Abstract
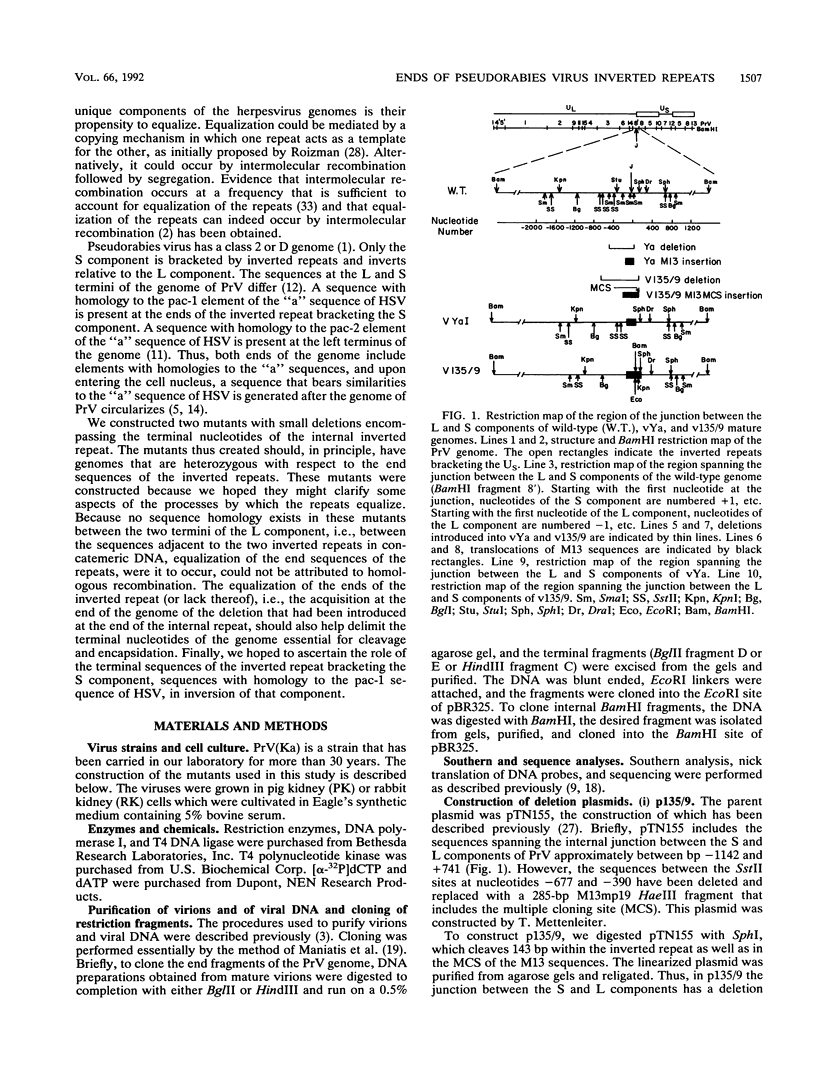
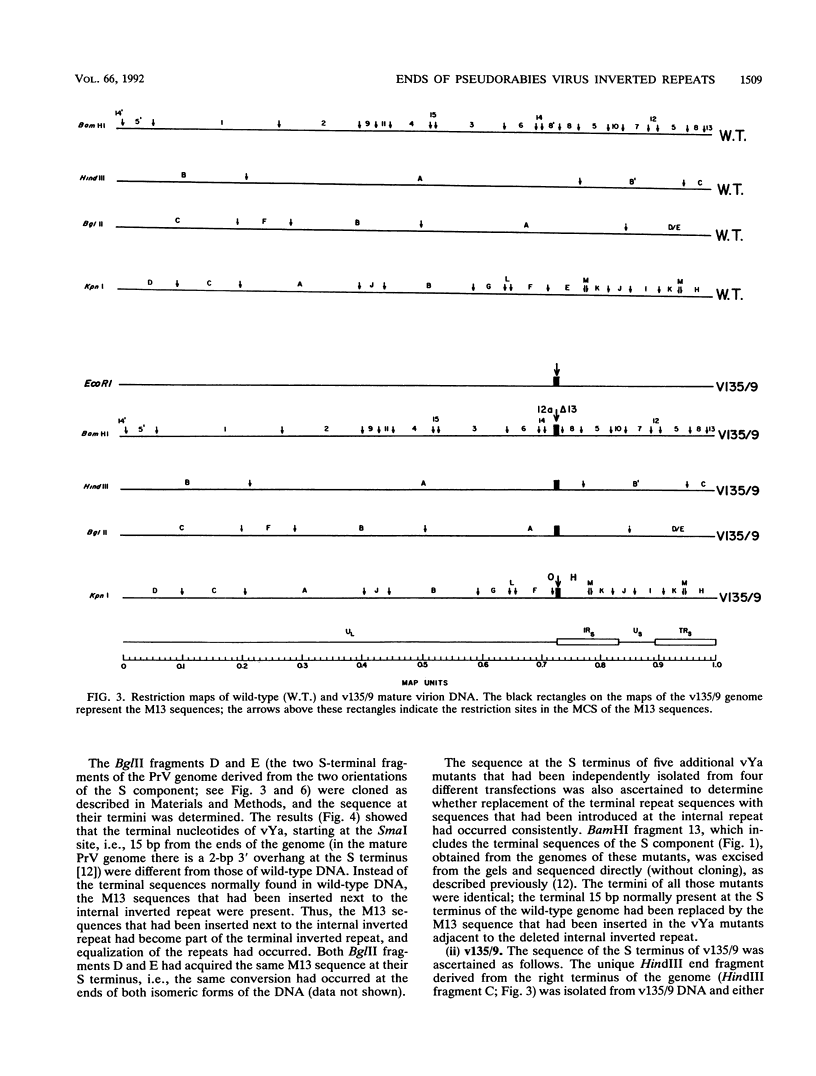
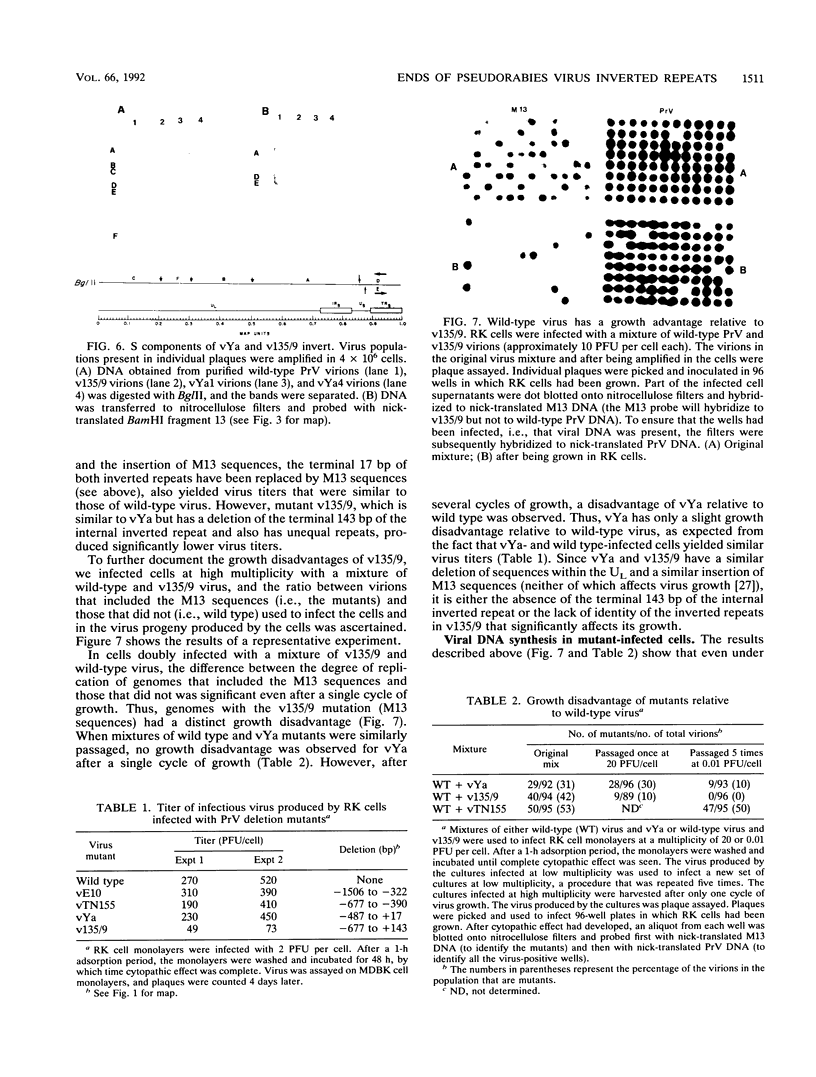
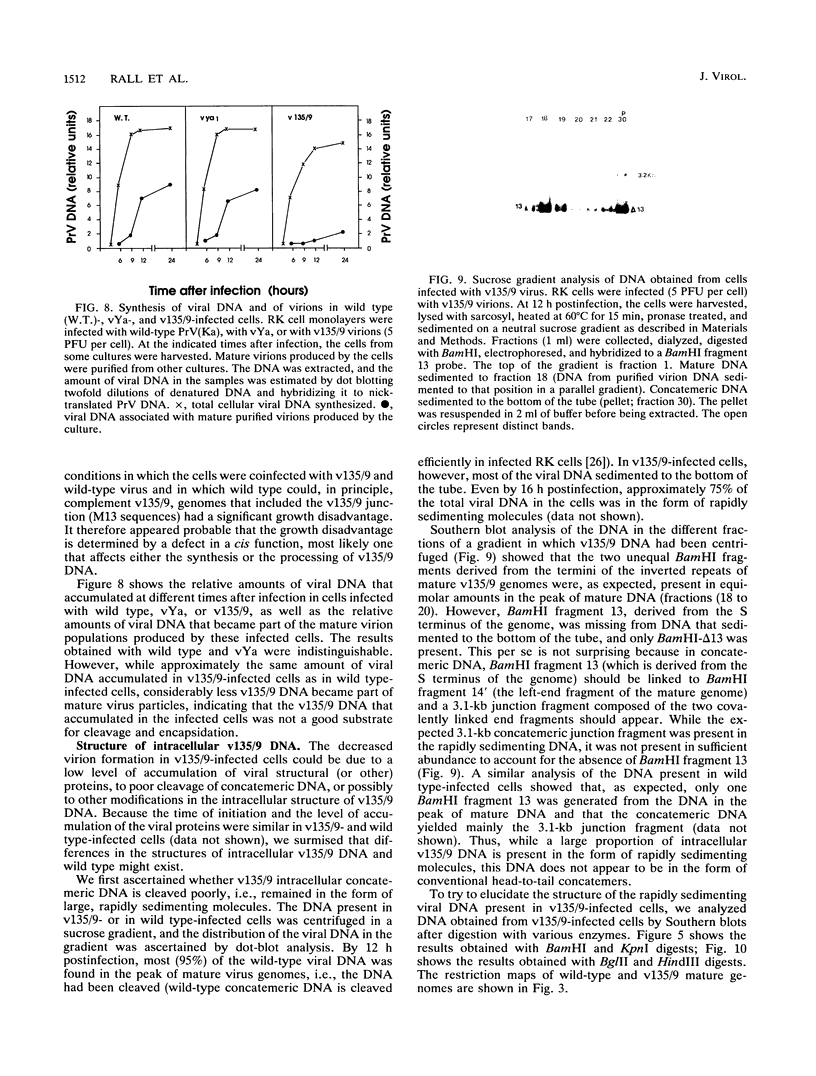
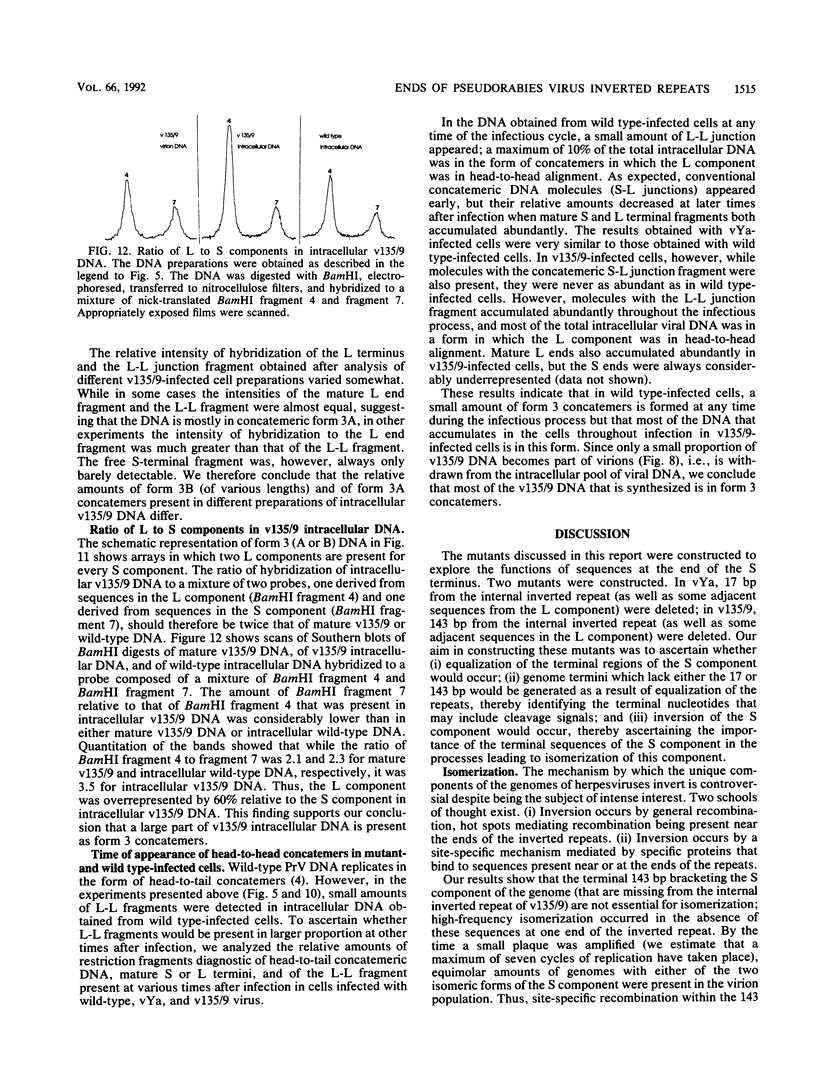
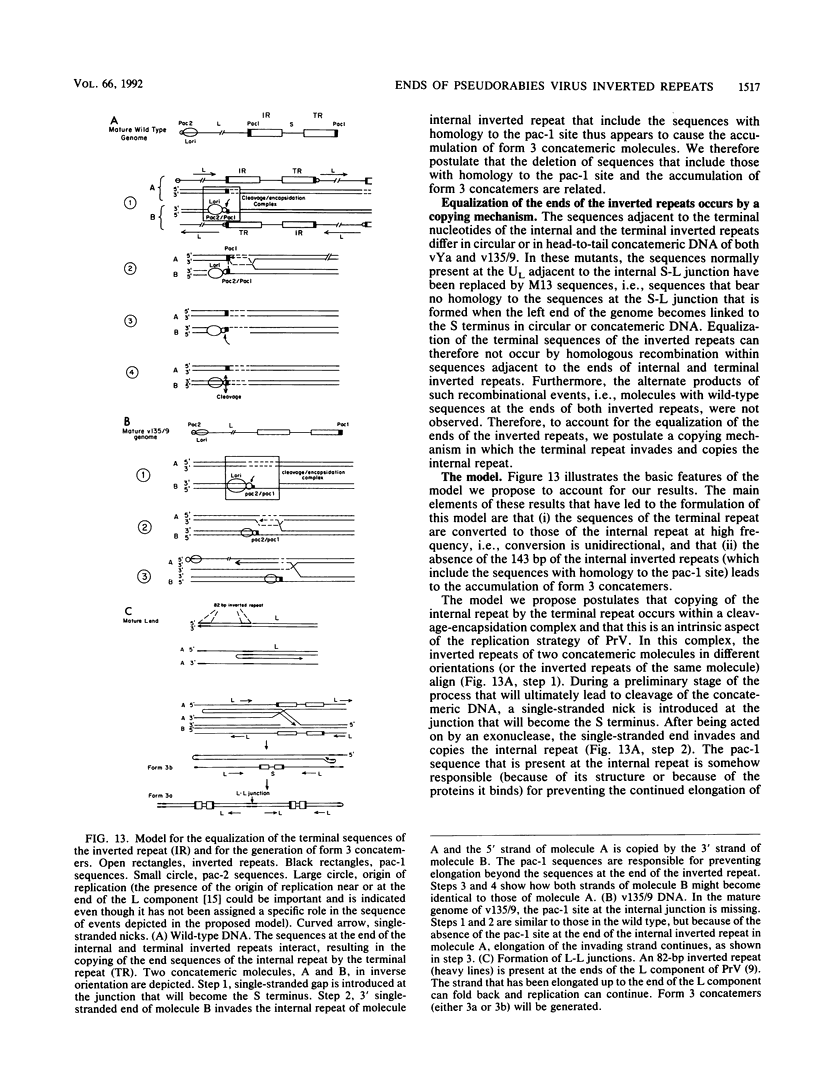
Two mutants were constructed to explore the functions of the sequences at the end of the S terminus of pseudorabies virus (PrV). In mutant vYa, 17 bp from the internal inverted repeat, as well as adjacent sequences from the L component, were deleted. In mutant v135/9, 143 bp from the internal inverted repeat (including sequences with homology to the pac-1 site of herpes simplex virus), as well as adjacent sequences from the L component, were deleted. Our aim in constructing these mutants was to ascertain whether equalization of the terminal regions of the S component would occur, whether genome termini that lack either the terminal 17 or 143 bp would be generated as a result of equalization of the repeats (thereby identifying the terminal nucleotides that may include cleavage signals), and whether inversion of the S component would occur (thereby ascertaining the importance of the deleted sequences in this process). The results obtained show the following (i) The removal of the terminal 17 or 143 bp of the internal S component, including the sequences with homology to the pac-1 site, does not affect the inversion of the Us. (ii) The equalization of both the vYa and the v135/9 inverted repeats occurs at high frequency, the terminal repeats being converted and becoming similar to the mutated internal inverted repeat. (iii) Mutants in which the 17 terminal base pairs (vYa) have been replaced by unrelated sequences are viable. However, the 143 terminal base pairs appear to be essential to virus survival; concatemeric v135/9 DNA with equalized, mutant-type, inverted repeats accumulates, but mature virions with such equalized repeats are not generated at high frequency. Since concatemeric DNA missing the 143 bp at both ends of the S component is not cleaved, the terminal 143 bp that include the sequences with homology to the pac-1 site are necessary for efficient cleavage. (iv) v135/9 intracellular DNA is composed mainly of arrays in which one S component (with two equalized inverted repeats both having the deletion) is bracketed by two L components in opposite orientations and in which two L components are in head-to-head alignment.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ben-Porat T., Deatly A., Veach R. A., Blankenship M. L. Equalization of the inverted repeat sequences of the pseudorabies virus genome by intermolecular recombination. Virology. 1984 Jan 30;132(2):303–314. doi: 10.1016/0042-6822(84)90037-0. [DOI] [PubMed] [Google Scholar]
- Ben-Porat T., Demarchi J. M., Kaplan A. S. Characterization of defective interfering viral particles present in a population of pseudorabies virions. Virology. 1974 Sep;61(1):29–37. doi: 10.1016/0042-6822(74)90239-6. [DOI] [PubMed] [Google Scholar]
- Ben-Porat T. Replication of herpesvirus DNA. Curr Top Microbiol Immunol. 1981;91:81–107. doi: 10.1007/978-3-642-68058-8_4. [DOI] [PubMed] [Google Scholar]
- Ben-Porat T., Rixon F. J. Replication of herpesvirus DNA. IV: analysis of concatemers. Virology. 1979 Apr 15;94(1):61–70. doi: 10.1016/0042-6822(79)90438-0. [DOI] [PubMed] [Google Scholar]
- Ben-Porat T., Veach R. A., Ladin B. F. Replication of herpesvirus DNA. VI. Virions containing either isomer of pseudorabies virus DNA are infectious. Virology. 1980 Apr 30;102(2):370–380. doi: 10.1016/0042-6822(80)90104-x. [DOI] [PubMed] [Google Scholar]
- Ben-Porat T., Veach R. A. Origin of replication of the DNA of a herpesvirus (pseudorabies). Proc Natl Acad Sci U S A. 1980 Jan;77(1):172–175. doi: 10.1073/pnas.77.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J., Roizman B. Isomerization of herpes simplex virus 1 genome: identification of the cis-acting and recombination sites within the domain of the a sequence. Cell. 1985 Jul;41(3):803–811. doi: 10.1016/s0092-8674(85)80061-1. [DOI] [PubMed] [Google Scholar]
- DeMarchi J. M., Lu Z. Q., Rall G., Kupershmidt S., Ben-Porat T. Structural organization of the termini of the L and S components of the genome of pseudorabies virus. J Virol. 1990 Oct;64(10):4968–4977. doi: 10.1128/jvi.64.10.4968-4977.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deiss L. P., Chou J., Frenkel N. Functional domains within the a sequence involved in the cleavage-packaging of herpes simplex virus DNA. J Virol. 1986 Sep;59(3):605–618. doi: 10.1128/jvi.59.3.605-618.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt W., Ludwig H., Buhk H. J. Specificity of cleavage in replicative-form DNA of bovine herpesvirus 1. J Virol. 1988 Apr;62(4):1355–1363. doi: 10.1128/jvi.62.4.1355-1363.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper L., Demarchi J., Ben-Porat T. Sequence of the genome ends and of the junction between the ends in concatemeric DNA of pseudorabies virus. J Virol. 1986 Dec;60(3):1183–1185. doi: 10.1128/jvi.60.3.1183-1185.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honess R. W., Watson D. H. Unity and diversity in the herpesviruses. J Gen Virol. 1977 Oct;37(1):15–37. doi: 10.1099/0022-1317-37-1-15. [DOI] [PubMed] [Google Scholar]
- Jean J. H., Ben-Porat T. Appearance in vivo of single-stranded complementary ends on parental herpesvirus DNA. Proc Natl Acad Sci U S A. 1976 Aug;73(8):2674–2678. doi: 10.1073/pnas.73.8.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupershmidt S., DeMarchi J. M., Lu Z. Q., Ben-Porat T. Analysis of an origin of DNA replication located at the L terminus of the genome of pseudorabies virus. J Virol. 1991 Nov;65(11):6283–6291. doi: 10.1128/jvi.65.11.6283-6291.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longnecker R., Roizman B. Generation of an inverting herpes simplex virus 1 mutant lacking the L-S junction a sequences, an origin of DNA synthesis, and several genes including those specifying glycoprotein E and the alpha 47 gene. J Virol. 1986 May;58(2):583–591. doi: 10.1128/jvi.58.2.583-591.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z. Q., DeMarchi J. M., Harper L., Rall G. F., Ben-Porat T. Nucleotide sequences at recombinational junctions present in pseudorabies virus variants with an invertible L component. J Virol. 1989 Jun;63(6):2690–2698. doi: 10.1128/jvi.63.6.2690-2698.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Site-specific inversion sequence of the herpes simplex virus genome: domain and structural features. Proc Natl Acad Sci U S A. 1981 Nov;78(11):7047–7051. doi: 10.1073/pnas.78.11.7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Structure and role of the herpes simplex virus DNA termini in inversion, circularization and generation of virion DNA. Cell. 1982 Nov;31(1):89–97. doi: 10.1016/0092-8674(82)90408-1. [DOI] [PubMed] [Google Scholar]
- Moyer R. W., Graves R. L. The mechanism of cytoplasmic orthopoxvirus DNA replication. Cell. 1981 Dec;27(2 Pt 1):391–401. doi: 10.1016/0092-8674(81)90422-0. [DOI] [PubMed] [Google Scholar]
- Nasseri M., Mocarski E. S. The cleavage recognition signal is contained within sequences surrounding an a-a junction in herpes simplex virus DNA. Virology. 1988 Nov;167(1):25–30. doi: 10.1016/0042-6822(88)90050-5. [DOI] [PubMed] [Google Scholar]
- Pogue-Geile K. L., Lee G. T., Spear P. G. Novel rearrangements of herpes simplex virus DNA sequences resulting from duplication of a sequence within the unique region of the L component. J Virol. 1985 Feb;53(2):456–461. doi: 10.1128/jvi.53.2.456-461.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rall G. F., Kupershmidt S., Lu X. Q., Mettenleiter T. C., Ben-Porat T. Low-level inversion of the L component of pseudorabies virus is not dependent on sequence homology. J Virol. 1991 Dec;65(12):7016–7019. doi: 10.1128/jvi.65.12.7016-7019.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rall G. F., Lu Z. Q., Sugg N., Veach R. A., Ben-Porat T. Acquisition of an additional internal cleavage site differentially affects the ability of pseudorabies virus to multiply in different host cells. J Virol. 1991 Dec;65(12):6604–6611. doi: 10.1128/jvi.65.12.6604-6611.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly L. M., Rall G., Lomniczi B., Mettenleiter T. C., Kuperschmidt S., Ben-Porat T. The ability of pseudorabies virus to grow in different hosts is affected by the duplication and translocation of sequences from the left end of the genome to the UL-US junction. J Virol. 1991 Nov;65(11):5839–5847. doi: 10.1128/jvi.65.11.5839-5847.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B. The structure and isomerization of herpes simplex virus genomes. Cell. 1979 Mar;16(3):481–494. doi: 10.1016/0092-8674(79)90023-0. [DOI] [PubMed] [Google Scholar]
- Sheldrick P., Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- Smiley J. R., Duncan J., Howes M. Sequence requirements for DNA rearrangements induced by the terminal repeat of herpes simplex virus type 1 KOS DNA. J Virol. 1990 Oct;64(10):5036–5050. doi: 10.1128/jvi.64.10.5036-5050.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smiley J. R., Fong B. S., Leung W. C. Construction of a double-jointed herpes simplex viral DNA molecule: inverted repeats are required for segment inversion, and direct repeats promote deletions. Virology. 1981 Aug;113(1):345–362. doi: 10.1016/0042-6822(81)90161-6. [DOI] [PubMed] [Google Scholar]
- Varmuza S. L., Smiley J. R. Signals for site-specific cleavage of HSV DNA: maturation involves two separate cleavage events at sites distal to the recognition sequences. Cell. 1985 Jul;41(3):793–802. doi: 10.1016/s0092-8674(85)80060-x. [DOI] [PubMed] [Google Scholar]
- Varmuza S. L., Smiley J. R. Unstable heterozygosity in a diploid region of herpes simplex virus DNA. J Virol. 1984 Feb;49(2):356–362. doi: 10.1128/jvi.49.2.356-362.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber P. C., Challberg M. D., Nelson N. J., Levine M., Glorioso J. C. Inversion events in the HSV-1 genome are directly mediated by the viral DNA replication machinery and lack sequence specificity. Cell. 1988 Jul 29;54(3):369–381. doi: 10.1016/0092-8674(88)90200-0. [DOI] [PubMed] [Google Scholar]
- Weber P. C., Levine M., Glorioso J. C. Recombinogenic properties of herpes simplex virus type 1 DNA sequences resident in simian virus 40 minichromosomes. J Virol. 1990 Jan;64(1):300–306. doi: 10.1128/jvi.64.1.300-306.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. A., Harper L., Ben-Porat T. cis Functions involved in replication and cleavage-encapsidation of pseudorabies virus. J Virol. 1986 Aug;59(2):318–327. doi: 10.1128/jvi.59.2.318-327.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]