Abstract
Insect sensory arrestins act to desensitize visual and olfactory signal transduction pathways, as evidenced by the phenotypic effects of mutations in the genes encoding both Arr1 and Arr2 in Drosophila melanogaster. To assess whether such arrestins play similar roles in other, more medically relevant dipterans, we examined the ability of Anopheles gambiae sensory arrestin homologues AgArr1 and AgArr2 to rescue phenotypes associated with an olfactory deficit observed in D. melanogaster arrestin mutants. Of these, only AgArr1 facilitated significant phenotypic rescue of the corresponding Drosophila arr mutant olfactory phenotype, consistent with the view that functional orthology is shared by these Arr1 homologues. These results represent the first step in the functional characterization of AgArr1, which is highly expressed in olfactory appendages of An. gambiae in which it is likely to play an essential role in olfactory signal transduction. In addition to providing insight into the common elements of the peripheral olfactory system of dipterans, this work validates the importance of AgArr1 as a potential target for novel anti-malaria strategies that focus on olfactory-based behaviors in An. gambiae.
Keywords: arrestin, desensitization, olfaction, Anopheles gambiae, Drosophila melanogaster
1. Introduction
Olfaction plays a significant role in mediating a variety of critical behaviors in insects (Hallem et al., 2006). This olfactory dependence is particularly relevant in host seeking and other behaviors of the mosquito Anopheles gambiae (An. gambiae), which is the principal Afro-tropical vector for human malaria (Zwiebel and Takken, 2004). In this mosquito, as in other insects, odorants first encounter the peripheral olfactory system through pores on sensory hairs, known as sensilla, which populate head appendages (the antennae, maxillary palps and proboscis). It is here that they contact the dendrites of olfactory receptor neurons (ORNs), and the components of signal transduction pathways translate chemical information from the environment into neuronal activity (Steinbrecht, 1996). Largely as a result of studies in the insect genetic model system Drosophila melanogaster, many of the key players presumed to be involved in odorant receptor (OR) activation and subsequent olfactory signal transduction have been identified. Indeed, a novel family of putative seven-transmembrane G-protein coupled receptors (GPCRs) have been identified in D. melanogaster (Clyne et al., 1999; Gao and Chess, 1999; Vosshall et al., 1999) and An. gambiae (Hill et al., 2002; Robertson et al., 2003); many of these proteins have been subsequently shown to function as bona fide ORs (Dobritsa et al., 2003; Hallem et al., 2004b; Lu et al., 2007).
Several components downstream of odorant-activated OR signaling pathways have been implicated as playing a role in Drosophila olfactory signal transduction. These include genes encoding G-protein (Kalidas and Smith, 2002), phospholipase C (Riesgo-Escovar et al., 1995), phosphatidylinositol transfer protein (Riesgo-Escovar et al., 1994), cyclic AMP (cAMP) phosphodiesterase (Gomez-Diaz et al., 2004), cyclic nucleotide and voltage-gated ion channels (Dubin et al., 1998), and, from our own work, sensory arrestins (Merrill et al., 2002; Merrill et al., 2005). While the roles of these downstream elements have not been fully elucidated, they are all consistent with the overall paradigm of GPCR-mediated signal transduction. There are, however, emerging indications that insect olfactory transduction may not embrace canonical GPCR signaling (Benton et al., 2006), suggesting that a comprehensive model for olfactory signal transduction in D. melanogaster and other insects still remains undefined.
As crucial as receptor activation and primary signal transduction are to the facilitation of detection of olfactory cues, integration of the signal in space and time is reliant on the appropriate termination of the transduction cascade, a process known as deactivation. Homologous GPCR desensitization, which ultimately results in reduced receptor responses, was originally studied in vertebrate systems and specifically occurs subsequent to receptor activation, involving the rapid uncoupling of a receptor from its partner G-protein (Krupnick and Benovic, 1998). Under this paradigm, ligand binding is followed by activation-dependent receptor phosphorylation (Wilden and Kuhn, 1982; Kuhn et al., 1984). While this step slightly diminishes signaling, subsequent binding of arrestin proteins are necessary for full GPCR deactivation (Kuhn and Wilden, 1987). Arrestins, which display a high specificity toward the activated, phosphorylated form of the receptor (Gurevich and Gurevich, 2004), mediate this process by functionally competing with G-proteins for binding sites within the receptor to prevent further signal transduction (Ferguson and Caron, 1998). The first arrestins to be identified were isolated from the vertebrate visual systems (Wilden et al., 1986) and have accordingly been characterized as visual arrestins.
A second class of vertebrate arrestins encompass the non-visual subtypes (Krupnick and Benovic, 1998) and are known as β-arrestins because they were first described according to their role in the desensitization of β-adrenergic receptors (Lohse et al., 1990; Attramadal et al., 1992). They have since been shown also to regulate vertebrate olfactory signal transduction cascades (Dawson et al., 1993; Mashukova et al., 2006). Since that time, a plethora of these and other studies have led to the well-accepted paradigm whereby arrestin proteins facilitate a diversity of processes pertaining to GPCR-mediated signal transduction. These include desensitization processes linked to endocytic pathways that function in receptor internalization as well as recycling and degradation (Prossnitz, 2004). Moreover, the β-arrestins have been shown to mediate other cellular processes via interactions with secondary signal transduction cascades by means of recruitment and activation of MAP kinase and other effector proteins (Lefkowitz and Shenoy, 2005; Gurevich and Gurevich, 2006).
Arrestins have also been identified and characterized in several other insect species, most notably the fruit fly D. melanogaster, in which two visual arrestins, DmArr1 and DmArr2, were originally identified based on sequence homology (Smith et al., 1990; Hyde et al., 1990; LeVine et al., 1990) and function (Dolph et al., 1993) to vertebrate visual arrestins. Not surprisingly, these arrestins have also been shown to play a role in the internalization of rhodopsin in Drosophila (Satoh and Ready, 2005; Orem et al., 2006). DmArr1 and DmArr2 are also expressed in D. melanogaster olfactory tissues, leading to their reclassification as sensory arrestins (Merrill et al., 2002).
More recently, a comprehensive evaluation of the in vivo physiological and behavioral roles of D. melanogaster sensory arrestins in olfactory function via mutant analysis revealed decreased responsiveness to a diverse panel of odorants in both single and double arr11 and arr25 mutants in a concentration- and odorant-specific manner (Merrill et al., 2005). Moreover, arr11 mutant phenotypes were definitively linked to the arr1 locus by means of functional rescue via expression of a wild-type DmArr1 transgene in arr11 mutant backgrounds. The results of this work firmly established an odorant-dependent role for sensory arrestins as they act in peripheral olfactory signal transduction in this system.
These findings led us to extend our analysis of olfactory arrestins to economically and medically relevant insects—the malaria vector mosquito An. gambiae. In an initial examination of An. gambiae arrestins, we characterized AgArr1, which is homologous to DmArr1 and not surprisingly similarly expressed in both photoreceptors and olfactory tissues (Merrill et al., 2002). In a subsequent study, three additional An. gambiae arrestins were characterized. AgArr2 is highly homologous to DmArr2 and, likewise, is expressed in multiple sensory systems. AgArr3 is homologous to the non-visual Kurtz arrestin gene in Drosophila (DmKrz) (Roman et al., 2000) and is similarly ubiquitously expressed, while AgArr4, which is also widely expressed, belongs to a divergent arrestin class and has an unknown function (Merrill et al., 2003). Accordingly, AgArr1 and AgArr2 have been preliminarily characterized as insect sensory arrestins, whereas AgArr3 and AgArr4 represent non-sensory and atypical insect arrestins, respectively.
As the first step in the direct functional characterization of sensory arrestins, we have examined the hypothesis that AgArr genes act as bona fide sensory arrestins and true DmArr1 and DmArr2 orthologs by asking whether transgenic expression of AgArr1 or AgArr2 in Drosophila arr mutant ORNs can functionally rescue the olfactory phenotypes associated with each mutant background. The results reported here demonstrate that while AgArr1 can functionally replace DmArr1 by restoring olfactory physiology to wild-type levels, there is a general lack of functional complementarity between putative interspecific Arr2 orthologs as well as all Arr1 and Arr2 paralogs. This, therefore, is a de facto validation of the hypothesis that AgArr1 is able to function as a true olfactory arrestin, thereby leading to the logical conclusion that it serves similar roles in An. gambiae ORNs. Thus, further exploration of the role of AgArr1 and other sensory arrestins may be carried out in An. gambiae to provide insight into the mechanisms controlling olfactory perception in this important disease-vector insect. This will likely foster more informed approaches toward devising olfactory-based, vector-control strategies that may be utilized to reduce the transmission of malaria and other vector-borne diseases.
2. Experimental Procedures
2.1. Flies and cultures
The wild-type flies used as controls in this experiment were Oregon R, obtained from Dr. C. Desai (Vanderbilt University). Hypomorphic arr11 cn mutant flies were kindly provided by Dr. P. Dolph (Dartmouth College) and have been well described (Dolph et al., 1993). Flies were grown in Erlenmeyer flask-shaped plastic bottles on a standard cornmeal, molasses, agar and sugar medium with yeast. All flies were cultured at 25°C, on a 12-h:12-h light:dark cycle.
2.2. Odorants
Five odorants that have been shown to elicit diminished responses in arr11 cn mutant flies (Merrill et al., 2005) were used for the physiological assays: 1-butanol, heptanoic acid and octyl acetate (Aldrich, Milwaukee, WI), and 2-heptanone and 1-octanol (Sigma, St. Louis, Missouri). Odorants were diluted in paraffin oil (Sigma, St. Louis, Missouri) to the concentrations indicated in figure legends.
2.3. Electroantennogram physiology
The electroantennogram (EAG) responses of wild-type and arrestin mutant flies were recorded in a similar manner to those in previous studies (Alcorta, 1991; Riesgo-Escovar et al., 1994; Merrill et al., 2005). Briefly, 2- to 7-day-old adult female Drosophila were immobilized in the narrow end of a pipette tip, such that only the anterior portion of the head protruded. Flies were placed 5 mm from the tip of a constant stream of clean humidified air provided by a stimulus control device (CS-05, Syntech, The Netherlands). Odorants were delivered by passing 0.5-s pulses of air through a glass Pasteur pipette containing a 1.5-cm-diameter filter disk (VWR International, West Chester, Pennsylvania) saturated with 20 μl diluted odorant into the constant air stream. The stimulus control device delivered a continuous flow rate of ∼340 cc/min and a pulse rate of ∼350 cc/min. The difference in air velocity between the continuous air stream and stimulus pulse did not produce a significant EAG response, as evidenced by the fact that odor-free (oil-alone) responses are consistently low and reflect only minor background noise. Glass microelectrodes filled with 0.1 M KCl transmitted electrical responses to odorant stimulation via silver-chloride wires to a signal acquisition system (IDAC232, Syntech, The Netherlands). Data were collected at 25 Hz, amplified 10X and converted from analog to digital, then displayed on a Gateway PC computer. EAG analysis was performed using EAG2000 software (Syntech, The Netherlands). The reference electrode was inserted into the back of the head while the recording electrode was placed on the anterior dorso-medial surface of the 3rd antennal segment to establish electrical contact. The amplitude of response, which represents the peak voltage deflection in response to odorant presentation, was recorded for analysis.
2.4. D. melanogaster germline transformation and generation of transgenic rescue lines
All techniques were performed as previously described (Merrill et al., 2005). Strains of D. melanogaster with either mutant DmArr1 (arr11 cn) or DmArr2 (arr25 veh) background were used as parentals to generate the appropriate rescue lines. The Gal4-UAS yeast transcriptional system, originally described by Brand and Perrimon (Brand and Perrimon, 1993), was utilized to drive expression of the AgArr1, AgArr2 and DmArr2 transgenes in the DmArr1 mutant (arr11 cn ) background; expression of the AgArr1, AgArr2, DmArr1 and DmArr2 transgenes was also driven in the DmArr2 mutant (arr25 veh) background. The arr11 and arr25 mutations were crossed into the Gal4-C155 (Lin and Goodman, 1994) line, in which the yeast Gal4 transcription factor is expressed under control of the pan-neuronal elav promoter (Robinow and White, 1988): w, elavGal4c155; arr11cn/arr11cn, henceforth denoted as C155/11, or w, elavGal4c155; +; arr25 ve h. For these experiments, a variety of UAS responder lines were used, in which the specific transgenes were positioned downstream of five UAS Gal4 binding sites. The arr11 cn and arr25 mutations were crossed into the AgArr1, AgArr2, DmArr1 and DmArr2 transgene backgrounds. Rescue lines to be examined were generated by crossing the Gal4 driver line in the arr11 mutant background with the responder lines containing the AgArr1, AgArr2 and DmArr1 transgenes, also in the arr11 mutant background. Similarly, the Gal4/arr25 line was crossed with responder lines containing the AgArr1, AgArr2, DmArr1 and DmArr2 transgenes, which were also in the arr25 mutant background. A specific genotypic description of the UAS-AgArr1 line used in this report is as follows: w/w; arr11 cn/arr11cn; p[UAS-AgArr11A]/p[UAS-AgArr11A], henceforth called 11/1A. Crossing virgin female w, elavGal4c155; arr11cn with male w; arr11cn; p[UAS-AgArr11A] generated progeny that expressed Gal4 and AgArr1 neuronally in the arr11cn background, henceforth C155/11/1A.
2.5. In situ hybridization
Fluorescence in situ hybridization was performed using a modified version of previously reported methodology (Vosshall et al., 1999; Vosshall et al., 2000). Briefly, digoxygenin (DIG)-labeled AgArr1 and DmArr1 and fluorescein (FITC)-labeled DmOR83b sense and antisense riboprobes were generated using standard kit reagents and protocol (Roche Applied Sciences, Indianapolis, Indiana). For each experiment, antennae from approximately 25 4- to 8-day-old female flies were dissected directly into Tissue-Tek O.C.T. compound-embedding medium (Sakura Finetek U.S.A, Torrence, California). Cryo-sections of 15 μm were generated and applied to Superfrost Plus VWR Microslides (VWR International, West Chester, Pennsylvania), then allowed to air dry for 3 h. A 10-min fixation in 4% paraformaldehyde [4% paraformaldehyde/1X phosphate buffer saline (PBS)] was followed by three 5-min washes with 1X PBS, a 10-min acetylation application and then three additional 5-min washes with 1X PBS (all washes performed at room temperature). Pre-hybridization and hybridization steps were carried out using hybridization solution as follows: 50% formamide, 5X SSC, 5X Denhardt's solution, 250 μg/ml salmon sperm DNA, 50 μg/ml heparin, 2.5 mM EDTA, 0.1% Tween-20. Pre-hybridization was carried out for 2 h at 55°C, and hybridization for 21 h at 55°C.
Subsequently, one 10-min 5X SSC wash (55°C), three 20-min 0.2X SSC washes (55°C) and one 10-min 1X PBS-tw wash (1X PBS, 0.1% Tween) were sequentially carried out prior to blocking and antibody labeling. Blocking was carried out for 2 h at room temperature with B2 sheep solution [10% Normal Sheep Solution (Jackson ImmunoResearch, West Grove, Pennsylvania), 1X PBS-tw]. For antibody labeling, sheep anti-DIG-POD (1:200) and sheep anti-FITC-AP (1:500) (both Roche Applied Sciences, Indianapolis, Indiana) were diluted in B2 sheep solution, and applied to slides for 21 h at 4°C.
Five 5-min 1X PBS-tw washes were performed at room temperature. For visualization of DmArr1 and AgARR1 label, diluted tyramide-FITC reagent (Perkin Elmer, Waltham, Massachusetts) was applied for a 10-min incubation at room temperature. For visualization of DmOR83b, two 10-min equilibration washes with 0.1 M Tris-HCl (pH 8.2) were followed by a 30-min incubation with applied Fast Red tablet (Roche Applied Sciences, Indianapolis, Indiana) dissolved in 2 ml 0.1 M Tris-HCl (pH 8.2) and three subsequent 5-min 1X PBS-tw washes. Nuclear staining was performed with TOTO-3 (Molecular Probes, Eugene Oregon) reagent diluted in 1X PBS-tw (1:1200), applied for 15 min at room temperature, followed by three 5-min washes with 1X PBS-tw. Sections were mounted with Vectashield reagent (Vector Laboratories, Burlingame, California) viewed on a Zeiss Inverted LSM510 Confocal Microscope (Carl Zeiss Microimaging, Inc., Thornwood, New York) and analyzed with Zeiss LSM Image Browser Software (Carl Zeiss Microimaging, Inc., Thornwood, New York).
2.6. RNA extractions
Antennae were dissected by hand and placed into 1.5-ml eppendorf tubes on dry ice. For each genotype, 120−150 antennae from female flies were dissected for RNA preparation. Total RNA was prepared using RNeasy Mini reagents, as described in the supplier's protocol (Qiagen Inc, Valencia, California), and then treated with DNase according to the DNA Free protocol (Ambion, Austin, Texas) to eliminate potential genomic DNA contamination. The total RNA preparation was then used for oligo-dT cDNA first-strand synthesis using Superscript III Reverse Transcriptase (Gibco/Invitrogen, Carlsbad, California), according to the manufacturer's instruction. For each genotype, 500 ng total RNA was used for each reverse transcription reaction. For each genotype, three independent RNA extractions were conducted, in parallel, for subsequent use in comparison of relative expression levels (see 2.8. and 3.3.).
2.7. Primer design
Real-time PCR primers were designed to span exon–intron boundaries, where appropriate, and were obtained from Integrated DNA Technologies (IDT, Coralville, Iowa). In all cases, the following oligonucleotide primers were selected with an ideal annealing temperature of 60°C and optimized to minimize hindrance on account of hairpin formation and primer dimerization:.ribosomal protein rp49 (forward) GTCGCACAAATGGCGCAAGC and rp49 (reverse) TGCTGCCCACCGGATTCAAG produce a 133-bp cDNA product; DmArr1 (forward) ACTCAGGTGGAACCCATTGATGGAA and (reverse) TTTCCGATATGGGCGCGAGG yield a 109-bp product; AgArr1 (forward) CGATCGATGGTATCGTCGTCCTC and (reverse) CGAAGAGGACGAGGTGATGGG yield a 112-bp product.
2.8. Quantitative real-time PCR
For these experiments, an ABI 7300 Real Time PCR Instrument was used (Applied Biosystems, Foster City, California) together with the Qiagen QuantiTect SYBR Green PCR kit (Qiagen Inc, Valencia, California). For each reaction, 11.5 μl of 1X master-mix, containing 0.5 μM forward and reverse primers diluted with RNase Free water, was added to individual wells of a 96-well plate to which 1 μl cDNA was added as a template for the reaction. The ABI 7300 experimental protocol used was—activation (50°C, 2 min; 95°C, 10 min), amplification (95°C, 15 s; 60°C, 1 min) repeated 40 times, and a melting curve (60−95°C at 1°C/s). For each genotype and primer set, reactions were run in triplicate, and average fluorescence Ct values were obtained. Amplification efficiencies were calculated as described previously (Bohbot and Vogt, 2005), and relative gene expression ratios were determined using the Pfaffl method of analysis (Pfaffl, 2001).
3. Results
The objective of the studies reported here was to validate a functional role for the An. gambiae sensory arrestins AgArr1 and AgArr2 in peripheral olfactory processes in an insect system. In order to address this, and in the absence of appropriate genetic tools (e.g., mutants and a robust system for germline transformation) in An gambiae, we have taken advantage of previously characterized olfactory phenotypes in D. melanogaster arr11 and arr25 mutants, of which arr11 mutants can be functionally rescued via transgene expression of the endogenous DmArr1 arrestin-encoding gene (Merrill et al., 2005). We tested the hypothesis that the D. melanogaster arr mutant phenotypes could also be rescued by AgArr1 and AgArr2 transgenes, which would demonstrate true functional orthology for these genes, thereby providing support for a similar role for these sensory arrestins in olfactory signaling in An. gambiae. Amino acid sequence analysis supports this hypothesis, because AgArr1 has been shown to be 67% identical to DmArr1 (Merrill et al., 2002) and AgArr2 74% identical to DmArr2 (Merrill et al., 2003) (Fig. 1). It is worth noting here that the sensory arrestin family of proteins, consisting of DmArr1, AgArr1, DmArr2 and AgArr2, share two conserved elements—four of five charged polar core residues (Hirsch et al., 1999) and two amino-terminal lysine residues (Vishnivetskiy et al., 2000)—both of which are demonstratively essential in receptor-phosphorylation-state recognition (Gurevich and Gurevich, 2004). Of the two putative Src Homology 3 (SH3) domains (Boxes, Fig. 1), the presence of which suggests potential interactions with downstream MAP kinase signaling pathways (Luttrell et al., 1999), only one is conserved among all four insect sensory arrestins.. Moreover, a putative C-terminal AP2 interaction domain, which has been shown to facilitate interactions between arrestin proteins and cellular endocytic machinery (Laporte et al., 2000), is conserved among the Arr2 proteins but absent from the Arr1 proteins, which overall are 46% and 53% identical, respectively, in D. melanogaster and An. gambiae.
Figure 1. Sensory arrestin protein sequence alignment.
D. melanogaster Arr1 (DmArr1) protein sequence (GenBank accession no. NP 476681), An. gambiae Arr1 (AgArr1) protein sequence (GenBank accession no. AAG54081), D. melanogaster Arr2 (DmArr2) protein sequence (GenBank accession no. NP 523976), An. gambiae Arr2 (AgArr2) protein sequence (GenBank accession no. DAA00888). Shaded boxes indicate conserved phosphorylation detection residues (Vishnivetskiy et al., 2000; Gurevich and Gurevich, 2004). Open boxes indicate conserved putative SH3, PxxP recognition domains (Luttrell et al., 1999). Underlined residues are putative AP2 interaction domains (Laporte et al., 2000), which while conserved in arr2 proteins are lacking in the Arr1 orthologs.
Recent studies demonstrating the functionality of AgORs within the context of the olfactory system of D. melanogaster (Hallem et al., 2004a; Jones et al., 2005) lend credence to the notion of compatibility between the olfactory systems of An. gambiae and D. melanogaster. Indeed, successful rescue of function in these studies provides strong support for the experimental paradigm that is tested here, i.e., whether the mosquito sensory arrestins, AgArr1 and/or AgArr2, are capable of functionally restoring peripheral olfactory responses in the context of arrestin deficits in D. melanogaster. Validation of function in this regard would lend strong support to the view that these AgArrs are likely to function in the same manner in mosquito olfactory pathways. This result would foster the rationale that in vivo targeting of AgArr1 or AgArr2 function in An. gambiae should result in olfactory deficits in the mosquito, and would be expected to impact its vectorial capacity.
3.1 Electrophysiology data
As in previous studies (Merrill et al., 2005), we utilized the bipartite Gal4/UAS system (Brand and Perrimon, 1993) to drive expression of a wild-type copy of the AgArr1 gene across all ORNs in arr11 mutant D. melanogaster. This was accomplished by crossing fly lines expressing the yeast Gal4 transcription factor under regulatory control of the pan-neuronal elav promoter (Campos et al., 1987; Robinow and White, 1988) with lines containing a UAS-AgArr1 transgene in an arr11 mutant background. Olfactory signaling was then assayed within the context of EAG analysis of Oregon R wild-type and rescue flies, as well as the parental mutant lines which, on their own, should not express the AgArr1 transgene. In these assays, we have examined olfactory responses to several representative class-I and class-II odorants previously observed to elicit diminished responses in arr11 mutant flies, and for which rescue of function was observed when tested in the DmArr1 transgenic flies (Merrill et al., 2005).
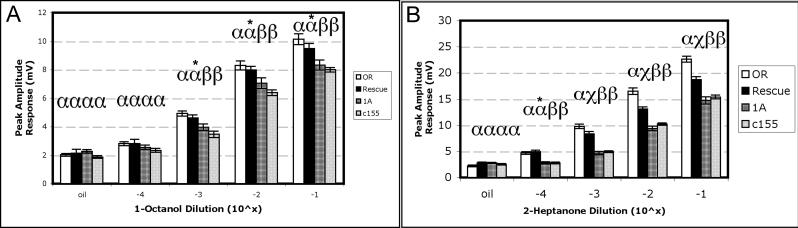
In these studies, class-I odorants, which include 1-octanol and 2-heptanone, elicited diminished responses in arrestin mutant fruit flies at all concentrations examined (Fig. 2). In the case of 1-octanol (Fig. 2A), rescue of function was apparent for C155/11/1A flies at the three highest concentrations (10−1 through 10−3 dilutions) when compared with wild-type flies, while both parental Gal4 and UAS arr11 mutant lines responded at significantly lower amplitudes, consistent with our previous studies (Merrill et al., 2005). At the lowest concentration examined (10−4 dilution), wild-type and rescue responses were statistically indistinguishable from mutant responses, and all responses at this dose were no different from those to the paraffin oil solvent. For 2-heptanone (Fig. 2B), wild-type responses at all concentrations were significantly higher than those of the mutant counterparts, and rescue flies responded at an intermediate level relative to wild-type and mutant flies at the three highest concentrations, indicative of a partial rescue. Only at the lowest concentration (10−4 dilution) were responses observed to be statistically indistinguishable from wild-type responses.
Figure 2. AgArr1-mediated rescue of function observed for class-I odorants.
A) Restoration of olfactory sensitivity to 1-octanol. B) For the odorant 2-heptanone, at all but the lowest concentration, partial rescue of function is observed wherein olfactory responses are intermediary to those of wild type and parental Gal4 and UAS mutant strains. Genotypes: OR: wild-type Oregon R, c155: w, elavGal4c155; arr11, 1A: w; arr11; UAS-AgArr11A, Rescue: w, elavGal4c155; arr11; UAS-AgArr11A/+. α, β, and χ represent statistically distinct groups; where differences exist, p < 0.05. For all genotypes, n= 10−15 flies. One-way ANOVA was used for statistical analyses.
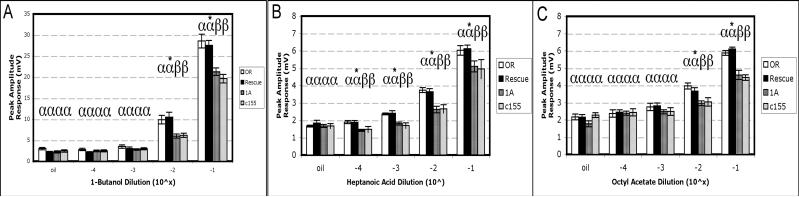
Class-II odorants, which include 1-butanol, heptanoic acid and octyl acetate, generally elicit diminished EAG responses in arrestin mutant flies only at the highest odorant concentrations (Merrill et al., 2005). For 1-butanol (Fig. 3A), olfactory sensitivity in AgArr1 transgenic rescue flies was restored to wild-type levels at the two highest concentrations examined, while parental Gal4 and UAS lines consistently responded at significantly lower levels. At the two lower concentrations, olfactory responses were indistinguishable from the oil solvent and across all genotypes. For heptanoic acid (Fig. 3B), AgArr1 rescue flies’ responses were insignificantly different from wild-type flies at all concentrations tested. For octyl acetate (Fig. 3C), rescue flies expressing AgArr1 responded at wild-type levels at the two highest concentrations examined, whereas responses at the two lowest concentrations were statistically indistinguishable across all fly lines as well as from responses to the paraffin oil solvent. Together, these data provide strong support of AgArr1's capacity to nearly completely rescue the arr11 mutant olfactory phenotype when transgenically expressed in D. melanogaster arr11 background.
Figure 3. AgArr1-mediated rescue of function observed for class-II odorants.
A) Restoration of olfactory sensitivity to 1-butanol. B) Heptanoic acid: at all concentrations, olfactory sensitivity is restored for rescue flies. C) For the odorant octyl acetate, full rescue of function is observed at the two highest concentrations. Labels and genotypes are as presented in Fig. 2. For all genotypes, n= 10−15 flies. One-way ANOVA was used for statistical analyses.
To further explore this phenomenon, we considered the possibility that, given the use of the Elavc155-Gal4 promoter, transgenic expression levels of AgArr1 may not precisely mimic expression levels of endogenous DmArr1 in wild-type flies. To that end and to assess the phenotypic effects of alterations in DmArr1 levels, EAG response levels were examined in arr11cn heterozygous flies. Thus, Oregon R virgin females were crossed with w; arr11cn mutant males, and responses were recorded for 2-heptanone and other odorants (described above). In all cases, wild-type response levels that were indistinguishable from those of Oregon R flies were observed (data not shown). In consideration of this, we suggest that a threshold be established wherein the heterozygous condition (i.e., approximately 50% of wild-type DmArr1 expression levels) sufficiently confers wild-type levels of olfactory sensitivity to the fly. This model however does not address the possibility that over-expression of AgArr1 may in fact be selectively responsible for the phenotype observed in response to olfactory stimulation with 2-heptanone.
Previously, we utilized a simple larval mobility assay to examine the ability of DmArr1 transgenes to functionally rescue defects in olfactory-mediated behavior in Drosophila arr11 mutant's responses to 1-octanol (Merrill et al., 2005). In a similar manner, we used these behavioral paradigms to examine the ability of transgenic AgARR1 to rescue arr11-linked larval olfactory deficits when presented with 1-octanol and several other odorants toward which arr11 mutants displayed reduced attraction relative to that of wild-type counterparts (Merrill et al., 2005). Behavioral assays across multiple AgArr1 transgenic lines were carried out to test larval responsiveness to multiple odorant concentrations, and in all cases a statistically significant level of functional rescue was not observed (data not shown).
In order to fully understand the functional relationships between An. gambiae and D. melanogaster sensory arrestins, we examined the ability of AgArr2 and DmArr2 transgenes to rescue the olfactory-deficit phenotype associated with the arr25 mutant allele in D. melanogaster (Merrill et al., 2005). Furthermore, similar efforts were undertaken to rescue the arr11 mutant phenotype with DmArr2 and AgArr2 transgenes. In all of these scenarios, involving multiple independent lines of inserts for each transgene, restoration of olfactory sensitivity to wild-type levels was never observed (data not shown).
3.2. Localization of transgenic AgArr1 in the antennae of rescue flies
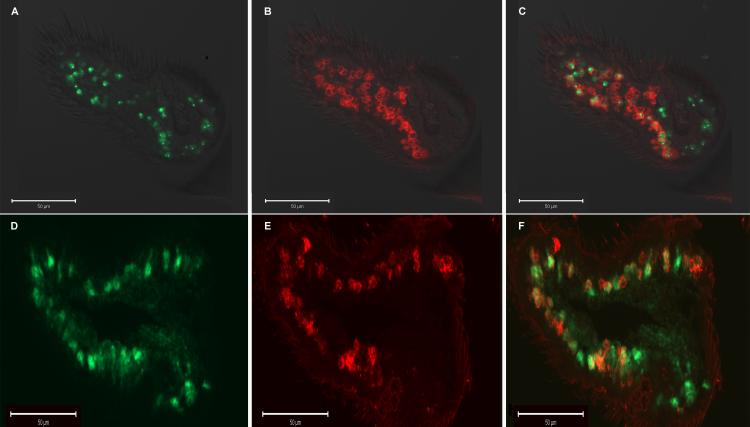
In order to confirm the neuronal expression of the AgArr1 transgene in ORNs as an underlying basis for transgenic rescue, we applied the in situ hybridization (Fig. 4) technique in an examination of the Gal4/UAS rescue flies. In these studies, the broadly expressed co-receptor, Or83b, which plays an obligatory role in olfactory sensory physiology, acts as a de facto ORN marker for the majority (70−80%) of these cell types (Larsson et al., 2004). For these experiments, co-expression of AgArr1 and Or83b is observed in a majority of Or83b-positive olfactory sensory neurons, indicative of the presence of transgenic AgArr1 in the ORNs (Fig. 4). Furthermore, these results are comparable to expression patterns of transgenic DmArr1, being driven by the same elav-Gal4 construct, for which we have previously demonstrated rescue of function in the arr11 mutant background (Merrill et al., 2005) (Figs. 4D-4F). In situ hybridization control studies were also carried out in parental UAS-AgArr1 and UAS-DmArr1 lines, in which transgenic AgArr1 and DmArr1 expression is precluded, respectively. In both these instances, expression of transgenic AgArr1 or DmArr1 was undetectable in these flies (data not shown).
Figure 4. Transgenic AgArr1 is expressed in or83b positive olfactory sensory neurons.
A) Fluorescence image of antisense DIG-labeled AgArr1 probe in arr11 mutant antennae. B) Fluorescence image of antisense FITC-labeled Or83b probe. C) Merge of A and B, indicating co-expression of Or83b and AgArr1 in most Or83b positive olfactory sensory neurons. D) Fluorescence image of antisense DIG-labeled DmArr1 probe in Gal4/UAS-DmArr1 flies. E) Fluorescence image of antisense FITC-labeled OR83b probe. F) Merge of D and E, indicating co-expression of OR83b and transgenic DmArr1.
3.3. Real-time PCR analysis of AgArr1 mRNA levels in transgenic rescue animals
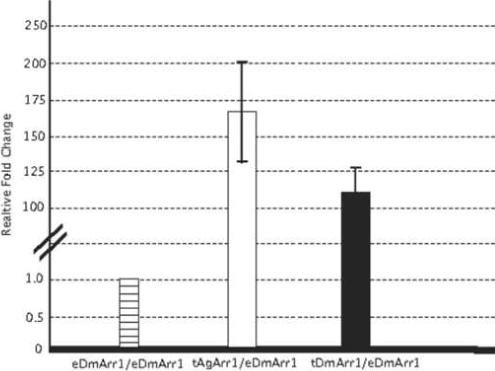
In order to further validate transgene expression levels, we employed quantitative real-time PCR to assess antennal AgArr1 expression levels relative to both endogenous and transgenic DmArr1 in the antennae of Gal4/UAS-DmArr1 rescue flies. In these studies, in which expression levels of the ribosomal protein gene rp49 were examined to serve as a calibrator for total RNA levels (Fig. 5), both DmArr1 and AgArr1 transgenes were expressed at markedly higher levels than the endogenous DmArr1 gene in wild-type Oregon R flies. Furthermore, we observed only an overall modest difference in levels of AgArr1 transgene expression (1.5-fold increase) relative to similarly expressed DmArr1 transgenes in arr11 mutant backgrounds, which is consistent with the similar patterns of phenotypic rescue in these lines. In all cases, endogenous levels of DmArr1 were undetectable in the arr11 mutant background (data not shown), consistent with its characterization as a hypomorphic arrestin allele (Smith et al., 1990).
Figure 5. Expression of transgenic arrestin is quantitatively greater than that of endogenous wild-type arrestin.
Relative expression levels of transgenic AgArr1 (tAgArr1) and transgenic DmArr1 (tAgArr1) in Gal4/UAS rescue flies, normalized to endogenous rp49 expression levels, compared with expression levels of endogenous DmArr1 (eDmArr1) in wild-type Oregon R flies. Error bars are indicative of standard error, calculated from variability of relative expression levels for three independent experiments.
4. Discussion
In light of our observations that both DmArr1/DmArr2 and AgArr1/AgArr2 are expressed in the antennae of D. melanogaster and An. gambiae, respectively (Merrill et al., 2002), where they may be reasonably assumed to be active in olfactory processes, we initially focused on a broad examination of the potential combinations for transgenic rescue of olfactory arr mutant phenotypes. Accordingly, DmArr1, AgArr1, DmArr2 and AgArr2 transgenes were assayed for the ability to restore olfactory sensitivity in arr11 and arr25 mutant fruit flies under the control of the pan-neuronal elav-Gal4 (Lin and Goodman, 1994) driver line. This driver was chosen rather than other ORN-biased Gal4 lines, such as those based on DOR83b promoters, based on its ability to act over the complete spectrum and not just a simple majority of ORNs. In electrophysiological assays, both DmArr1 and AgArr1 transgenes were able to robustly rescue Drosophila arr11 mutations insofar as peripheral olfactory signal transduction, while none of the Drosophila or Anopheles transgenes was able to restore olfactory sensitivity associated with arr25 mutations. In addition, DmArr2/AgArr2 transgenes failed to rescue the arr11 olfactory deficit phenotype, consistent with the hypothesis that Arr1 and Arr2 play distinct and non-complementary roles in olfactory signal transduction. Furthermore, in contrast to electrophysiological studies, AgArr1 transgenes failed to functionally rescue larval behavioral deficits linked to the arr11 mutation.
Although intriguing, the ability to restore peripheral olfactory sensitivity while failing to rescue behavioral phenotypes may, in part, reflect uncharacterized aspects of transgenic expression as well as the complex nature of the behavioral output being considered. In the first assay, the electrophysiological responses to odorants recorded in the antennae in the form of EAGs were specifically linked to signal transduction in the antennal ORNs that respond to the odorants (Alcorta, 1991). While utilization of the pan-neuronal elav-Gal4 driver to mediate expression of transgenic arrestins in non-ORN neurons should not impact the electrophysiological responses of the ORNs to the odorants, it is reasonable to speculate that ectopic over-expression of transgenic AgArr1 in all the peripheral as well as central processing neurons of the Drosophila larvae may have confounding effects on the olfactory behavioral output. Thus, a lack of behavioral rescue in arr1 mutant larvae must not necessarily be misconstrued to have a bearing on the impact of physiological rescue of peripheral ORN olfactory signal transduction in arr1 mutant adult flies.
While both DmArr1/AgArr1 and DmArr2/AgArr2 protein pairs display significant conservation relative to each other, there is considerable divergence at the carboxy terminus between Arr1 and Arr2 (Fig. 1). In light of the inability of cross-arrestin complementarity, this likely reflects significant divergence in the functional roles of these sensory arrestins. Indeed, recent reports have suggested a similar divergence of function of Arr1 and Arr2 within the context of the Drosophila visual system (Satoh and Ready, 2005; Orem et al., 2006). Elucidation of the relative expression patterns of endogenous DmArr1 and DmArr2 over antennal ORNs in D. melanogaster may shed further light on the specific roles that these two arrestin sub-types play in olfactory sensation.
Nevertheless, the results of this study confirm one of the central elements of our initial hypothesis in that we demonstrate that AgArr1 is a functional ortholog to DmArr1. Rescue of function of the physiological olfactory deficit phenotype observed in arr11cn mutant flies was observed for four of five odorants examined, the lone exception being 2-heptanone, for which a partial rescue was observed for high-end concentrations. In this instance, intermediate EAG amplitude response levels between the responses of the wild-type Oregon R and parental arrestin mutant flies were observed. This is especially pertinent, because it relates to our previously reported physiological rescue of function of olfactory phenotypes in arr11 mutant flies with DmArr1 transgene, where a complete rescue was observed in response to 2-heptanone at all concentrations examined (Merrill et al., 2005).
AgArr1 transgene localization studies suggest that there is not a strict one-to-one correspondence between the expression of the endogenous DmOR83b non-conventional OR and the transgenic AgArr1 protein. We interpret this in light of the observation that not all antennal neurons display the Or83b marker (Larsson et al., 2004), such that it is expected that AgArr1 under the control of the pan-neuronal elav promoter should be present in a number of cells lacking the Or83b marker. While this is precisely what was observed, we also noted the presence of a subpopulation of ORNs (15−20%) that display the Or83b marker and paradoxically lack the transgenic AgArr1 message. Without postulating as to the mechanism underlying this, it is reasonable to suggest that this lack of AgArr1 expression in OR83b-positive ORNs may also contribute to the partial rescue observed in response to the odorant 2-heptanone. We additionally examined mRNA expression levels of the AgArr1 and DmArr1 transgenes in relation to endogenous wild-type DmArr1 and observed a significant over-expression of the mRNA expression levels of each transgene relative to the endogenous wild-type DmArr1 transcript. This is consistent with numerous studies utilizing elav-Gal4 driver lines and apparently does not cause a phenotype in and of itself, as olfactory sensitivity is restored to wild-type levels when these transgenes are expressed in the arr11 mutant background. However, only a modest difference between expression levels of the AgArr1 and DmArr1 transgenes was observed. With all this in mind, we favor the view that a variety of factors, including differential mRNA or protein stability leading to subtle alterations in expression levels and/or spatial patterns, account for the data, rather than just a true functional divergence between DmArr1 and AgArr1. Such differences would reasonably be expected to be reflected in a broader contrast in phenotypic rescue, rather than a partial rescue in response to one specific odorant.
In conclusion, this study supports the view of both a broad requirement for sensory arrestins and a specific functional role for AgArr1 in insect olfaction. That said, a mechanistic understanding of the precise role of sensory arrestins within the context of insect olfactory signal transduction still remains unclear. Inasmuch as several studies (Dolph et al., 1993; Satoh and Ready, 2005; Orem et al., 2006) provide evidence that arrestins facilitate desensitization and receptor internalization in the Drosophila visual system, it has been assumed that Dm/AgArr1 and other sensory arrestins function in a similar manner within GPCR-based olfactory signal transduction paradigms in insect ORNs. Interestingly, it has recently been suggested that because Drosophila ORs manifest an inverted membrane topology when compared with conventional GPCRs (Benton et al., 2006), they may not act as canonical GPCRs in olfactory signal transduction. Therefore, insect sensory arrestins may affect peripheral olfactory sensitivity via cryptic interactions with other GPCRs or indeed other signaling pathways in ORNs.
Conversely, as the vertebrate β-arrestins are known to interact with classes of receptors unrelated to GPCRs (Lefkowitz et al., 2006), it may indeed be the case that the insect sensory arrestins play an essential role in the regulation of the functionality of insect ORs, irrespective of whether they are bona fide GPCRs. This study opens the possibility that the goal of disrupting proper olfactory signaling necessary for host-seeking and other behaviors underlying the vectorial capacity of An. gambiae may indeed be accomplished through targeting AgArr1 or other olfactory proteins. While still speculative, these efforts may ultimately help provide the basis for the design of novel approaches for anti-malarial programs that target olfactory-based mosquito behaviors.
Acknowledgements
We thank Dr. Hyung-Wook Kwon, Tan Lu and Yuanfeng Xia for valuable advice and help during the development of the FISH technique; Lujuan Sun for assistance with EAG experiments; Dr. David McCauley (Vanderbilt University, Department of Biological Sciences) for access to the ABI Real Time PCR equipment; Patricia Russell for transgenic fly-line establishment; and members of the Zwiebel laboratory for discussions and comments on the manuscript; and Dr. A.M. McAinsh for editorial assistance. This work was funded by grants from the NIH to L.J.Z.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- Alcorta E. Characterization of the electroantennogram in Drosophila melanogaster and its use for identifying olfactory capture and transduction mutants. Journal of Neurophysiology. 1991;65:702–714. doi: 10.1152/jn.1991.65.3.702. [DOI] [PubMed] [Google Scholar]
- Attramadal H, Arriza JL, Aoki C, Dawson TM, Codina J, Kwatra MM, Snyder SH, Caron MG, Lefkowitz RJ. Beta-arrestin2, a novel member of the arrestin/beta-arrestin gene family. Journal of Biological Chemistry. 1992;267:17882–17890. [PubMed] [Google Scholar]
- Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. Public Library of Science Biology. 2006;4:e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohbot J, Vogt RG. Antennal expressed genes of the yellow fever mosquito (Aedes aegypti L.); characterization of odorant-binding protein 10 and takeout. Insect Biochemistry and Molecular Biology. 2005;35:961–979. doi: 10.1016/j.ibmb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Campos AR, Rosen DR, Robinow SN, White K. Molecular analysis of the locus elav in Drosophila melanogaster: a gene whose embryonic expression is neural specific. European Molecular Biology Organization Journal. 1987;6:425–431. doi: 10.1002/j.1460-2075.1987.tb04772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron. 1999;22:327–338. doi: 10.1016/s0896-6273(00)81093-4. [DOI] [PubMed] [Google Scholar]
- Dawson TM, Arriza JL, Jaworsky DE, Borisy FF, Attramadal H, Lefkowitz RJ, Ronnett GV. Beta-adrenergic receptor kinase-2 and beta-arrestin-2 as mediators of odorant-induced desensitization. Science. 1993;259:825–829. doi: 10.1126/science.8381559. [DOI] [PubMed] [Google Scholar]
- Dobritsa AA, Van Der Goes Van Naters W, Warr CG, Steinbrecht RA, Carlson JR. Integrating the Molecular and Cellular Basis of Odor Coding in the Drosophila Antenna. Neuron. 2003;37:827–841. doi: 10.1016/s0896-6273(03)00094-1. [DOI] [PubMed] [Google Scholar]
- Dolph PJ, Ranganathan R, Colley NJ, Hardy RW, Socolich M, Zuker CS. Arrestin function in inactivation of G protein-coupled receptor rhodopsin in vivo. Science. 1993;260:1910–1916. doi: 10.1126/science.8316831. [DOI] [PubMed] [Google Scholar]
- Dubin AE, Liles MM, Seligman F, Le T, Tolli J, Harris GL. Involvement of genes encoding a K+ channel (ether a go-go) and a Na+ channel (smellblind) in Drosophila olfaction. Annals of the New York Academy of Sciences. 1998;855:212–222. doi: 10.1111/j.1749-6632.1998.tb10569.x. [DOI] [PubMed] [Google Scholar]
- Ferguson SS, Caron MG. G protein-coupled receptor adaptation mechanisms. Seminars in Cell and Developmental Biology. 1998;9:119–127. doi: 10.1006/scdb.1997.0216. [DOI] [PubMed] [Google Scholar]
- Gao Q, Chess A. Identification of candidate Drosophila olfactory receptors from genomic DNA sequence. Genomics. 1999;60:31–39. doi: 10.1006/geno.1999.5894. [DOI] [PubMed] [Google Scholar]
- Gomez-Diaz C, Martin F, Alcorta E. The cAMP transduction cascade mediates olfactory reception in Drosophila melanogaster. Behavior Genetics. 2004;34:395–406. doi: 10.1023/B:BEGE.0000023645.02710.fe. [DOI] [PubMed] [Google Scholar]
- Gurevich EV, Gurevich VV. Arrestins: ubiquitous regulators of cellular signaling pathways. Genome Biology. 2006;7:236. doi: 10.1186/gb-2006-7-9-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich VV, Gurevich EV. The molecular acrobatics of arrestin activation. Trends in Pharmacological Sciences. 2004;25:105–111. doi: 10.1016/j.tips.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Hallem E, Fox AN, Zwiebel LJ, Carlson JR. A mosquito odorant receptor tuned to a component of human sweat. Nature. 2004a;427:212–213. doi: 10.1038/427212a. [DOI] [PubMed] [Google Scholar]
- Hallem E, Ho MG, Carlson JR. The Molecular Basis of Odor Coding in the Drosophila Antenna. Cell. 2004b;117:965–979. doi: 10.1016/j.cell.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Dahanukar A, Carlson JR. Insect odor and taste receptors. Annual Review of Entomology. 2006;51:113–135. doi: 10.1146/annurev.ento.51.051705.113646. [DOI] [PubMed] [Google Scholar]
- Hill CA, Fox AN, Pitts RJ, Kent LB, Tan PL, Chrystal MA, Cravchik A, Collins FH, Robertson HM, Zwiebel LJ. G protein-coupled receptors in Anopheles gambiae. Science. 2002;298:176–178. doi: 10.1126/science.1076196. [DOI] [PubMed] [Google Scholar]
- Hirsch JA, Schubert C, Gurevich VV, Sigler PB. The 2.8 A crystal structure of visual arrestin: a model for arrestin's regulation. Cell. 1999;97:257–269. doi: 10.1016/s0092-8674(00)80735-7. [DOI] [PubMed] [Google Scholar]
- Hyde DR, Mecklenburg KL, Pollock JA, Vihtelic TS, Benzer S. Twenty Drosophila visual system cDNA clones: one is a homolog of human arrestin. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:1008–1012. doi: 10.1073/pnas.87.3.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones WD, Nguyen TA, Kloss B, Lee KJ, Vosshall LB. Functional conservation of an insect odorant receptor gene across 250 million years of evolution. Current Biology. 2005;15:R119–121. doi: 10.1016/j.cub.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Kalidas S, Smith DP. Novel genomic cDNA hybrids produce effective RNA interference in adult Drosophila. Neuron. 2002;33:177–184. doi: 10.1016/s0896-6273(02)00560-3. [DOI] [PubMed] [Google Scholar]
- Krupnick JG, Benovic JL. The role of receptor kinases and arrestins in G protein-coupled receptor regulation. Annual Review of Pharmacology and Toxicology. 1998;38:289–319. doi: 10.1146/annurev.pharmtox.38.1.289. [DOI] [PubMed] [Google Scholar]
- Kuhn H, Hall SW, Wilden U. Light-induced binding of 48-kDa protein to photoreceptor membranes is highly enhanced by phosphorylation of rhodopsin. Federation of European Biochemical Societies Letters. 1984;176:473–478. doi: 10.1016/0014-5793(84)81221-1. [DOI] [PubMed] [Google Scholar]
- Kuhn H, Wilden U. Deactivation of photoactivated rhodopsin by rhodopsin-kinase and arrestin. Journal of Receptor Research. 1987;7:283–298. doi: 10.3109/10799898709054990. [DOI] [PubMed] [Google Scholar]
- Laporte SA, Oakley RH, Holt JA, Barak LS, Caron MG. The interaction of beta-arrestin with the AP-2 adaptor is required for the clustering of beta 2-adrenergic receptor into clathrin-coated pits. Journal of Biological Chemistry. 2000;275:23120–23126. doi: 10.1074/jbc.M002581200. [DOI] [PubMed] [Google Scholar]
- Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, Vosshall LB. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43:703–714. doi: 10.1016/j.neuron.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Lefkowitz RJ, Rajagopal K, Whalen EJ. New roles for beta-arrestins in cell signaling: not just for seven-transmembrane receptors. Molecular Cell. 2006;24:643–652. doi: 10.1016/j.molcel.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Lefkowitz RJ, Shenoy SK. Transduction of receptor signals by beta-arrestins. Science. 2005;308:512–517. doi: 10.1126/science.1109237. [DOI] [PubMed] [Google Scholar]
- Levine HD, Smith DP, Whitney M, Malicki DM, Dolph PJ, Smith GF, Burkhart W, Zuker CS. Isolation of a novel visual-system-specific arrestin: an in vivo substrate for light-dependent phosphorylation. Mechanisms of Development. 1990;33:19–25. doi: 10.1016/0925-4773(90)90131-5. [DOI] [PubMed] [Google Scholar]
- Lin DM, Goodman CS. Ectopic and increased expression of Fasciclin II alters motoneuron growth cone guidance. Neuron. 1994;13:507–523. doi: 10.1016/0896-6273(94)90022-1. [DOI] [PubMed] [Google Scholar]
- Lohse MJ, Benovic JL, Codina J, Caron MG, Lefkowitz RJ. beta-Arrestin: a protein that regulates beta-adrenergic receptor function. Science. 1990;248:1547–1550. doi: 10.1126/science.2163110. [DOI] [PubMed] [Google Scholar]
- Lu T, Qiu YT, Wang G, Kwon JY, Rutzler M, Kwon HW, Pitts RJ, Van Loon JJ, Takken W, Carlson JR, Zwiebel LJ. Odor Coding in the Maxillary Palp of the Malaria Vector Mosquito Anopheles gambiae. Current Biology. 2007;17:1533–1544. doi: 10.1016/j.cub.2007.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttrell LM, Ferguson SS, Daaka Y, Miller WE, Maudsley S, Della Rocca GJ, Lin F, Kawakatsu H, Owada K, Luttrell DK, Caron MG, Lefkowitz RJ. Beta-arrestin-dependent formation of beta2 adrenergic receptor-Src protein kinase complexes. Science. 1999;283:655–661. doi: 10.1126/science.283.5402.655. [DOI] [PubMed] [Google Scholar]
- Mashukova A, Spehr M, Hatt H, Neuhaus EM. Beta-arrestin2-mediated internalization of mammalian odorant receptors. Journal of Neuroscience. 2006;26:9902–9912. doi: 10.1523/JNEUROSCI.2897-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill CE, Pitts RJ, Zwiebel LJ. Molecular characterization of arrestin family members in the malaria vector mosquito, Anopheles gambiae. Insect Molecular Biology. 2003;12:641–650. doi: 10.1046/j.1365-2583.2003.00450.x. [DOI] [PubMed] [Google Scholar]
- Merrill CE, Riesgo-Escovar J, Pitts RJ, Kafatos FC, Carlson JR, Zwiebel LJ. Visual arrestins in olfactory pathways of Drosophila and the malaria vector mosquito Anopheles gambiae. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:1633–1638. doi: 10.1073/pnas.022505499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill CE, Sherertz TM, Walker WB, Zwiebel LJ. Odorant-specific requirements for arrestin function in Drosophila olfaction. Journal of Neurobiology. 2005;63:15–28. doi: 10.1002/neu.20113. [DOI] [PubMed] [Google Scholar]
- Orem NR, Xia L, Dolph PJ. An essential role for endocytosis of rhodopsin through interaction of visual arrestin with the AP-2 adaptor. Journal of Cell Science. 2006;119:3141–3148. doi: 10.1242/jcs.03052. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prossnitz ER. Novel roles for arrestins in the post-endocytic trafficking of G protein-coupled receptors. Life Sciences. 2004;75:893–899. doi: 10.1016/j.lfs.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Riesgo-Escovar J, Raha D, Carlson JR. Requirement for a phospholipase C in odor response: overlap between olfaction and vision in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:2864–2868. doi: 10.1073/pnas.92.7.2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesgo-Escovar JR, Woodard C, Carlson JR. Olfactory physiology in the Drosophila maxillary palp requires the visual system gene rdgB. Journal of Comparative Physiology A. 1994;175:687–693. doi: 10.1007/BF00191841. [DOI] [PubMed] [Google Scholar]
- Robertson HM, Warr CG, Carlson JR. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(Suppl 2):14537–14542. doi: 10.1073/pnas.2335847100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinow S, White K. The locus elav of Drosophila melanogaster is expressed in neurons at all developmental stages. Developmental Biology. 1988;126:294–303. doi: 10.1016/0012-1606(88)90139-x. [DOI] [PubMed] [Google Scholar]
- Roman G, He J, Davis RL. kurtz, a Novel Nonvisual Arrestin, Is an Essential Neural Gene in Drosophila. Genetics. 2000;155:1281–1295. doi: 10.1093/genetics/155.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh AK, Ready DF. Arrestin1 mediates light-dependent rhodopsin endocytosis and cell survival. Current Biology. 2005;15:1722–1733. doi: 10.1016/j.cub.2005.08.064. [DOI] [PubMed] [Google Scholar]
- Smith DP, Shieh BH, Zuker CS. Isolation and structure of an arrestin gene from Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:1003–1007. doi: 10.1073/pnas.87.3.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbrecht RA. Structure and function of insect olfactory sensilla. Ciba Foundation Symposia. 1996;200:158–174. doi: 10.1002/9780470514948.ch13. [DOI] [PubMed] [Google Scholar]
- Vishnivetskiy SA, Schubert C, Climaco GC, Gurevich YV, Velez MG, Gurevich VV. An additional phosphate-binding element in arrestin molecule: implications for the mechanism of arrestin activation. Journal of Biological Chemistry. 2000 doi: 10.1074/jbc.M007159200. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96:725–736. doi: 10.1016/s0092-8674(00)80582-6. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Wong AM, Axel R. An olfactory sensory map in the fly brain. Cell. 2000;102:147–159. doi: 10.1016/s0092-8674(00)00021-0. [DOI] [PubMed] [Google Scholar]
- Wilden U, Hall SW, Kuhn H. Phosphodiesterase activation by photoexcited rhodopsin is quenched when rhodopsin is phosphorylated and binds the intrinsic 48-kDa protein of rod outer segments. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:1174–1178. doi: 10.1073/pnas.83.5.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilden U, Kuhn H. Light-dependent phosphorylation of rhodopsin: number of phosphorylation sites. Biochemistry. 1982;21:3014–3022. doi: 10.1021/bi00541a032. [DOI] [PubMed] [Google Scholar]
- Zwiebel LJ, Takken W. Olfactory Regulation of Mosquito-Host Interactions. Insect Biochemistry and Molecular Biology. 2004;34:645–652. doi: 10.1016/j.ibmb.2004.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]