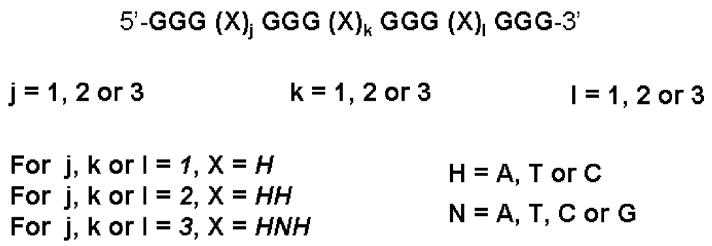Abstract
G-Rich sequences found within biologically important regions of the genome have been shown to form intramolecular G-quadruplexes with varied loop lengths and sequences. Many of these quadruplexes will be distinguishable from each other on the basis of their thermodynamic stabilities and folded conformations. It has been proposed that loop lengths can strongly influence the topology and stability of intramolecular G-quadruplexes. Previous studies have been limited to the analysis of quadruplex sequences with particular loop sequences, making it difficult to make generalizations. Here, we describe an original study that aimed to elucidate the effect of loop length on the biophysical properties of G-quadruplexes in a sequence-independent context. We employed UV melting and circular dichroism spectroscopy to examine and compare the properties of 21 DNA quadruplex libraries, each comprising partially randomized loop sequences with lengths ranging from one to three nucleotides. Our work supports a number of general predictions that can be made solely on the basis of loop lengths. In particular, the results emphasize the strong influence of single-nucleotide loops on quadruplex properties. This study provides a predictive framework that may help identify or classify biologically relevant G-quadruplex-forming sequences.
Genomic DNA is usually present as a canonical right-handed double helix, but it also has the potential to form other structures. Nucleic acid sequences rich in guanine (G)1 in particular are predisposed to form higher-order structures because of the capacity of guanine bases to self-associate via Hoogsteen hydrogen bonds to form planar G-quartets. Within a single G-rich DNA strand, several consecutive G-quartets can form and stack upon each other, leading to an intramolecular four-stranded structure, called a G-quadruplex. Such structures are further stabilized by the presence of monovalent cations, especially potassium (1, 2). Several G-rich DNA sequences identified in biologically important regions of the genome have been shown to form an intramolecular G-quadruplex under near-physiological conditions in vitro. Such sequences have been found in the telomeric repeats from a variety of organisms (3-6). More recently, G-quadruplex-forming sequences have been identified in the gene promoters of c-myc (7, 8), K-ras (9), c-kit (10, 11), VEGF (12), HIF-1α (13), Bcl-2 (14, 15), and RFP2 (16). Evidence of G-quadruplex formation in the regulatory sequences of muscle-specific genes (17) and at the 5′-end of the retinoblastoma gene (18) has also been reported. In addition to these examples, putative quadruplex-forming sequences seem to be prevalent in eukaryotic genomes. Genome-wide bioinformatic searches have revealed a significant number (∼376000) of putative quadruplex-forming sequences in the human genome (19, 20). In particular, the promoter regions of genes are significantly enriched in quadruplex-forming sequences relative to the rest of the genome, with >40% of human gene promoters containing one or more quadruplex motifs (21). Furthermore, it has been suggested that the potential for DNA quadruplex formation is correlated with gene function (22).
While there is accumulating evidence of the formation of DNA G-quadruplexes in vitro, more work is needed to elucidate the details of their existence and function in vivo. However, the discovery of proteins that interact specifically with G-quadruplexes has provided support for biological function (23-25). Recently, Paeschke et al. (26) have demonstrated that two ciliate telomere end-binding proteins cooperate, under the apparent control of a protein phosphorylation event, to regulate the formation of the DNA G-quadruplex at telomeres in vivo. Such telomeric DNA structures have been previously shown to act as caps that inhibit telomerase activity in vitro (27), which has inspired the development of quadruplex-based cancer therapeutic approaches (28-30).
All intramolecular G-quadruplexes share the property of containing stacked G-quartets; nevertheless, there is a considerable range of other structural variations, which can lead to differences in the thermodynamic and kinetic properties (31-34). Furthermore, a single quadruplex-forming sequence can exhibit an equilibrium between several different G-quadruplex conformations (35). Biological function and molecular recognition properties may well be associated with one predominant conformation, which has been suggested for the intramolecular c-myc quadruplex and also for the intermolecular quadruplex formed by ciliate telomeres (7, 26). A primary cause of structural polymorphism in G-quadruplexes is the relative arrangement of strands, which can have different polarities. They can all be parallel or a mixture of parallel and antiparallel strands (Figure 1A) (33, 34, 36).
Figure 1.
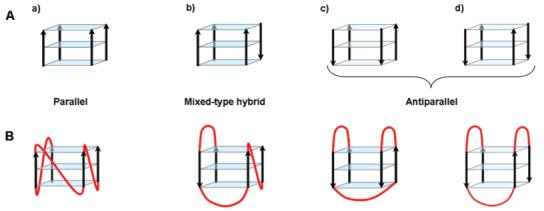
Arrows indicate 5′ to 3′ polarity. (A) Different arrangements of strand polarity for G-quadruplexes: (a) all strands parallel, (b) three parallel strands and one antiparallel strand, (c) two pairs of adjacent parallel strands, and (d) alternating antiparallel strands. (B) Examples of loop topologies leading to different arrangements of strand polarity.
Intramolecular G-quadruplex formation requires three loops to link between the top and bottom G-tetrads. Loops can take various conformations. They can adopt a double-chain-reversal conformation (or propeller-type), linking two adjacent parallel strands, or they can fold back, bridging either two diagonally opposite (diagonal loops) or two adjacent (edgewise loops) antiparallel strands (Figure 1B). A single intramolecular G-quadruplex structure can be based on loops with different conformations. Both loop length and sequence play a role in determining the folded conformation and thermodynamic stability of a quadruplex-forming sequence. It has been shown that single-base changes can affect the folding and stability of quadruplexes formed by human and Tetrahymena telomeric repeat sequences (37). It has also been observed for particular intramolecular G-quadruplexes that loop-loop interactions such as hydrogen bonding and stacking interactions can influence quadruplex properties (38-40). During the course of our study, two independent biophysical studies that aimed to investigate the sequence effects of short loops on quadruplex formation have been published (41, 42). Nonetheless, a comprehensive understanding of the effect of loop sequences on intramolecular quadruplex formation would still require more biophysical and, possibly, structural studies of a large number of examples. Loop lengths also play an important role in intramolecular quadruplex formation. There are a few reports on the effect of loop length on quadruplex folding and stability (40, 43-45). They have included oligo-dT loops (43, 44), punctual modifications in one or more loops of the thrombin binding aptamer (40), or unnatural polyethylene glycol loops (45). While they all are instructive, each of these studies has been limited to the analysis of quadruplex sequences with particular loop sequences and loop length arrangements. Here, we report a sequence-unbiased study that aims to elucidate general effects of loop lengths on quadruplex formation. We have addressed this goal by using UV melting and CD spectroscopy to examine the thermodynamic properties and folding topologies of 21 libraries of DNA G-quadruplex-forming sequences in which the loop sequences have been partially randomized.
MATERIALS AND METHODS
Oligonucleotides
Oligonucleotides were purchased from IBA. Stock solutions were made with molecular biology-grade water (Sigma-Aldrich). Concentrations were determined by UV absorption at 260 nm. Molar extinction coefficients were determined using OligoAnalyzer version 3.0, which makes use of a nearest-neighbor approximation (46) and is freely available (http://www.idtdna.com/analyzer/Applications/OligoAnalyzer/). For oligomers containing randomized base sites, extinction coefficients are predicted by averaging nearest-neighbor values at these sites.
UV Melting
UV melting curves were collected using a Varian Cary 100 Bio UV-visible spectrophotometer by measuring the spectral absorbance at 295 nm. Oligonucleotide library solutions were prepared at a final concentration of 5 μM in 10 mM sodium cacodylate (pH 7.0) containing 20 mM KCl. They were transferred to a 1 cm path length quartz cuvette, covered with a layer of mineral oil (Sigma-Aldrich), placed in the spectrophotometer, and equilibrated at 90 °C for 10 min. Samples were then cooled to 10 °C and heated to 90 °C twice consecutively at a rate of 0.25 °C/min, with data collection every 1 °C during both annealing and melting. For libraries L111, L112, L121, and L211 (where Tm > 70 °C), the melting-annealing experiments had an upper temperature of 100 °C (instead of 90 °C). A stream of dry nitrogen was gently applied through the sample compartment to prevent condensation of water on the cuvette at low temperatures.
Tm values were obtained from van’t Hoff analysis of the melting profiles as the temperature at which the folded fraction (θ) is 0.5 (47). To convert absorbance values into folded fraction, we manually chose linear upper and lower baselines as previously described (47). Using this method, uncertainty mainly arises because baseline determination remains subjective. All experiments were performed at least twice, and several different upper and lower baselines were chosen. Tm values differed by ≤2 °C. A prerequisite for the extraction of thermodynamic parameters from melting curves is that they be true equilibrium curves. A good criterion for this is the coincidence of the heating and annealing profiles. Under the heating-cooling rate (0.25 °C/min) and buffer conditions used in this work, all oligonucleotide libraries displayed superimposable and reproducible melting and annealing profiles. Temperature-independent van’t Hoff enthalpy and entropy changes (ΔHVH and ΔSVH) were calculated according to the equation ln K(T) = −ΔHVH/RT + ΔSVH/R, where K is the equilibrium constant for G-quadruplex formation and was determined assuming a two-state model for describing the single-strand to folded form process (47, 48). Plotting ln K(T) versus 1/T allowed us to access the values of ΔHVH and ΔSVH from the slope and intercept, respectively, of the linear regression in the interval 0.15 < θ < 0.85 (49). Free energies were calculated at 37 °C using the Gibbs equation ΔGVH(T) = ΔHVH - TΔSVH.
CD Spectroscopy
Circular dichroism experiments were conducted on a Chirascan spectrapolarimeter using a quartz cuvette with an optical path length of 1 mm. Oligonucleotide library solutions were prepared at 10 μM in 10 mM sodium cacodylate (pH 7.0) containing 20 mM KCl. The samples were annealed by being heated at 95 °C for 5 min and slowly cooled to room temperature on a heat block. Scans were performed over the range of 220-320 nm at 20 °C. Each trace is the result of the average of three scans taken with a step size of 1 nm, a time per point of 1 s, and a bandwidth of 0.5 nm. A blank sample containing only buffer was treated in the same manner and subtracted from the collected data. The data were zero-corrected at 320 nm.
Gel Electrophoresis
Nondenaturing gel electrophoresis was performed using a 20% polyacrylamide gel (16 cm × 20 cm, 19:1 acrylamide:bisacrylamide), prepared in 1× TBE “buffer which had been supplemented with 20 mM KCl. Electrophoresis was carried out at 80 V (constant) and 4 °C for 16 h, using a 1x TBE, 20 mM KCl buffer as a running buffer. The oligonucleotide concentration was 20 μM, and the samples were annealed in 10 mM sodium cacodylate (pH 7.0) containing 20 mM KCl by being heated at 95 °C for 5 min and slowly cooled to room temperature on a heat block. After electrophoresis, the gels were stained with Stains-all (Sigma).
RESULTS
Experimental Design
UV melting and CD spectroscopy, which are both recognized methods for the characterization of quadruplex formation, were employed to examine the thermodynamic stability and folding topology of DNA oligonucleotide libraries designed to contain G-quadruplexes with three G-quartets (Figure 2). G3-tracks were chosen because they are the shortest guanine runs that are reasonably stable for the formation of intramolecular DNA G-quadruplexes in aqueous solutions. We previously described a simple rule for predicting intramolecular quadruplex-forming sequences, d(G3+N1-7G3+N1-7G3+N1-7G3+), where N is A, T, C, or G, that we employed for a bioinformatic searching of the human genome (19). This study suggested that approximately half of the ∼376000 putative quadruplex-forming sequences identified in the human genome contain three loops each with a length of no more than three nucleotides (19). Here, we have focused on the subpopulations of G-quadruplex-forming sequences containing combinations of mono-, di-, and trinucleotide loops. To elucidate the effect of these loop lengths in a manner that is independent of their sequences, we designed and studied 21 libraries of G-quadruplex-forming sequences that include loops of partially random sequences (Figure 2). The general sequence of any mononucleotide and dinucleotide loop is H and HH, respectively, where H is A, T, or C, with equal probability; the general sequence of any trinucleotide loops is HNH, where N is A, T, C, or G, with equal probability. Each library has been named Ljkl, where j, k, and l represent the number of residues in the first, second, and third loops, respectively (Figure 2).
Figure 2.
General design of the quadruplex libraries (Ljkl) used for this study.
UV Melting
It has previously been demonstrated that G-quadruplex structures melt with a characteristic hypochromic shift at 295 nm (48). The oligonucleotide libraries studied here each exhibited clear hypochromic melting transitions at 295 nm, indicating that they include stable folded structures with a characteristic of quadruplex formation. As expected, transitions generally became broader as the total length of the loops increases (Figure 3). However, melting and annealing curves were superimposable for all quadruplex libraries, indicating reversible G-quadruplex formation with relatively fast kinetics. Previous investigations have shown that, generally, the kinetics of tetrameric quadruplex formation are extremely slow, leading to irreversible transitions, while dimeric quadruplexes form faster, but still slowly, leading to hysteresis between the melting and annealing curves (50-52). In contrast, intramolecular quadruplexes form quickly and reversibly (50, 52).
Figure 3.
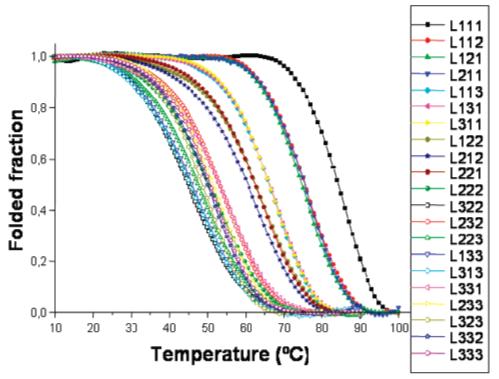
Fraction folded as a function of temperature for the 21 DNA quadruplex libraries.
Additionally, libraries of quadruplexes with a total length of loops of no more than five nucleotides migrate as a single band on a native polyacrylamide gel run in 20 mM KCl (Figure 4) and showed no indication of mixtures of monomeric quadruplexes and structures of higher molecularity, which might have been expected (42). Furthermore, melting and annealing profiles for these libraries were reversible and independent of oligonucleotide concentration from 2.5 to 20 μM. Taken together, these results are in good agreement with the formation of intramolecular G-quadruplex structures for the majority of sequences in each library.
Figure 4.
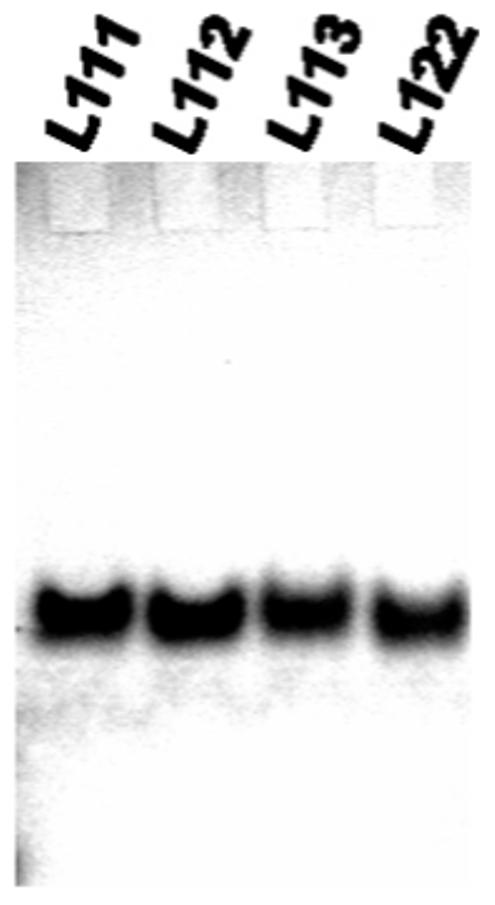
Nondenaturing gel electrophoresis of quadruplex libraries representative of total loop lengths of not more than five nucleotides.
Although the UV melting data cannot distinguish between the different sequences and conformations contained within quadruplex libraries, they provide an estimation of the overall stability of the different quadruplexes formed by a defined loop length arrangement. Melting temperature values obtained under the conditions used for this study are given in Table 1. Thermodynamic parameters (ΔHVH and ΔSVH) for the folding of each library, under the conditions used for this study, were estimated from van’t Hoff analysis of the melting profiles (47, 48). This two-state transition model led in each case to a straight line (r ≥ 0.995; data not shown), the slope of which is −ΔHVH/R and the y-intercept of which is ΔSVH/R. Values are presented in Table 1. They are in the same range of those previously reported for intramolecular G-quadruplex formation in potassium (39-41, 44, 50, 52, 53).
Table 1.
Melting Temperatures and van’t Hoff Thermodynamic Parameters for the Libraries of G-Quadruplex-Forming Sequencesa
| Tm (°C) | ΔHVH (kJ/mol) | ΔSVH (kJ mol−1 K−1) | ΔGVH(37 °C) (kJ/mol) | |
|---|---|---|---|---|
| L111 | 84/84 | −223 | −0.624 | −29.5 |
| L112 | 76/75 | −190 | −0.545 | −21.0 |
| L121 | 75/75 | −195 | −0.560 | −21.3 |
| L211 | 75/75 | −191 | −0.548 | −21.0 |
| L113 | 68/69 | −184 | −0.538 | −17.1 |
| L131 | 69/69 | −185 | −0.541 | −17.2 |
| L311 | 69/69 | −191 | −0.559 | −17.6 |
| L122 | 65/64 | −149 | −0.442 | −11.9 |
| L212 | 64/63 | −143 | −0.425 | −11.2 |
| L221 | 65/64 | −141 | −0.418 | −11.4 |
| L222 | 53/53 | −127 | −0.390 | −6.0 |
| L133 | 47/47 | −112 | −0.350 | −3.4 |
| L313 | 50/49 | −130 | −0.404 | −4.7 |
| L331 | 55/56 | −141 | −0.429 | −7.9 |
| L223 | 48/47 | −132 | −0.413 | −3.9 |
| L232 | 46/46 | −128 | −0.401 | −3.6 |
| L322 | 53/51 | −135 | −0.416 | −6.0 |
| L233 | 54/53 | −136 | −0.417 | −6.7 |
| L323 | 50/51 | −139 | −0.429 | −5.9 |
| L332 | 52/50 | −143 | −0.443 | −5.6 |
| L333 | 51/50 | −148 | −0.458 | −6.0 |
The samples were prepared at an oligonucleotide concentration of 5 μM in 10 mM sodium cacodylate buffer containing 20 mM KCl. They were heated and cooled at a rate of 0.25 °C/min. All experiments were performed at least twice. Tm, ΔHVH, and ΔSVH values were each the average of at least four determinations (two annealing and two melting profiles). The first Tm values were obtained from van’t Hoff analysis of the melting profiles as the temperature at which the folded fraction is 0.5. They are accurate to within 2 °C. ΔHVH and ΔSVH values are given with an experimental error of ±10%, determined on the basis of at least two independent experiments. It should be noted that ΔHVH and ΔSVH are not determined independently of each other. ΔGVH is calculated using the Gibbs equation ΔGVH(T) = ΔHVH − TΔSVH. As the equilibrium is intramolecular, K = 1 at Tm; thus, ΔGVH = 0 = ΔHVH − TmΔSVH, and therefore, Tm = ΔHVH/ΔSVH. The ratios of the calculated ΔHVH and ΔSVH values give second Tm values (in italics) in good agreement with the measured values.
A general trend emerged from the data indicating that thermodynamic stabilities of quadruplexes decrease as the total length of the loops increases (Table 1 and Figure 5). Quadruplex library L111, with a total length of the loops of three nucleotides, formed the most stable quadruplexes [Tm = 83 °C; ΔGVH(37 °C) = −29.5 kJ/mol]. Libraries with a total length of the loops equal to four nucleotides (i.e., L112, L121, and L211) also formed very stable quadruplexes, but their ΔGVH(37 °C) values increased by ∼8 kJ/mol. Interestingly, as the total length of the loops was increased from four to five nucleotides, the decrease in thermodynamic stability correlated with the number of single-nucleotide loops; libraries with two single-nucleotide loops and a remaining loop of three nucleotides (i.e., L113, L131, and L311) formed more stable quadruplexes [Tm = 69 °C; ΔGVH-(37 °C) ∼ −17.5 kJ/mol] as compared to libraries with only one single-nucleotide loop and a two dinucleotide loops (i.e., L122, L212, and L221) [Tm = 65 °C; ΔGVH(37 °C) ∼ −11.5 kJ/mol]. A further increase in the total loop length from five to six nucleotides resulted in an ∼5 kJ/mol increase in the ΔGVH(37 °C) of the quadruplex. Any subsequent increase in the total loop length beyond six nucleotides did not significantly alter the stability of the quadruplex, although minor variations were observed depending on the loop length arrangement.
Figure 5.
Changes in ΔGVH as a function of total loop length.
CD Spectroscopy
Several studies have demonstrated that the CD spectra of quadruplexes are a good indicator of their folding topology (36, 54-58). For G-quadruplexes, main characteristic CD signals arise from G-G stacking between G-quartets, the strength of which mainly depends on the conformation of guanine bases around the glycosylic bond (syn or anti) (36, 40). Restrictions apply to adjacent guanines involved in the same G-quartet. If they are on parallel strands, they must have the same glycosylic torsion conformation, and conversely, if they are on antiparallel strands, they must have opposite glycosylic torsion conformations (36, 40). As a result, quadruplex structures have been shown to exhibit different CD profiles depending on their parallel/antiparallel strand composition. A CD spectrum with a maximum around 265 nm and a minimum around 240 nm is generally indicative of a quadruplex with all strands being parallel (Figure 1A) (55-57). Antiparallel quadruplexes with either two pairs of adjacent parallel strands or alternating parallel strands (Figure 1A) typically display a maximum around 295 nm and a negative peak around 260 nm (54-56). Unstructured single-stranded DNA sequences exhibit neither of these characteristic CD signatures. Nevertheless, CD spectra with peaks at both 265 and 295 nm can indicate either the coexistence of distinct parallel and antiparallel folded species in solution or a single mixed-type hybrid quadruplex structure that contains three parallel strands and one antiparallel strand (Figure 1A). The latter has been recently exemplified by Yang and co-workers for a quadruplex-forming sequence identified within the promoter region of the human Bcl-2 proto-oncogene (14) and for an extended 26-nucleotide modified human telomeric sequence, d[A3(G3TTA)3G3A2] (59). Such a mixed-type hybrid G-quadruplex has also previously been described for the Tetrahymena telomeric repeat (5) d(T2G4)4 and for the human telomeric repeat d(T2AG3)4 upon induction by a small-molecule ligand (58).
Overlays of the CD spectra generated by each of the 21 oligonucleotide libraries, under the conditions used for this study, are presented in Figure 6. Libraries where a total length of the loops is not more than five nucleotides produce very similar CD spectra with single maxima at ∼265 nm (Figure 6A), suggesting that they all include quadruplexe with the same topology, which is likely to be the parallel form containing double-chain-reversal loops. When the total length of the loops increases to more than five nucleotides, CD profiles all display two positive peaks at ∼265 and ∼295 nm (Figure 6B). This suggests that these libraries are probably composed of mixtures of parallel, antiparallel, and mixed-type hybrid quadruplexes.
Figure 6.
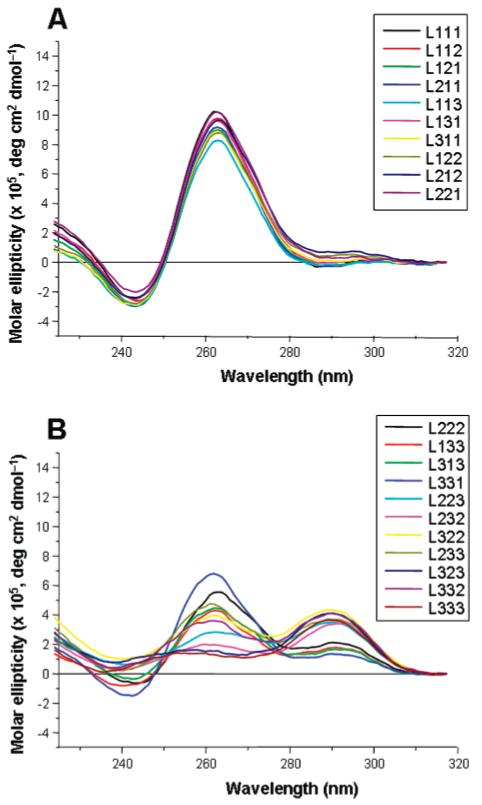
Overlays of the CD spectra generated by (A) quadruplex libraries with a total loop length of not more than five nucleotides and (B) quadruplex libraries with a total loop length of more than five nucleotides.
On the basis of results obtained from molecular dynamics simulations on quadruplexes with only oligo-dT loops, it was previously proposed that, due to steric constraints, a single-nucleotide thymidine loop bridging three G-tetrad layers can accommodate only a double-chain-reversal conformation (43). Moreover, recent biophysical and structural studies suggest that the conformation of single-nucleotide loops does not depend much on the sequence (14, 59-62). Our data support this assertion, since library L111 exhibits a CD spectrum characteristic of the parallel conformation of the quadruplex. On the other hand, the conformation of more flexible, longer (more than one nucleotide) loop is less predictable. It has been reported that a single quadruplex sequence with four G3-tracts and three T2-loops is likely to adopt a parallel structure (43). The CD spectrum of L222, which exhibits positive peaks at both 265 and 295 nm, now reveals that it might be different for other loop sequences. Interestingly, our data indicate that, in some cases, the conformation of long loops is strongly influenced by the length of the remaining loops. Indeed, the similarity between the CD spectrum of library L111 and the CD spectra of libraries with any two single-nucleotide loops and a remaining di- or trinucleotide loop is noteworthy (Figure 6A). Because of the flexibility of di- and trinucleotide loops, quadruplex-forming sequences with a 1:2(3):1 loop arrangement could in principle form both parallel and antiparallel quadruplexes, depending on the conformation of the longer, central loop (Figure 7A), and quadruplex-forming sequences with either a 1:1:2(3) or a 2(3):1:1 loop arrangement could in principle form both parallel and mixed-type hybrid quadruplexes (Figure 7B). That CD spectra of these libraries and the CD spectrum of L111 are essentially identical indicates that the presence of two single-nucleotide loops imposes an overall parallel fold upon the quadruplex, by constraining the longer loop into a double-chain-reversal orientation. It is also noteworthy that quadruplex libraries with two dinucleotide loops and a remaining single-nucleotide loop exhibit the same CD profile as L111, whereas quadruplexes with two trinucleotide loops and a remaining single-nucleotide do not. This might indicate a higher propensity of two-nucleotide loops to adopt the double-chain-reversal conformation compared to the three-nucleotide loop.
Figure 7.
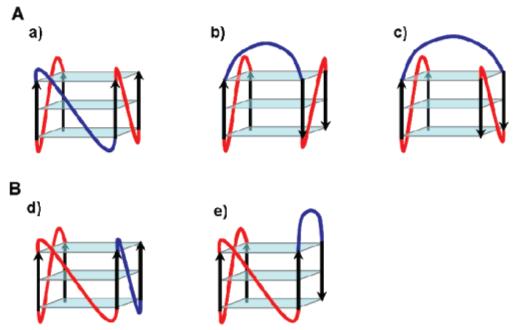
Arrows indicate 5′ to 3′ polarity. (A) Possible folds for quadruplex-forming sequences with two single-nucleotide loops (red) and a di- or trinucleotide central loop (blue) that (a) can adopt a double-chain-reversal conformation or can fold back, bridging either (b) two adjacent or (c) two diagonally opposite antiparallel strands [even though molecular dynamics simulations have previously shown that thymidine dinucleotide loops might be too short to span the diagonal of the G-tetrad (43)]. (B) Possible folds for quadruplex-forming sequences with two consecutive single-nucleotide loops (red) and trinucleotide loops (blue) that (d) can adopt a double-chain-reversal conformation or (e) can fold back to bridge two adjacent antiparallel strands.
DISCUSSION
This study investigated the effect of loop length on the thermodynamic stability and topology of intramolecular DNA G-quadruplexes. Related studies have employed G-quadruplex-forming oligonucleotides with defined loop sequences (40, 42-44) or non-nucleosidic linkers (45). Moreover, they were limited to certain loop length combinations. While such studies were insightful, they did not address the general case in which loop sequences, loop lengths, and loop length arrangements can vary significantly. Our study is the first attempt to establish a sequence-independent relationship between the lengths of the loops and the stability and topology of quadruplexes.
We have recorded the UV melting profiles and CD spectra of a set of 21 G-quadruplex-forming DNA oligonucleotide libraries. Each of them was designed to include G-quadruplex structures with different loop sequences but similar loop length arrangements. We excluded G in both the first and last positions in each loop to unambiguously define loop lengths. This experimental design precludes quadruplex-forming sequences which contain one (or more) G4+-tract(s) and will exclude some potentially unexpected quadruplex structures, such as the c-kit quadruplex published recently (63). Under the conditions used for this study, CD spectra of the libraries all exhibit clear positive peak(s) at ∼265 and/or ∼295 nm, and UV melting profiles, recorded at 295 nm, show reversible, superimposable hypochromic transitions, which are independent of oligonucleotide concentration. Besides, a comprehensive analysis also reveals several interesting connections between the CD and UV melting data.
Effect of Loop Length on the Topology of Intramolecular DNA G-Quadruplexes
The CD profiles recorded for quadruplex-forming libraries with a total loop length of more than five nucleotides indicate that they are mixtures of quadruplexes with different topologies: parallel, antiparallel, or mixed-type hybrid. This can reflect both sequence effects, i.e., different sequences leading to the formation of quadruplexes with different conformations, and the polymorphism of certain individual sequences. In any case, this points out of the conformational flexibility of loops with more than one nucleotide.
However, an important outcome of our study arises from the observation that the presence of two single-nucleotide loops within a quadruplex-forming sequence constrains the structure to a parallel fold, which is independent of the length of the remaining loop (two or three nucleotides). There are examples in the literature that support this observation for particular quadruplex-forming sequences containing four G3-tracts separated by two single-nucleotide loops and a remaining loop with at least two nucleotides. During the course of this work, Rachwal et al. (44) reported that intramolecular DNA quadruplexes with two single T-loops and a remaining T4-loop (in either a central or a lateral position) display CD spectra typical of a parallel topology. In addition, NMR solution structures of myc-2345 and myc-1245 G-quadruplexes, which have a 1:2:1 and a 1:6:1 loop length arrangement, respectively (Table 2), revealed that they both form a parallel-stranded structure where all loops, including the di- and hexanucleotide central loops, adopt a double-chain-reversal orientation (62). On the basis of CD and/or DMS footprinting data, the VEGF (12) and HIF-1α (13) promoter G-quadruplexes with 1:4:1 and 1:6:1 loop arrangements, respectively (Table 2), have been proposed to fold into a parallel-stranded topology similar to that of c-myc-1245. Using a combination of spectroscopic methods, we have recently shown that a 21-nucleotide G-rich sequence upstream of the c-kit transcription initiation site forms a mixture of quadruplex structures under near-physiological conditions (10). However, a mutant sequence (c-kit21T) with a 1:5:1 loop arrangement, and a CD spectrum consistent with parallel G-quadruplex conformation, was shown to be the predominant species (Table 2).
Table 2.
Sequences and Topologies of Some Biologically Relevant G-Quadruplexes with Two Single-Nucleotide Loops
| Name | Sequence | Topology | ||
|---|---|---|---|---|
| myc-2345 | GGG T | GGG GA | GGG T GGG | Parallela |
| myc-1245 | GGG A | GGG TTTTTA | GGG T GGG | Parallela |
| VEGF | GGG C | GGG CCGG | GGG C GGG | Parallelb |
| HIF-1α | GGG A | GGG GAGAGG | GGG C GGG | Parallelc |
| c-kit21T | GGG C | GGG CGCGA | GGG A GGG | Paralleld |
Determined by NMR in the presence of 90 mM K+.
Determined by CD in the presence of 100 mM K+.
Determined by DMS footprinting in the presence of 140 mM K+ and by CD in the presence of 25 mM K+.
Determined by CD in the presence of 100 mM K+.
Relationship between Loop Length and Thermodynamic Stability
Examination of the Tm and thermodynamic values obtained for the 21 DNA quadruplex libraries studied here reveals that loop lengths have a major effect on the stability of quadruplexes with a total loop length of not more than five nucleotides. For these libraries, a trend is apparent with an increase in the total loop length that involves a significant decrease in both the Tm and the ΔGVH at 37 °C for each one-nucleotide addition (Figure 5 and Table 1). This is consistent with the outcome of previous studies restricted to defined loop sequences (43, 44). For quadruplex libraries with a total loop length of more than five nucleotides, any increase in loop lengths does not significantly affect the stability.
Our CD data suggest that all quadruplex libraries containing any two single-nucleotide loops include quadruplexes with the same topology, which is likely to be the parallel form containing double-chain-reversal loops. However, UV melting experiments reveal that they have different thermodynamic stabilities, depending of the size of the remaining loop. Indeed, the ΔGVH(37 °C) of L111 is ∼8 kJ/mol lower than the ΔGVH(37 °C) of libraries with two one-nucleotide loops and a remaining two-nucleotide loop, and ∼12 kJ/mol lower than the ΔGVH(37 °C) of libraries with two one-nucleotide loops and a remaining three-nucleotide loop. It is also noteworthy that for quadruplex libraries with a total loop length of five nucleotides, which all exhibit a similar CD profile with a single peak at 265 nm, the thermodynamic stability correlates with the number of single-nucleotide loops. Libraries with two single-nucleotide loops and a remaining loop of three nucleotides (i.e., L113, L131, and L311) formed more stable quadruplexes [ΔGVH(37 °C) ∼ −17.5 kJ/mol] as compared to libraries with only one single-nucleotide loop and a two dinucleotide loops (i.e., L122, L212, and L221) [ΔGVH(37 °C) ∼ −11.5 kJ/mol]. These data are consistent with single-nucleotide loops being the most stable double-chain-reversal loops. This agrees with the suggestion made as a consequence of the NMR study of myc-1245 and myc-2345 G-quadruplexes (Table 2), which both form a parallel quadruplex with three double-chain-reversal loops (62). The main difference between myc-1245 and myc-2345 is the length of the central loop, six and two nucleotides, respectively. The melting temperature of myc-1245 has been shown to be 15 °C lower than that of myc-2345 (62).
Biological Significance
With the exception of the single-stranded 3′-overhang of telomeres, G-quadruplex-forming sequences in the genome are present with their complementary C-rich strands, which generate competing duplex structures. If quadruplex formation is biologically relevant, then in some circumstances it must be favored over duplex formation. The quadruplex to duplex equilibrium depends on many factors that will include their relative thermodynamic stability. The higher stability of G-quadruplex structures containing short loops makes the existence of this structure more likely, especially given that shorter loop lengths will result in a shorter competing duplex with a correspondingly lower stability. Remarkably, with the exception of the human telomere region, most intramolecular quadruplex-forming genomic sequences reported so far in the literature contain at least one single-nucleotide loop (7, 10, 12-15, 17). It is also noteworthy that for both c-myc and Bcl-2 promoters, in which multiple runs of guanine occur, in each case the predominant G-quadruplex species contains either one or two single-nucleotide loops (7, 14, 15, 59, 62). We recently reported that human gene promoters are enriched in quadruplex-forming sequence with single-nucleotide loops (21). Of the 3087 quadruplex-forming sequences identified in the first 100 bases upstream of the transcription start site, 78% of them have at least one single-base loop. This proportion gradually declines toward the genome average (64%) when one moves away from the transcription start site, suggesting that, proximal to the transcription start site, evolutionary selective pressure has favored quadruplex motifs with stabilizing loop lengths. Risitano et al. have examined the quadruplex-duplex equilibrium for several DNA oligonucleotides that have been shown to form quadruplexes (39). They showed that the human telomere repeat sequence d[G3(TTAG3)3], with a 3:3:3 loop length arrangement, predominantly forms a duplex in the presence of the complementary C-rich strand under physiological conditions, whereas a sequence related to the c-myc promoter, d(G4AG3T)2, with a 1:2:1 loop length arrangement, preferentially adopts the quadruplex form. If quadruplex stability proves to be an indicator of biological function, this work may provide a basis for helping to identify biologically relevant G-quadruplex-forming sequences on the basis of loop length. So far, models have been developed to predict stabilities of nucleic acid secondary structures with only Watson–Crick pairs and/or some non-Watson–Crick regions (such as G·U base pairs, hairpin loops, internal loops, and junctions), but there is no model for predicting the thermodynamics of quadruplex formation. Our study could be considered as a starting point for the development of such a model.
Footnotes
This work was supported by the BBSRC and Cancer Research UK. S.B. is a BBSRC Development Research Fellow.
- G
- guanine
- UV
- ultraviolet
- CD
- circular dichroism
- NMR
- nuclear magnetic resonance
- Tm
- melting temperature
- ΔHVH
- van’t Hoff enthalpy change
- ΔSVH
- van’t Hoff entropy change
- ΔGVH
- van’t Hoff free energy change.
SUPPORTING INFORMATION AVAILABLE
Extinction coefficients and UV melting curves for the 21 quadruplex libraries. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Williamson JR, Raghuraman MK, Cech TR. Monovalent cation-induced structure of telomeric DNA: The G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 2.Sen D, Gilbert W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature. 1990;344:410–414. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- 3.Henderson E, Hardin CC, Walk SK, Tinoco I, Jr., Blackburn EH. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs. Cell. 1987;51:899–908. doi: 10.1016/0092-8674(87)90577-0. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Patel DJ. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Patel DJ. Solution structure of the Tetrahymena telomeric repeat d(T2G4)4 G-tetraplex. Structure. 1994;2:1141–1156. doi: 10.1016/s0969-2126(94)00117-0. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Patel DJ. Solution structure of the Oxytricha telomeric repeat d[G4(T4G4)3] G-tetraplex. J. Mol. Biol. 1995;251:76–94. doi: 10.1006/jmbi.1995.0417. [DOI] [PubMed] [Google Scholar]
- 7.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simonsson T, Pecinka P, Kubista M. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998;26:1167–1172. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cogoi S, Xodo LE. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006;34:2536–2549. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernando H, Reszka AP, Huppert J, Ladame S, Rankin S, Venkitaraman AR, Neidle S, Balasubramanian S. A Conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene. Biochemistry. 2006;45:7854–7860. doi: 10.1021/bi0601510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rankin S, Reszka AP, Huppert J, Zloh M, Parkinson GN, Todd AK, Ladame S, Balasubramanian S, Neidle S. Putative DNA quadruplex formation within the human c-kit oncogene. J. Am. Chem. Soc. 2005;127:10584–10589. doi: 10.1021/ja050823u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun D, Guo K, Rusche JJ, Hurley LH. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005;33:6070–6080. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Armond R, Wood S, Sun D, Hurley LH, Ebbinghaus SW. Evidence for the presence of a guanine quadruplex forming region within a polypurine tract of the hypoxia inducible factor 1α promoter. Biochemistry. 2005;44:16341–16350. doi: 10.1021/bi051618u. [DOI] [PubMed] [Google Scholar]
- 14.Dai J, Dexheimer TS, Chen D, Carver M, Ambrus A, Jones RA, Yang D. An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution. J. Am. Chem. Soc. 2006;128:1096–1098. doi: 10.1021/ja055636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dexheimer TS, Sun D, Hurley LH. Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the bcl-2 P1 promoter. J. Am. Chem. Soc. 2006;128:5404–5415. doi: 10.1021/ja0563861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skoblov M, Shakhbazov K, Oshchepkov D, Ivanov D, Guskova A, Ivanov D, Rubtsov P, Prasolov V, Yankovsky N, Baranova A. Human RFP2 gene promoter: Unique structure and unusual strength. Biochem. Biophys. Res. Commun. 2006;342:859–866. doi: 10.1016/j.bbrc.2006.01.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yafe A, Etzioni S, Weisman-Shomer P, Fry M. Formation and properties of hairpin and tetraplex structures of guanine-rich regulatory sequences of muscle-specific genes. Nucleic Acids Res. 2005;33:2887–2900. doi: 10.1093/nar/gki606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y, Sugiyama H. Formation of the G-quadruplex and i-motif structures in retinoblastoma susceptibility genes (Rb) Nucleic Acids Res. 2006;34:949–954. doi: 10.1093/nar/gkj485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eddy J, Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34:3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arthanari H, Bolton PH. Functional and dysfunctional roles of quadruplex DNA in cells. Chem. Biol. 2001;8:221–230. doi: 10.1016/s1074-5521(01)00007-2. [DOI] [PubMed] [Google Scholar]
- 24.Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat. Struct. Mol. Biol. 2006;13:1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- 25.Shafer RH, Smirnov I. Biological aspects of DNA/RNA quadruplexes. Biopolymers. 2000;56:209–227. doi: 10.1002/1097-0282(2000/2001)56:3<209::AID-BIP10018>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 26.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 27.Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 28.Mergny JL, Riou JF, Mailliet P, Teulade-Fichou MP, Gilson E. Natural and pharmacological regulation of telomerase. Nucleic Acids Res. 2002;30:839–865. doi: 10.1093/nar/30.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neidle S, Parkinson G. Telomere maintenance as a target for anticancer drug discovery. Nat. Rev. Drug Discovery. 2002;1:383–393. doi: 10.1038/nrd793. [DOI] [PubMed] [Google Scholar]
- 30.Rezler EM, Bearss DJ, Hurley LH. Telomeres and telomerases as drug targets. Curr. Opin. Pharmacol. 2002;2:415–423. doi: 10.1016/s1471-4892(02)00182-0. [DOI] [PubMed] [Google Scholar]
- 31.Davis JT. G-quartets 40 years later: From 5′-GMP to molecular biology and supramolecular chemistry. Angew. Chem., Int. Ed. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 32.Keniry MA. Quadruplex structures in nucleic acids. Biopolymers. 2000;56:123–146. doi: 10.1002/1097-0282(2000/2001)56:3<123::AID-BIP10010>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 33.Phan AT, Kuryavyi V, Patel DJ. DNA architecture: From G to Z. Curr. Opin. Struct. Biol. 2006;16:288–298. doi: 10.1016/j.sbi.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simonsson T. G-quadruplex DNA structures: Variations on a theme. Biol. Chem. 2001;382:621–628. doi: 10.1515/BC.2001.073. [DOI] [PubMed] [Google Scholar]
- 35.Ying L, Green JJ, Li H, Klenerman D, Balasubramanian S. Studies on the structure and dynamics of the human telomeric G quadruplex by single-molecule fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. U.S.A. 2003;100:14629–14634. doi: 10.1073/pnas.2433350100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williamson JR. G-quartet structures in telomeric DNA. Annu. Rev. Biophys. Biomol. Struct. 1994;23:703–730. doi: 10.1146/annurev.bb.23.060194.003415. [DOI] [PubMed] [Google Scholar]
- 37.Miyoshi D, Karimata H, Sugimoto N. Drastic effect of a single base difference between human and tetrahymena telomere sequences on their structures under molecular crowding conditions. Angew. Chem., Int. Ed. 2005;44:3740–3744. doi: 10.1002/anie.200462667. [DOI] [PubMed] [Google Scholar]
- 38.Keniry MA, Owen EA, Shafer RH. The contribution of thymine-thymine interactions to the stability of folded dimeric quadruplexes. Nucleic Acids Res. 1997;25:4389–4392. doi: 10.1093/nar/25.21.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Risitano A, Fox KR. Stability of intramolecular DNA quadruplexes: Comparison with DNA duplexes. Biochemistry. 2003;42:6507–6513. doi: 10.1021/bi026997v. [DOI] [PubMed] [Google Scholar]
- 40.Smirnov I, Shafer RH. Effect of loop sequence and size on DNA aptamer stability. Biochemistry. 2000;39:1462–1468. doi: 10.1021/bi9919044. [DOI] [PubMed] [Google Scholar]
- 41.Vorlickova M, Bednarova K, Kejnovska I, Kypr J. Intramolecular and intermolecular guanine quadruplexes of DNA in aqueous salt and ethanol solutions. Biopolymers. 2007;86:1–10. doi: 10.1002/bip.20672. [DOI] [PubMed] [Google Scholar]
- 42.Rachwal PA, Brown T, Fox KR. Sequence effects of single base loops in intramolecular quadruplex DNA. FEBS Lett. 2007;581:1657–1660. doi: 10.1016/j.febslet.2007.03.040. [DOI] [PubMed] [Google Scholar]
- 43.Hazel P, Huppert J, Balasubramanian S, Neidle S. Loop-length-dependent folding of G-quadruplexes. J. Am. Chem. Soc. 2004;126:16405–16415. doi: 10.1021/ja045154j. [DOI] [PubMed] [Google Scholar]
- 44.Rachwal PA, Findlow IS, Werner JM, Brown T, Fox KR. Intramolecular DNA quadruplexes with different arrangements of short and long loops. Nucleic Acids Res. 2007;(12):4214–4222. doi: 10.1093/nar/gkm316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Risitano A, Fox KR. Influence of loop size on the stability of intramolecular DNA quadruplexes. Nucleic Acids Res. 2004;32:2598–2606. doi: 10.1093/nar/gkh598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cantor CR, Warshaw MM, Shapiro H. Oligonucleotide interactions. 3. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers. 1970;9:1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- 47.Mergny JL, Lacroix L. Analysis of thermal melting curves. Oligonucleotides. 2003;13:515–537. doi: 10.1089/154545703322860825. [DOI] [PubMed] [Google Scholar]
- 48.Mergny JL, Phan AT, Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 49.Puglisi JD, Tinoco I., Jr. Absorbance melting curves of RNA. Methods Enzymol. 1989;180:304–325. doi: 10.1016/0076-6879(89)80108-9. [DOI] [PubMed] [Google Scholar]
- 50.Hardin CC, Perry AG, White K. Thermodynamic and kinetic characterization of the dissociation and assembly of quadruplex nucleic acids. Biopolymers. 2000;56:147–194. doi: 10.1002/1097-0282(2000/2001)56:3<147::AID-BIP10011>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 51.Mergny JL, De Cian A, Ghelab A, Sacca B, Lacroix L. Kinetics of tetramolecular quadruplexes. Nucleic Acids Res. 2005;33:81–94. doi: 10.1093/nar/gki148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sacca B, Lacroix L, Mergny JL. The effect of chemical modifications on the thermal stability of different G-quadruplex-forming oligonucleotides. Nucleic Acids Res. 2005;33:1182–1192. doi: 10.1093/nar/gki257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rachwal PA, Brown T, Fox KR. Effect of G-tract length on the topology and stability of intramolecular DNA quadruplexes. Biochemistry. 2007;46:3036–3044. doi: 10.1021/bi062118j. [DOI] [PubMed] [Google Scholar]
- 54.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Balagurumoorthy P, Brahmachari SK. Structure and stability of human telomeric sequence. J. Biol. Chem. 1994;269:21858–21869. [PubMed] [Google Scholar]
- 56.Balagurumoorthy P, Brahmachari SK, Mohanty D, Bansal M, Sasisekharan V. Hairpin and parallel quartet structures for telomeric sequences. Nucleic Acids Res. 1992;20:4061–4067. doi: 10.1093/nar/20.15.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jin R, Gaffney BL, Wang C, Jones RA, Breslauer KJ. Thermodynamics and structure of a DNA tetraplex: A spectroscopic and calorimetric study of the tetramolecular complexes of d(TG3T) and d(TG3T2G3T) Proc. Natl. Acad. Sci. U.S.A. 1992;89:8832–8836. doi: 10.1073/pnas.89.18.8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rezler EM, Seenisamy J, Bashyam S, Kim MY, White E, Wilson WD, Hurley LH. Telomestatin and diseleno sapphyrin bind selectively to two different forms of the human telomeric G-quadruplex structure. J. Am. Chem. Soc. 2005;127:9439–9447. doi: 10.1021/ja0505088. [DOI] [PubMed] [Google Scholar]
- 59.Ambrus A, Chen D, Dai J, Jones RA, Yang D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 60.Phan AT, Kuryavyi V, Gaw HY, Patel DJ. Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter. Nat. Chem. Biol. 2005;1:167–173. doi: 10.1038/nchembio723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Phan AT, Kuryavyi V, Ma JB, Faure A, Andreola ML, Patel DJ. An interlocked dimeric parallel-stranded DNA quadruplex: A potent inhibitor of HIV-1 integrase. Proc. Natl. Acad. Sci. U.S.A. 2005;102:634–639. doi: 10.1073/pnas.0406278102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Phan AT, Modi YS, Patel DJ. Propeller-type parallel-stranded G-quadruplexes in the human c-myc promoter. J. Am. Chem. Soc. 2004;126:8710–8716. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Phan AT, Kuryavyi V, Burge S, Neidle S, Patel DJ. Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter. J. Am. Chem. Soc. 2007;129:4386–4392. doi: 10.1021/ja068739h. [DOI] [PMC free article] [PubMed] [Google Scholar]