and the development of a monoclonal antibody that inhibits these activities in vitro suggest potential strategies for risk assessment, prevention, and therapy of C. albicans bloodstream infection in susceptible populations.
Acknowledgments
We thank Gerald Fink, Dana Davis, and Aaron Mitchell for Candida albicans reference strains, disruption mutants, and plasmids pGEM-URA3 and pRS-ARG4ΔSpe1.
Financial support: National Institutes of Health (grant AI50813) and March of Dimes (research grant 6FY02-151 to M.K.H.).
APPENDIX
SUPPLEMENTARY MATERIALS AND METHODS
PCR-mediated disruption of INT1
Disruption of both alleles of INT1 at nucleotide 64 of the open reading frame was performed in C. albicans strain BWP17. Table 4 shows the PCR primers used to amplify the DNA constructs for gene disruption. For gene disruption, PCR mixtures contained 1 mL of miniprep template DNA (pGEM-URA3 and pRS-ARG4ΔSpe1), 2 μL of a 10-mmol/L solution of deoxynucleoside triphosphates, 2 mL of a 10-mmol/L stock of each (forward and reverse) primer, and 0.75 mL of a 100 mmol/L solution of MgSO4; water was inserted until the mixture reached a volume of 50 mL, after which 0.5 mL of a 1 U/mL solution of Vent enzyme (New England BioLabs) was added. The PCR cycling conditions were as follows: 94°C for 5 min followed by 30 cycles of 94°C for 45 s, 50°C for 1 min and 72°C for 3 min with a final extension step of 72°C for 8 min. Strain BWP17 was grown to mid-exponential phase (∼0.5 at A600 nm) in 50 mL YPD at 30°C. After collection by centrifugation, the cells were washed in 5 mL of LATE buffer (0.1 mol/L of lithium acetate, 10 mmol/L of Tris · HCl [pH 8.0], and 1 mmol/L of EDTA) at room temperature. After centrifugation, the cells were resuspended in 0.5 mL of LATE. One hundred microliters of cells were mixed with 45 mL of each PCR product and 5 mL of carrier DNA (Sigma calf thymus DNA at 10 mg/mL). The transformation mixture was incubated at room temperature for 30 min, mixed with 0.7 mL of PLATE buffer (40% of PEG 3350, 0.1 mol/L of lithium acetate, 10 mmol/L of Tris-HCl [pH 8.0], and 1 mmol/L of EDTA), and incubated at room temperature overnight. The transformation mixture was heated to 42°C for 1 h and centrifuged for 3 min at 2000 g. The pelleted cells were gently washed twice in ∼0.4 mL of YPD without resuspension. The cells were suspended in 0.2 mL of YPD and spread on CSM agar without arginine and uridine (1.7 g/L of yeast nitrogen base without amino acids, 5 g/L of ammonium sulfate, 2% glucose, and 0.72 g/L of Bio 101 CSM dropout mix minus arginine and uridine). The plates were supplemented with 50 mL of uridine at 50 μg/mL in the first round of disruption, since the ARG4-mediated disruptions were done first. A rapid extraction protocol provided template DNA for verifying genotypes of resultant Arg-positive and Arg-positive/Ura-positive colonies [20]. Genotyping PCRs confirmed INT1 gene disruptions in strain VBIDM2 (figure 9), using 1.5 mL of DNA with locus-specific detection primers (table 4) and the cycling conditions described earlier. The primer combination of INT1U, 5Det-INT1, and 3Det-INT1 was used to detect wild-type and disrupted INT1 alleles.
Table 4.
Supplementary primers used in this study.
| Primer | Sequence, 5′-3′ |
|---|---|
| Targeted gene disruption of INT1 | |
| INT1FKO | ATGAACTCAACTCCAAGTAAATTATTACCGATAGATAAACATTCTCATTTACAATT ACAGCCTGTTTTCCCAGTCACGACGTT |
| INT1RKO | TTGAAGCATCAAATTTAGCCATGGTTGACGTCTGAACCGATTTCTATAGATAATTT CTTGTAAATTGTGGAATTGTGAGCGGATA |
| Detection of targeted INT1 | |
| 5Det-INT1 | TTCTCCATCTATCCATTCCTC |
| 3Det-INT1 | CAAAATGGGCATATATTTGCC |
| INT1U | GTGCGGGTTCTAAACCAA |
| Construction of Pep263 expression vectors | |
| DDC11F | ATAGAATTCACGGATTAGAAGCCGCCGAGCGGGTGAGAGC |
| DDC11R | AGCAAGCTTGATATCGATCGCATGCTTCTTTTAACAATGGATC |
| DDC15F | ATAGCATGCACCACCATCACCATCACATGAACTCAACTCCAAGTAAATTATTACCG |
| DDC15R | AGCAAGCTTTCAGTGCTTTAATTTCATATCTGTCAATGCCTCTGGTTCCGAATTGAC |
| DDC13F | AGCAAGCTTGTTTAAACCCGCTGATCTGATAACAA |
| DDC1R | TAAGTCGACAATTCTCTTAGGATTCGATTCACATTC |
| DDC43F | CCCCGGATCCATTGTTAAAAGAAGCATGAACTCAACTCCAAGTAAATTATTACCG |
| DDC29R | CCCCAAGCTTTCAATGGTGATGGTGATGGTGCTTTAATTTCATATCTGTCAATGC |
| CUG to CTG mutagenesis | |
| DCMUT2F | CAACGAAGTCAATTCGGAACCAGAG |
| DCMUT2R | CTCTGGTTCCGAATTGACTTCGTTG |
| Pep263 COOH deletion mutants | |
| DDC44F | CCCCGGATCCATTGTTAAAAGAAGCATGCACCACCATCACCATCACATGAACTCA |
| DDC33R | CCCCAAGCTTTCATTTCATATCTGTCAATGCCTCTGGTTCCGAATTGACTTC |
| DDC34R | CCCCAAGCTTTCATAATTTCATTGTCAATGCCTCTGGTTCCGAATTGAC |
| DDC35R | CCCCAAGCTTTCACTTTAATTTCATATCTGTCAATGCCTCTGGTTCCGAATT |
| Amino acid 263 substitution | |
| DDC43F | CCCCGGATCCATTGTTAAAAGAAGCATGAACTCAACTCCAAGTAAATTATTACCG |
| DDC36R | CCCCAAGCTTTCAATGGTGATGGTGATGGTGCGCCTTTAATTTCATATCTGTCAA |
| DDC37R | CCCCAAGCTTTCAATGGTGATGGTGATGGTGGCGCTTTAATTTCATACTTGTCAA |
| DDC40R | CCCCAAGCTTTCAATGGTGATGGTGATGGTGATCCTTTAATTTCATACTTGTCAA |
| Sequencing | |
| T3 | AATTAACCCTCACTAAAGGG |
| T7 | GTAATACGACTCACTATAGGGC |
NOTE. Mutated codons are in bold font.
Figure 9.
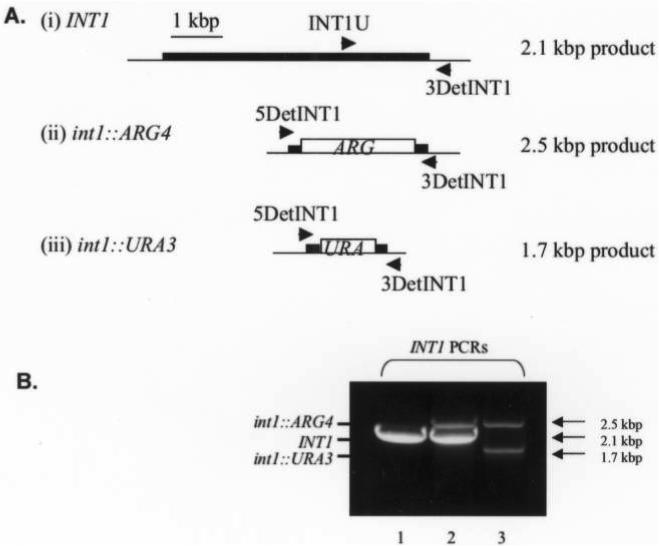
A, Strategy for polymerase chain reaction genotyping of INT1 alleles by use of forward primers INT1U and 5det-INT1 and reverse primer 3det-INT1. B, Genomic DNA from wild-type BWP17 (lane 1), VBIDM1 (INT1/int1::ARG4; lane 2), and VBIDM2 (int1::ARG4/int1::URA3; lane 3).
Construction of NH2-tagged 6XHis-Pep263 expression vector
pBluescript SK phagemid (Stratagene) was used to assemble the GAL1 promoter and the sequence coding for the first 263 amino acids of Int1 tagged with 6 histidines at either the amino or carboxy terminus, followed by the MATα transcription termination sequence. Plasmid pCG01 [14] containing the GAL1:10 promoter and the entire INT1 open reading frame served as a template for PCR amplification with primers DDC11F and DDC11R to yield product PCR1. The GAL1 promotor flanked by a 5' EcoRI site and a 3' BamHI site followed by 13 base pairs of the INT1 5' untranslated region (UTR) ending in sequential SphI and HindIII sites was excised from PCR1 and cloned into pBSIISK as an EcoRI/HindIII fragment to generate pBS · PCR1. Primers DDC15F and DDC15R generated PCR2, which was cloned into the SphI/HindIII sites of pBS · PCR1 to generate pBS · PCR1,2, thereby placing the signal methionine codon followed by the 6XHIS-Pep263 coding sequence and stop codon immediately downstream of the GAL1 promoter and 5' INT1 UTR.
The MATα transcription termination sequence was PCR amplified from vector pYD1 (Invitrogen), using primers DDC13F and DDC1R. The HindIII/SalI fragment was cloned into pBS · PCR1,2 to yield pBS · PCR1,2,3. The sequence of the entire EcoRI/SalI insert was confirmed using T3 and T7 primers (HHMI/Keck Foundation Biotechnology Resource Laboratory).
Site-directed mutagenesis of Pep263 codon 251
The CUG codon is translated as serine in Candida albicans and as leucine in S. cerevisiae [21, 22]. INT1 contains 6 CUG codons, one of which lies within the first 789 nucleotides of INT1. To correct for the difference in the product of the CUG codon between C. albicans and S. cerevisiae, 1 round of site-directed mutagenesis was performed using the Quickchange XL site-directed mutagenesis kit (Stratagene) to convert the C. albicans CTG serine codon at amino acid 251 to TCG, such that expression in S. cerevisiae substituted a serine residue for leucine at this position. Briefly, gel-purified mutagenesis primers DCMUT2F and DCMUT2R were used to PCR amplify pBS · PCR1,2,3, according to the manufacturer's instructions. The manufacturer's recommended temperatures were used for denaturation and annealing. After 18 cycles, 10 U of restriction enzyme DPN1 were added to the PCR reaction to digest the methylated template DNA. XL-10 Gold competent cells were transformed with digest. DNA preparations from ampicillin-resistant colonies were sequenced with T3 and T7 primers to confirm the codon change. Expression vector p6XH-Pep263 was created by cloning the 1600-bp EcoRI/Sal I fragment derived from pBS · PCR1,2,3 with TCG at codon 251 into Yep357 (ATCC 37732), a yeast shuttle vector containing the URA3 selectable marker and a 2-μm replicon to maintain high copy number in S. cerevisiae.
Construction of COOH-tagged Pep263-6XHis expression vector
The 452-bp GAL1 promoter excised from pBS · PCR1,2,3 by EcoRI/ BamHI digestion was cloned into the corresponding restriction sites of Yep357. p6XH-Pep263 served as template for PCR amplification of the 5' UTR of INT1 followed by the sequence encoding Pep263-6XHis, using primers DDC43F and DDC29R. The BamHI/Hind III digest of this PCR product along with the HindIII/SalI digest of the MATα transcription termination PCR product were cloned into the BamHI/Sal I sites of pBSIISK. Sequence was confirmed using the T3 and T7 primers. This BamHI/SalI fragment was cloned into the corresponding restriction sites of Yep357 that contained the GAL1 promoter to yield expression vector pPep263-6XH.
Expression of Pep263-6XHis
Protease-deficient S. cerevisiae strain BJ3501 (table 3) was transformed with expression vector p6XH-Pep263 or pPep263-6XH by of use lithium acetate and was plated on complete medium containing 2% glucose minus uridine. Transformants were grown to saturation in liquid medium containing 2% raffinose and 0.01% glucose and subcultured at an OD600 of 0.1 in CM minus uridine with 2% raffinose and 2% galactose. Expression of 6XHis-Pep263 or Pep263-6XHis was induced by galactose for 18 h at 30° C with shaking at 250 rpm. Yeast cells were washed twice with dH2O and resuspended at 200 mg wet cell paste per mL of Y-Per dialyzable yeast protein extraction reagent (Pierce). Cells were lysed for 4 h at 30°C with shaking at 250 rpm. Lysates collected after centrifugation of cell debris were filtered through a 0.45-μm cellulose membrane to remove all particulates and were stored at −80°C, pending purification.
Purification of Pep263
6XHis-tagged Pep263 was purified by metal chelate affinity chromatography, using HiTrap chelating HP columns (Amersham Biosciences) charged with NiSO4. Lysates diluted with an equal volume of 2× column buffer (40 mmol/L of phosphate, 1 mol/L of NaCl, and 40 mmol/L imidazole [pH 7.4]) were applied to Ni2+ columns equilibrated with 1× column buffer. Columns were washed extensively with the same buffer before elution of Pep263 with column buffer that contained 300 mmol/L of imidazole. Pep263 eluates were equilibrated with PBS (pH 7.4 [Invitrogen]), using 5000-molecular weight-cutoff centrifugal filter devices (Amicon). Protein concentration was determined using the 2-D Quant kit (Amersham). Pep263 preparations were run on 12% SDS-PAGE gels and either stained with the SilverXpress kit (Invitrogen) to access purity or transferred to nitrocellulose for Western blotting with PentHisHRP mAb (Qiagen) at 1:2000 or with multiple mAbs (including mAb 163.5) at 10 μg/mL followed by anti-mouse IgGHRP at 1:5000 (Amersham). Blots were developed with Supersignal West Pico chemiluminescent substrate, according to the manufacturers' instructions.
Generation and purification of Pep263 truncation constructs
Vector p6XH-Pep263 served as template for PCR amplification of the 5' INT1 UTR and coding sequences of Pep262, Pep261, and Pep260, using primer DDC44F paired with primers DDC35R, DDC34R, and DDC33R, respectively. Constructs substituting the histidine codon at amino acid 263 with the codons for alanine, arginine, or aspartic acid were generated using pPep263-6XH as a template and primer DDC43F paired with primers DDC36R, DDC37R, and DDC40R, respectively. BamHI/HindIII digests of PCR products were cloned upstream of the MATα transcription termination sequence inserted into pBSIISK as a BamHI/SalI fragment. Following sequence confirmation, BamHI/SalI digests of each construct were cloned downstream of the GAL1 promoter previously inserted into Yep357, yielding expression vectors p6XH-Pep262, p6XH-Pep261, p6XH-Pep260, pPep263A-6XH, pPep263R-6XH, and pPep263D-6XH. Truncation constructs were purified after expression in S. cerevisiae, using metal chelate affinity chromatography, as described above.
Biotinylation of Pep263
One milligram of purified Pep263 was incubated with a 10-fold molar excess of biotin in 1 mL of PBS (pH 7.4) for 2 h at 4°C, according to the instructions of the Sulfo-NHS-LC-biotinylation kit (Pierce). Biotinylated Pep263 was desalted on a Zeba column by centrifugation at 1000 g for 2 min. Biotinylated Pep263 was confirmed as functionally active by its ability to increase expression of the IL-2 receptor and to expand the T lymphocytes expressing Vβ2 and 14 subsets, as described above. Biotinylated Pep263 was analyzed by Western immunblotting with a 1:5000 dilution of HRP-conjugated streptavidin-PE.
Footnotes
Potential conflicts of interest: M.K.H. had a research agreement with Inhibitex, which ended in 2005. All other authors: none reported.
Presented in part: Maxwell Finland Lecture of the Infectious Diseases Society of America, San Francisco, CA, October 2001.
References
- 1.Nucci M, Colombo AL, Silveira F, et al. Risk factors for death in patients with candidemia. Infect Control Hosp Epidemiol. 1998;19:846–50. doi: 10.1086/647743. [DOI] [PubMed] [Google Scholar]
- 2.Pfaller MA, Jones RN, Doern GV, et al. Bloodstream infections due to Candida species: SENTRY antimicrobial surveillance program in North America and Latin America, 1997−1998. Antimicrob Agents Chemother. 2000;44:747–51. doi: 10.1128/aac.44.3.747-751.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edmond MB, Wallace SE, McClish DK, Pfaller MA, Jones RN, Wenzel RP. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin Infect Dis. 1999;29:239–44. doi: 10.1086/520192. [DOI] [PubMed] [Google Scholar]
- 4.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004;39:309–17. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 5.Beck-Sague C, Jarvis WR. Secular trends in the epidemiology of nosocomial fungal infections in the United States, 1980−1990. National Nosocomial Infections Surveillance System. J Infect Dis. 1993;167:1247–51. doi: 10.1093/infdis/167.5.1247. [DOI] [PubMed] [Google Scholar]
- 6.Zaoutis TE, Argon J, Chu J, Berlin JA, Walsh TJ, Feudtner C. The epidemiology and attributable outcomes of candidemia in adults and children hospitalized in the United States: a propensity analysis. Clin Infect Dis. 2005;41:1232–9. doi: 10.1086/496922. [DOI] [PubMed] [Google Scholar]
- 7.Pappas PG, Rex JH, Lee J, et al. A prospective observational study of candidemia: epidemiology, therapy, and influences on mortality in hospitalized adult and pediatric patients. Clin Infect Dis. 2003;37:634–43. doi: 10.1086/376906. [DOI] [PubMed] [Google Scholar]
- 8.Braude AI, Rock JA. The syndrome of acute disseminated moniliasis in adults. AMA Arch Intern Med. 1959;104:91–100. doi: 10.1001/archinte.1959.00270070093012. [DOI] [PubMed] [Google Scholar]
- 9.Presterl E, Lassnigg A, Mueller-Uri P, El-Menyawi I, Graninger W. Cytokines in sepsis due to Candida albicans and in bacterial sepsis. Eur Cytokine Netw. 1999;10:423–30. [PubMed] [Google Scholar]
- 10.Faix RG. Invasive neonatal candidiasis: comparison of albicans and parapsilosis infection. Pediatr Infect Dis J. 1992;11:88–93. [PubMed] [Google Scholar]
- 11.Kupfahl C, Ruppert T, Dietz A, Geginat G, Hof H. Candida species fail to produce the immunosuppressive secondary metabolite gliotoxin in vitro. FEMS Yeast Res. 2007;7:986–92. doi: 10.1111/j.1567-1364.2007.00256.x. [DOI] [PubMed] [Google Scholar]
- 12.Walsh P, Norris DA, Abe J, Martin DK, Giorno R, Leung DY. Candida albicans induces selective expansion of human T lymphocytes expressing the T-cell receptor variable region Vβ 5.1. J Dermatol Sci. 1996;12:140–6. doi: 10.1016/0923-1811(95)00473-4. [DOI] [PubMed] [Google Scholar]
- 13.Gale C, Finkel D, Tao N, et al. Cloning and expression of a gene encoding an integrin-like protein in Candida albicans. Proc Natl Acad Sci U S A. 1996;93:357–61. doi: 10.1073/pnas.93.1.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gale CA, Bendel CM, McClellan M, et al. Linkage of adhesion, filamentous growth, and virulence in Candida albicans to a single gene, INT1. Science. 1998;279:1355–8. doi: 10.1126/science.279.5355.1355. [DOI] [PubMed] [Google Scholar]
- 15.Cole BC, Knudtson KL, Oliphant A, et al. The sequence of the Mycoplasma arthritidis superantigen, MAM: identification of functional domains and comparison with microbial superantigens and plant lectin mitogens. J Exp Med. 1996;183:1105–10. doi: 10.1084/jem.183.3.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson RB, Davis D, Mitchell AP. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol. 1999;181:1868–4. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–28. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu H, Kohler J, Fink GR. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–6. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- 19.Davis D, Wilson RB, Mitchell AP. RIM101-dependent and -independent pathways govern pH responses in Candida albicans. Molec Cell Biol. 2000;20:971–8. doi: 10.1128/mcb.20.3.971-978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffman CS, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–72. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 21.Ohama T, Suzuki T, Mori M, et al. Non-universal decoding of the leucine codon CUG in several Candida species. Nucleic Acids Res. 1993;21:4039–45. doi: 10.1093/nar/21.17.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santos MA, Tuite MF. The CUG codon is decoded in vivo as serine and not leucine in Candida albicans. Nucleic Acids Res. 1995;23:1481–6. doi: 10.1093/nar/23.9.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfaller MA, Bale M, Buschelman B, et al. Quality control guidelines for National Committee for Clinical Laboratory Standards recommended broth macrodilution testing of amphotericin B, fluconazole, and flucytosine. J Clin Microbiol. 1995;33:1104–7. doi: 10.1128/jcm.33.5.1104-1107.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wanger A, Mills K, Nelson PW, Rex JH. Comparison of Etest and National Committee for Clinical Laboratory Standards broth macrodilution method for antifungal susceptibility testing: enhanced ability to detect amphotericin B–resistant Candida isolates. Antimicrob Agents Chemother. 1995;39:2520–2. doi: 10.1128/aac.39.11.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moreno J, Lipsky PE. Differential ability of fixed antigen-presenting cells to stimulate nominal antigen-reactive and alloreactive T4 lymphocytes. J Immunol. 1986;136:3579–87. [PubMed] [Google Scholar]
- 26.Kappler J, Kotzin B, Herron L, et al. V[H9252]-specific stimulation of human T cells by staphylococcal toxins. Science. 1989;244:811–3. doi: 10.1126/science.2524876. [DOI] [PubMed] [Google Scholar]
- 27.Fleischer B, Gerardy-Schahn R, Metzroth B, Carrel S, Gerlach D, Kohler W. An evolutionary conserved mechanism of T cell activation by microbial toxins: evidence for different affinities of T cell receptor–toxin interaction. J Immunol. 1991;146:11–7. [PubMed] [Google Scholar]
- 28.Norrby-Teglund A, Chatellier S, Low DE, McGeer A, Green K, Kotb M. Host variation in cytokine responses to superantigens determine the severity of invasive group A streptococcal infection. Eur J Immunol. 2000;30:3247–55. doi: 10.1002/1521-4141(200011)30:11<3247::AID-IMMU3247>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 29.Cameron SB, Nawijn MC, Kum WW, Savelkoul HF, Chow AW. Regulation of helper T cell responses to staphylococcal superantigens. Eur Cytokine Netw. 2001;12:210–22. [PubMed] [Google Scholar]
- 30.Levitz SM, North EA. Gamma interferon gene expression and release in human lymphocytes directly activated by Cryptococcus neoformans and Candida albicans. Infect Immun. 1996;64:1595–9. doi: 10.1128/iai.64.5.1595-1599.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Al-Daccak R, Mehindate K, Damdoumi F, et al. Staphylococcal enterotoxin D is a promiscuous superantigen offering multiple modes of interactions with the MHC class II receptors. J Immunol. 1998;160:225–32. [PubMed] [Google Scholar]
- 32.Hoffman M, Tremaine M, Mansfield J, Betley M. Biochemical and mutational analysis of the histidine residues of staphylococcal enterotoxin A. Infect Immun. 1996;64:885–90. doi: 10.1128/iai.64.3.885-890.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kokan-Moore NP, Bergdoll MS. Effect of chemical modification of histidine and tyrosine residues in toxic shock syndrome toxin 1 on the serologic and mitogenic activities of the toxin. Infect Immun. 1989;57:1901–5. doi: 10.1128/iai.57.7.1901-1905.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kotzin BL, Leung DY, Kappler J, Marrack P. Superantigens and their potential role in human disease. Adv Immunol. 1993;54:99–166. doi: 10.1016/s0065-2776(08)60534-9. [DOI] [PubMed] [Google Scholar]
- 35.Mu HH, Humphreys J, Chan FV, Cole BC. TLR2 and TLR4 differentially regulate B7−1 resulting in distinct cytokine responses to the myco-plasma superantigen MAM as well as to disease induced by Mycoplasma arthritidis. Cell Microbiol. 2006;8:414–26. doi: 10.1111/j.1462-5822.2005.00630.x. [DOI] [PubMed] [Google Scholar]
- 36.Earhart CA, Mitchell DT, Murray DL, et al. Structures of five mutants of toxic shock syndrome toxin-1 with reduced biological activity. Biochemistry. 1998;37:7194–202. doi: 10.1021/bi9721896. [DOI] [PubMed] [Google Scholar]
- 37.Gale C, Gerami-Nejad M, McClellan M, Vandoninck S, Longtine MS, Berman J. Candida albicans Int1p interacts with the septin ring in yeast and hyphal cells. Mol Biol Cell. 2001;12:3538–49. doi: 10.1091/mbc.12.11.3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Netea MG, Gow NA, Munro CA, et al. Immune sensing of Candida albicans requires cooperative recognition of mannans and glucans by lectin and Toll-like receptors. J Clin Invest. 2006;116:1642–50. doi: 10.1172/JCI27114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Timpel C, Strahl-Bolsinger S, Ziegelbauer K, Ernst JF. Multiple functions of Pmt1p-mediated protein O-mannosylation in the fungal pathogen Candida albicans. J Biol Chem. 1998;273:20837–46. doi: 10.1074/jbc.273.33.20837. [DOI] [PubMed] [Google Scholar]
- 40.Timpel C, Zink S, Strahl-Bolsinger S, Schroppel K, Ernst J. Morphogenesis, adhesive properties, and antifungal resistance depend on the Pmt6 protein mannosyltransferase in the fungal pathogen Candida albicans. J Bacteriol. 2000;182:3063–71. doi: 10.1128/jb.182.11.3063-3071.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hogan RJ, VanBeek J, Broussard DR, Surman SL, Woodland DL. Identification of MHC class II–associated peptides that promote the presentation of toxic shock syndrome toxin-1 to T cells. J Immunol. 2001;166:6514–22. doi: 10.4049/jimmunol.166.11.6514. [DOI] [PubMed] [Google Scholar]
- 42.Li H, Llera A, Malchiodi EL, Mariuzza RA. The structural basis of T cell activation by superantigens. Annu Rev Immunol. 1999;17:435–66. doi: 10.1146/annurev.immunol.17.1.435. [DOI] [PubMed] [Google Scholar]
- 43.Tada H, Nemoto E, Shimauchi H, et al. Saccharomyces cerevisiae– and Candida albicans– derived mannan induced production of tumor necrosis factor alpha by human monocytes in a CD14- and Toll-like receptor 4 – dependent manner. Microbiol Immunol. 2002;46:503–12. doi: 10.1111/j.1348-0421.2002.tb02727.x. [DOI] [PubMed] [Google Scholar]
- 44.Wheeler RT, Fink GR. A drug-sensitive genetic network masks fungi from the immune system. PLoS Pathog. 2006;2:e35. doi: 10.1371/journal.ppat.0020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Netea MG, Van Der Graaf CA, Vonk AG, Verschueren I, Van Der Meer JW, Kullberg BJ. The role of toll-like receptor (TLR) 2 and TLR4 in the host defense against disseminated candidiasis. J Infect Dis. 2002;185:1483–9. doi: 10.1086/340511. [DOI] [PubMed] [Google Scholar]
- 46.Riddell LA, Pinching AJ, Hill S, et al. A phase III study of recombinant human interferon gamma to prevent opportunistic infections in advanced HIV disease. AIDS Res Hum Retroviruses. 2001;17:789–97. doi: 10.1089/088922201750251981. [DOI] [PubMed] [Google Scholar]
- 47.Kotb M, Norrby-Teglund A, McGeer A, et al. An immunogenetic and molecular basis for differences in outcomes of invasive group A streptococcal infections. Nat Med. 2002;8:1398–404. doi: 10.1038/nm1202-800. [DOI] [PubMed] [Google Scholar]


