Abstract
Chromaffin cells of the adrenal medulla elaborate and secrete catecholamines and neuropeptides for hormonal and paracrine signaling in stress and during inflammation. We have recently documented the action of the cytokine TNF-α on neuropeptide secretion and biosynthesis in isolated bovine chromaffin cells. Here, we demonstrate that the type 2 TNF-α receptor (TNF-R2) mediates TNF-α signaling in chromaffin cells via activation of nuclear factor (NF)-κB. Microarray and suppression subtractive hybridization have been used to identify TNF-α target genes in addition to those encoding the neuropeptides galanin, vasoactive intestinal polypeptide, and secretogranin II in chromaffin cells. TNF-α, acting through the TNF-R2, causes an early up-regulation of NF-κB, long-lasting induction of the NF-κB target gene inhibitor κB (IκB), and persistent stimulation of other NF-κB-associated genes including mitogen-inducible gene-6 (MIG-6), which acts as an IκB signaling antagonist, and butyrate-induced transcript 1. Consistent with long-term activation of the NF-κB signaling pathway, delayed induction of neuropeptide gene transcription by TNF-α in chromaffin cells is blocked by an antagonist of NF-κB signaling. TNF-α-dependent signaling in neuroendocrine cells thus leads to a unique, persistent mode of NF-κB activation that features long-lasting transcription of both IκB and MIG-6, which may play a role in the long-lasting effects of TNF-α in regulating neuropeptide output from the adrenal, a potentially important feedback station for modulating long-term cytokine effects in inflammation.
DURING INFLAMMATION, reciprocal interactions between the immune and the neuroendocrine systems occur, leading to activation of integrated physiological circuits. Various signaling messengers including cytokines, hormones, and neuropeptides contribute to this coordinated organismic response (1). In particular, cytokines such as TNF-α play a central role in mediating the inflammatory response, by direct actions, mainly via activation of the type 1 TNF-α receptor (TNF-R1), on immune modulatory cells including lymphocytes and macrophages. Because the actions of TNF-α (and other cytokines) are potentially deleterious to the host, TNF-α also is capable of shutting down its own production via activation of immunosuppressive glucocorticoids in the adrenal cortex. TNF-α may also participate in an autoregulatory loop in the adrenal gland itself by activating catecholamine and neuropeptide biosynthesis and secretion, which directly affect antiinflammatory glucocorticoid production in the adjacent adrenal cortex (2).
We have recently characterized TNF-α signaling to neuropeptide target genes in cultured chromaffin cells of the adrenal medulla, and identified activator protein-1 (AP-1), and the MAPKs ERK1/2 and p38 as components required for TNF-mediated regulation of galanin (GAL), secretogranin II (SgII), and vasoactive intestinal polypeptide (VIP) biosynthesis. Signaling initiated by TNF-α in chromaffin cells, unlike TNF-α signaling in immunocytes, does not require activation of c-Jun N-terminal kinase (3). In addition, TNF-α signaling occurs exclusively through type 2 receptors and is responsible for a rather prolonged effect on neuropeptide gene expression that requires persistent signaling over a period of more than 24 h. These properties appear to be unique to TNF-R2-mediated neuroendocrine signaling and may be required for the role of TNF-α receptor activation in adrenal medulla as being physiologically counterregulatory to the main inflammatory effects of this cytokine. As postulated by Bornstein and others (2,4), neuropeptides secreted from the adrenal medulla, including GAL and VIP, function to augment glucocorticoid biosynthesis in the adjacent adrenal cortex. In this case, delayed regulation by the proinflammatory cytokine TNF-α may coordinate sensing of serum and/or monocyte TNF-α levels in chronic or after acute inflammation and, transducing this, via neuropeptide production and secretion, into a graded and prolonged glucocorticoid counterregulatory response. Furthermore, use of the chromaffin cell as a model allows investigation of important questions about the mechanism of TNF-α signaling through the type 2 receptor in general that are relevant to TNF-α’s counterinflammatory effects not only in the peripheral but also in the central nervous system.
It is known that both TNF-R1 and TNF-R2 receptor activation can lead to stimulation of nuclear factor (NF)-κB in a variety of cells, but by different mechanisms (5,6,7,8). However, the mechanisms mediating TNF-R2 signaling in neuroendocrine cells are not known. In neurons, for example, TNF-R2 signaling underlying neuroprotection from glutamate toxicity is mediated through NF-κB, but via an Akt-dependent signaling pathway (9). Specifically, we wondered whether TNF-α signaling in chromaffin cells requires NF-κB in either the early phase of TNF-α response (within the first several hours, as in immune/inflammatory cells) or the late phase, which leads to neuropeptide gene regulation after prolonged exposure. Here, we employed microarray and suppression subtractive hybridization (SSH) to identify a specifically neuroendocrine transcriptomic response to TNF-α. Signaling for induction of neuropeptide gene expression by TNF-α appears to depend upon a unique mode of persistent activation of NF-κB and NF-κB-dependent genes leading to delayed and sustained activation of neuropeptide gene expression. Thus, in addition to the previously identified involvement of MAPK and AP-1 (3), NF-κB is a prominent component in TNF-α regulation of neuropeptide gene expression in chromaffin cells. These properties of TNF-α signaling through the type 2 TNF-α receptor in neuroendocrine cells may determine the counterregulatory actions of TNF-α during chronic inflammation or inflammation accompanied by activation of the stress axis.
Materials and Methods
Materials
Human recombinant TNF-α was purchased from Eurobio (Courtaboeuf, France). Indomethacin, NG-monomethyl-l-arginine (l-NMMA), pyrrolidine dithiocarbamate (PDTC), and poly-l-lysine were obtained from Sigma-Aldrich (Saint-Quentin Fallavier, France).
Cell culture and treatments
Bovine adrenal glands were collected from the local slaughterhouse with the approval of our regional veterinary services (agreement no. 7627601). Primary cultures of bovine chromaffin cells (BCCs) were obtained after retrograde perfusion of bovine adrenal glands with 0.1% collagenase (Sigma-Aldrich) and 30 U/ml DNase (Sigma-Aldrich), followed by dissociation of the digested adrenal medulla. The cells were cultured in DMEM (Sigma-Aldrich) supplemented with 5% fetal calf serum (Eurobio) and 100 U/ml penicillin, 100 μg/ml streptomycin, and 0.25 μg/ml fungizone (Invitrogen, Cergy-Pontoise, France). Chromaffin cells were purified by differential plating as described previously (10). Cells were then plated in the same medium as above in 75-ml flasks or at a density of 106 cells/ml in poly-l-lysine-coated 12-well plates and treated with TNF-α in the presence or absence of NF-κB inhibitor, cyclooxygenase inhibitor, or nitric oxide (NO) synthase inhibitor. The inhibitors were dissolved in ethanol (indomethacin), in dimethylsulfoxide (l-NMMA), or directly in the medium (PDTC) and were added 30 min before the onset of secretagogue treatment.
RNA extraction and RT-PCR
Total RNA was harvested and extracted from individual BCC culture wells by adding Tri-Reagent (Sigma-Aldrich) using the method of Chomczynski and Sacchi (11), and approximately 5 μg of purified RNA was reverse transcribed by using an oligo(dT)12–18 primer and the superscript II reverse transcriptase RNase H– (Life Technologies, Cergy-Pontoise, France). PCR amplification was performed with 5 U Taq DNA polymerase (Life Technologies) and 50 pmol of the sense (TNF-R2-S, 5′-CTCGACCAGCAGCACGGA-3′) and antisense (TNF-R2-AS, 5′-GCTGGCGTCTGTGTCCCTGG-3′) TNF-R2 primers or 50 pmol of the sense (bCgA6, 5′-TCTGCCTCGATGGCTGACAA-3′) and antisense (bCgA7, 5′-GCAGAAGAAGCACAGCAGTTA-3′) chromogranin A (CgA) primers in the GeneAmp PCR System 9700 (Applied Biosystems, Norwalk, CT).
In situ hybridization histochemistry
Adrenal glands were collected, immediately frozen in isopentane (−30 C), and stored at −70 C until use. Frozen lateral sections (14 μm thick) were cut in a cryostat (Frigocut, Reicher-Jung, Germany) and collected onto poly-l-lysine-coated slides. Tissue sections were hybridized with 107 cpm/ml 35S-labeled TNF-R2 riboprobe and digoxigenin-labeled CgA as previously described (12). Sense and antisense riboprobes were prepared by in vitro transcription of bovine TNF-R2 and CgA cDNA fragments subcloned into pGEMT vector (Promega, Charbonnières, France), in the presence of [35S]UTP (Amersham Pharmacia Biotech, Les Ulis, France) or digoxigenin-11-UTP, respectively, and T7 or SP6 RNA polymerase (Promega). Tissue sections were dehydrated and exposed onto Hyperfilm β-max (Amersham Pharmacia Biotech) for 4 d. Tissue slices were subsequently dipped into Kodak NTB2 liquid emulsion at 40 C, exposed for 15 d, and developed. Anatomical structures were identified by staining tissue slices with hematoxylin and eosin.
Quantitative RT-PCR (Q-RT-PCR)
Approximately 2 μg total RNA extracted as described above was submitted to DNase I (RNase-free; Promega) digestion and reverse transcribed using random hexamers pdN6 (Invitrogen) and SuperScript II RNase H– reverse transcriptase (Invitrogen). Gene-specific forward and reverse primers were chosen using the Primer Express 2 software (PE Applied Biosystems, Courtaboeuf, France) or Primer 3 Input (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi), to amplify in each case across an exon-exon junction to avoid generation of same-sized amplicons from contaminating genomic DNA as follows: 5′-TCCTCTCAATCCTGCGACATC-3′ and 5′-TCCTCGTAACTGCTGTGCTTCTT-3′ for CgA; 5′-GACAGCCACAGGTCATTTCAA-3′ and 5′-GCCGGGCTTCGTCTTCA-3′ for GAL; 5′-CTCAGAGGTTGACTTAGACCATCCA-3′ and 5′-GCATGACCAGGTGCTT-TGG-3′ for SgII; 5′-ACCTGATGGTTACATGTCACTCA-3′ and 5′-GGGAAGGTTATTCAAAATTTGATGA-3′ for inhibitor κB (IκB); 5′-TTCAAAGACTCGGGCTGCAA-3′ and 5′-AACTGCCAATCTTCCAGATG-3′ for TRAF4; 5′-TGCTACGACACTCGGAACTG-3′ and 5′-GGAGCAAAATTGGAACCAGA-3′ for TNFAIP3; 5′-GCGGCAGTACTTTGAAGAGG-3′ and AGGTTCGGGGAAATAATTGG-3′ for DUSP10; 5′-GAGATTGGCTTCTCCCCTTC-3′ and 5′-CTGCCCAGGCTCTCGTTGCT-3′ for NOD1. Real-time PCR (Q-RT-PCR) was performed in a premade reaction mix (PE Applied Biosystems) in the presence of the transcribed cDNA and 300 nm specific primers, using the SYBR green chemistry and an ABI Prism 7000 (PE Applied Biosystems). Relative amounts of mRNA were determined from standard curves generated using dilutions of the cDNA and by normalizing against a nonvariable control gene, CgA, that was analyzed in parallel on the same RT reaction.
EMSA
Chromaffin cells were treated with 10 nm TNF-α for 2 h, and nuclear extracts were prepared as described previously (10). A double-stranded oligonucleotide containing consensus NF-κB response element, 5′-AGTTGAGGGGACTTTCCCAG-3′, was labeled by fill-in reaction using [α-32P]dCTP and the Klenow enzyme and used in gel-supershifting studies. Chromaffin cell nuclear extracts (2 μg) were incubated at room temperature for 30 min in a 10-μl volume reaction containing 10 mm MgCl2, 0.5 mm EDTA, 0.5 mm dithiothreitol, 4% Ficoll, and 0.1 μg/μl poly(dI-dC) (Amersham Pharmacia Biotech) and in the presence or absence of specific antibodies (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). The 32P-labeled NF-κB oligonucleotide (30,000 cpm per reaction) was added, and the incubation continued for 20 min. In competition studies, the nuclear extracts were incubated with the unlabeled homologous probe before the labeled probe. In the supershift EMSA, the antibodies used (1 μl per reaction) were affinity-purified IgG fractions recognizing p50, p52, p65, RelB, and cRel, all known subunits composing NF-κB transcription factor. The DNA-protein complexes were analyzed on 3.75% nondenaturing polyacrylamide gels in 0.25× TBE (10 mm Tris-borate, pH 8.3; 1 mm EDTA). The gels were then dried and exposed to fluorescent screens that were subsequently scanned using a STORM Phosphor Imager 840 system (Amersham Pharmacia Biotech).
Oligo microarray, hybridization, and data analysis
A glass microarray containing 32,000 mouse oligo probes (see http://oligos.qiagen.com/arrays/oligosets_mouse.php for more details) was used in this study. Labeled input cDNA was prepared from total RNA isolated from cultured BCCs treated for 6 or 48 h with TNF-α, using RNeasy Mini Spin Columns (QIAGEN, Valencia, CA) and quantified by spectrophotometry. Labeling and hybridization were performed according to standard National Human Genome Research Institute protocols (http://research.nhgri.nih.gov/ma/protocols.shtml). Briefly, 15–20 μg RNA was reverse transcribed with Superscript II reverse transcriptase Rnase H– (Invitrogen) in the presence of random hexamers and aminoallyl-dUTP/dNTP. Then, purified probes were labeled with NHS-Ester dye Cy3 and Cy5 (Amersham Pharmacia Biotech). After denaturation, purified Cy3/Cy5-labeled probes were hybridized to the coverslipped array in Agilent gaskets in a rotisserie hybridization oven at 65 C overnight in Agilent 2× hybridization buffer. Before scanning at 532 nm for Cy3 and 633 nm for Cy5 (Agilent Technologies, Foster City, CA), slides were successively washed at room temperature in 0.5× standard saline citrate (SSC)/0.05% SDS for 5 min, 0.5× SSC for 5 min (twice), and 0.06× SSC for 5 min. The two fluorescent images obtained from the scanner were analyzed with IPLab software (Scanalytics, Fairfax, VA). The data from three independent experiments were analyzed after entry into mAdb (NIH Center for Information Technology).
SSH
Total RNA was extracted from untreated and TNF-α-treated BCCs as described above, and poly(A)+ RNA was isolated with the polyATract mRNA Isolation System (Promega). cDNAs were synthesized from 2 μg poly(A)+ RNA, and subtractive hybridization was performed using the PCR Selected cDNA subtraction kit (BD Biosciences, Saint-Quentin en Yvelines, France). To isolate TNF-α-induced transcripts, cDNAs from TNF-α-treated cells were ligated to oligonucleotide linkers and hybridized with excess cDNAs from untreated cells. After hybridization, differentially expressed transcripts were selectively amplified by suppression PCR (13). Amplified cDNAs were introduced in TOPO vector (Invitrogen) and electroporated into DH10B cells to generate a subtractive library.
Macroarray preparation and hybridization
Clones obtained by SSH were amplified by PCR with T7 and SP6 primers using Taq DNA polymerase (Invitrogen) in a GeneAmp PCR system 9700 (Applied Biosystems, Norwalk, CT) and used as probes to make a macroarray. The PCR products contained in a 384-well plate were directly printed on Hybond NX membranes (Amersham Pharmacia Biotech) using a ChipWriter system (Virtek, Waterloo, Canada). These filters were denatured with 0.4 m NaOH, 0.1 m NaCl solution for 5 min and neutralized with 40 mm Na2HPO4/NaH2PO4 solution (pH 7.2) for 5 min. RNA derived from untreated and TNF-α-treated chromaffin cells was reverse transcribed with Superscript II reverse transcriptase Rnase H– (Invitrogen) in the presence of [α-33P]dCTP (Amersham Pharmacia Biotech) and used to hybridize the macroarray membranes at 42 C in a solution containing 50% formamide, 5× SSC, 5× Denhardt’s, 200 μg/ml salmon sperm DNA, 50 μg/ml yeast tRNA, 0.1% SDS, and 50 mm phosphate buffer (pH 6.5). The membranes were washed four times in 2× SSC, 0.1% SDS at room temperature and twice in 0.1× SSC, 0.1% SDS for 15 min at 50 C. The membranes were analyzed using the STORM Phosphor Imager system (Amersham Pharmacia Biotech), and the images corresponding to hybridization with untreated and TNF-α-treated chromaffin cell targets were quantified with the XDotsReader software (Cose, Dugny, France). Hybridization signals were normalized to those of a CgA probe that was printed at several locations of the macroarray.
Immunocytochemistry
Chromaffin cells were plated at a density of 5 × 105 cells/ml on poly-l-lysine-coated glass coverslips and incubated overnight in serum-free medium. The cells were washed twice with PBS at room temperature and fixed with 4% paraformaldehyde in PBS for 20 min. After two washes, the cells were permeabilized for 5 min in a solution of PBS containing 0.1% BSA and 0.5% Triton X-100 and then blocked with 3% normal goat serum in PBS containing 0.1% BSA during 20 min at room temperature to reduce nonspecific staining. After a wash, coverslips were incubated with the anti-IκB antibody (Santa Cruz Biotechnology) at a 1:100 dilution in PBS containing 0.1% BSA overnight at 4 C. The cells were washed with PBS and then incubated for 1 h at room temperature with goat antirabbit Igs coupled to Alexa-488 diluted 1:100. Finally, the coverslips were washed with PBS, rinsed with distilled water, mounted in PBS-glycerol (1:1), and examined using a confocal laser scanning microscope (Leica, Heidelberg, Germany) equipped with a diaplan DMRXA2 optical system and an argon ion laser. To verify the specificity of the immunoreaction, the primary or secondary antibodies were substituted with PBS.
Results and Discussion
Chromaffin cells express the TNF-R2
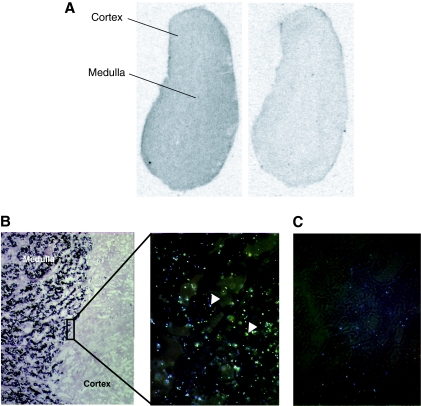
We examined the expression and distribution of TNF-R2 mRNA in the bovine adrenal gland in situ to establish that TNF-R2 expression in BCCs is indeed representative of the parenchymal tissue of this gland in vivo. In situ hybridization was performed using an input cDNA obtained by cloning of the bovine TNF-R2 from BCC mRNA and confirmed by sequencing as TNF-R2. mRNA encoding TNF-R2 complementary to probe was found widely and uniformly distributed in the adrenal gland, in both medulla and cortex (Fig. 1A). Colocalization studies using CgA mRNA labeled with digoxigenin showed the presence of TNF-R2 mRNA labeled with 35S evenly distributed over adrenomedullary chromaffin cells (Fig. 1B). When a sense probe was used for both TNF-R2 and CgA, no hybridization signal was observed (Fig. 1C). These results are consistent with our previous demonstration by RT-PCR that exclusively TNF-R2 mRNA is present in freshly cultured chromaffin cells derived from the bovine adrenal gland (3).
Figure 1.
Adrenochromaffin cells express the TNF-R2. A, Autoradiogram of a bovine adrenal gland transverse section hybridized with 35S-labeled bovine antisense TNF-R2 riboprobe showing expression of TNF-R2 in both cortex and medulla (left panel). Hybridization with the sense probe was used as a negative control (right panel). B, Colocalization studies using a bovine antisense CgA riboprobe labeled with digoxigenin (dark staining in the medulla) and 35S-labeled TNF-R2 riboprobe (arrowheads in cortex and medulla) showed that TNF-R2 is expressed in chromaffin cells. C, Hybridization with TNF-R2 and CgA sense probes showing only background labeling.
Characterization of signaling effectors for TNF-α induction of neuropeptide gene expression
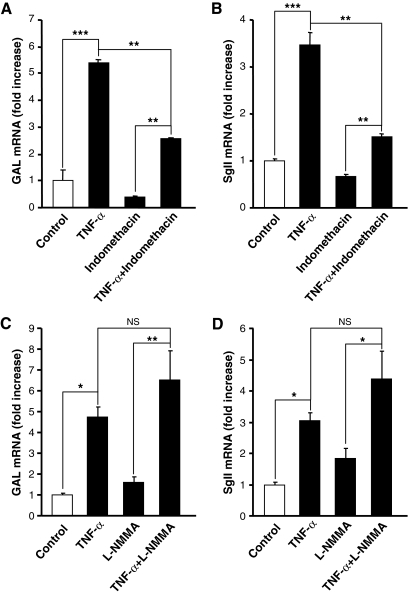
Induction of neuropeptide biosynthesis in adrenochromaffin cells by cytokines including TNF-α occurs only several hours after initiation of treatment and requires de novo protein synthesis during the first 12–24 h (3), raising the possibility that TNF-α’s action could be carried out by the generation of a soluble mediator, such as prostaglandins (PGs) or NO, key effectors in neuroimmune communication whose production is regulated by proinflammatory cytokines in other cell types (14,15,16). Indeed, several studies have shown that both primary chromaffin cells and pheochromocytomas are capable of the induction of cyclooxygenase 2 or neuronal NO synthase (17,18) and the subsequent production of PGs or NO under various conditions besides cytokine stimulation (19,20,21). To assess the possibility that TNF-α induction of neuropeptide gene expression occurs through an indirect mechanism involving generation of either PGs or NO, we assessed the effect of the cyclooxygenase inhibitor indomethacin and the NO synthase inhibitor l-NMMA on TNF-α-induced GAL and SgII gene expression (Fig. 2). Incubation of chromaffin cells during 48 h in the presence of indomethacin (1 μm) had no effect on the fold induction of GAL mRNA by TNF-α (Fig. 2A), although net GAL mRNA levels were significantly lower in the presence of indomethacin plus TNF-α than after TNF-α alone. These results suggest that the effect of TNF-α itself on GAL gene expression is not mediated by PGs, although PGs may well have a role in neuropeptide regulation during inflammation, by influencing basal neuropeptide expression. SgII mRNA was likewise induced by TNF-α in both the presence and absence of indomethacin, although there was a reduction in both the fold induction by TNF-α and the net levels of SgII mRNA after TNF-α due to pretreatment with indomethacin (Fig. 2B). These data indicate that PGs on their own may have a role in neuropeptide regulation during inflammation by influencing basal neuropeptide gene expression in those cells and are consistent with previous studies in which treatment of chromaffin cells with PGE2 was shown to elevate SgII and proenkephalin A (Pro-Enk A) mRNA and secretoneurin peptide levels, acting mainly via the calcium/calmodulin signaling pathway (22,23). Exposure of chromaffin cells to the NO synthase inhibitor l-NMMA (2 mm) was without effect on either basal levels of GAL or SgII mRNA or on the magnitude of TNF-α induction of neuropeptide gene expression (Fig. 2, C and D), ruling out a role for NO in TNF-α induction of neuropeptide expression.
Figure 2.
Effect of inhibiting prostaglandin or nitric oxide production on TNF-α induction of neuropeptide gene expression. Chromaffin cells were incubated for 48 h in control conditions or with 10 nm TNF-α, in the absence or presence of 1 μm indomethacin or 2 mm l-NMMA (C and D) as described in Materials and Methods. GAL (A) and SgII (B) mRNA levels, determined by Q-RT-PCR, are expressed as fold increase over corresponding control values, and represent means ± sem of four determinations for each condition from one experiment representative of three different experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 vs. the corresponding control (one-way ANOVA test, Bonferroni posttest). NS, Not significant.
TNF-α activates NF-κB transcription factor activity in adrenochromaffin cells
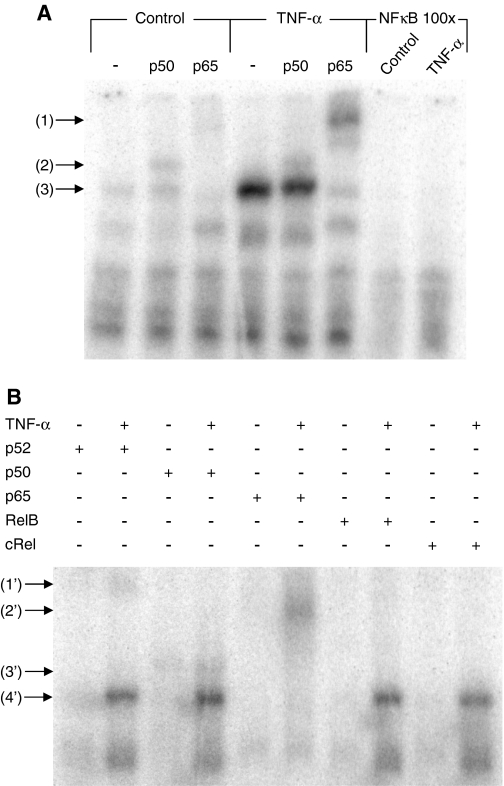
We have previously shown that TNF-α induces the binding activity of NF-κB transcription factor by an ERK 1/2-independent mechanism in chromaffin cells (3). To identify the type of NF-κB complex proteins induced by TNF-α, a supershift assay was performed using antibodies directed against five distinct subunits, p52, p50, p65, RelB, and cRel, which are all potential components of NF-κB. As shown in Fig. 3, only incubation with an anti-p65 antibody completely abolished the binding of chromaffin cell nuclear proteins to the NF-κB site. There appeared to be only a minor supershifting of this DNA-protein complex with either p52 or p50 antibodies, and none was detectable with anti-RelB or anti-cRel antibodies (Fig. 3). NF-κB is normally found in the cytoplasm in a complex with IκB and other components, and upon phosphorylation, ubiquitinylation, and degradation of IκB, NF-κB is liberated to move to the nucleus and bind to target genes, enhancing or inhibiting their transcription (24). NF-κB is actually a hetero- or homodimer consisting in members of the NF-κB/Rel family of pleiotropic transcription factors, which are well characterized in the context of regulation of multiple genes involved in immune and inflammatory responses as well as cell proliferation, oncogenesis, differentiation, apoptosis, and tumorigenesis (25,26). Homodimers of p50 or p52 are inhibitors of transcription, because they lack a functional transcriptional activation domain (26). Although p65-p50 and p65-p52 heterodimers are the most common functional form of NF-κB stimulating transcription (27), formation and transcriptional activity of p65 homodimer complexes have been demonstrated (28,29). Our data suggest that a p65 homodimer is likely the most common NF-κB species present in chromaffin cells and is therefore likely to mediate TNF-α’s NF-κB-dependent effects, although minor contributions of p52 or p50 species cannot be excluded.
Figure 3.
TNF-α activates NF-κB transcription factor in adrenochromaffin cells. A, Chromaffin cells were incubated with TNF-α (10 nm) for 2 h, and nuclear extracts were prepared and assayed for their binding to the 32P-labeled NF-κB site. Supershift assays of the NF-κB-bound complex were obtained in the presence of vehicle-treated or TNF-α-treated nuclear extracts, using antibodies directed against two NF-κB subunits, p50 and p65. The symbol (−) denotes control supershift reactions with no antibody. The binding specificity of the NF-κB site to vehicle-treated or TNF-α-treated nuclear extracts was verified by adding a 100-fold excess of unlabeled NF-κB oligonucleotide. The supershifted complexes formed by p65 and p50 are indicated by the arrows 1 and 2, whereas arrow 3 indicates the shift of NF-κB. B, Supershift assays of the NF-κB-bound complex obtained in the presence of vehicle-treated or TNF-α-treated nuclear extracts, using antibodies directed against five subunits p52, p50, p65, RelB, and cRel. The supershifted p52, p50, and p65 complexes are indicated by the arrows 1′, 2′, and 3′, respectively. The arrow 4′ indicates the shift of NF-κB.
Microarray analysis indicates long-term activation of NF-κB by TNF-α in BCCs
To further understand the mechanisms involved in the effect of TNF-α on chromaffin cell gene transcription, we analyzed gene expression changes in BCCs both early and late after TNF-α treatment. In the absence of a bovine cDNA or oligonucleotide microarray, we initially performed cross-species hybridization (CSH) of BCC cDNA from TNF-treated and untreated cells to a mouse oligonucleotide microarray. Microarray analysis at 6 and 48 h demonstrated most strikingly a cohort of NF-κB-related transcripts up-regulated early and persistently by TNF-R2 activation by TNF-α in BCCs (Tables 1 and 2). One target of NF-κB up-regulated early (6 h) and persistently (48 h) by TNF-α in BCCs was IκB. The IκB gene is activated by NF-κB in all cells examined to date. Its induction represents a negative feedback loop for NF-κB signaling because translation of the IκB protein results in reestablishment of NF-κB inactivation as a complex with IκB in the cytoplasm, inhibition of NF-κB signaling, and subsequent down-regulation of the NF-κB-mediated IκB gene transcription. For this reason, elevation of IκB mRNA levels represents a real-time monitor of ongoing NF-κB activation in the cell (30,31). Consistent with persistent up-regulation of the IκB/NF-κB pathway, another NF-κB-regulated transcript, TNF-α-induced protein 3, also known as A20 (32), was also up-regulated early and persistently by TNF-α treatment. In contrast, neuroendocrine-specific transcripts such as those encoding the catecholamine biosynthetic enzyme phenylethanolamine N-methyltransferase and the neuropeptide SgII were unchanged at 6 h but up-regulated significantly at 48 h. Thus, the microarray analysis revealed a general pattern of persistent up-regulation of transcripts encoding NF-κB-associated protein signaling factors and only later expression of multiple chromaffin cell-specific genes (Tables 2 and 3). These results suggested that signaling through NF-κB might occur through a novel neuroendocrine cell-specific mechanism and stimulated us to search for a set of bovine- and chromaffin cell-specific transcripts that might shed additional light on the properties of TNF-α signaling mediated through NF-κB specifically in chromaffin cells.
Table 1.
Named genes induced in 6-h TNF-α-treated chromaffin cells vs. control based on microarray analysis
| Gene | Ratio | Accession no. | GeneID |
|---|---|---|---|
| Signal transduction | |||
| Nuclear factor of κ-light chain gene enhancer in B-cells inhibitor, α | 5.69 ± 0.77 | NM_010907 | 18035 |
| Guanine nucleotide binding protein (G protein), γ5 subunit | 3.33 ± 0.04 | NM_010318 | 14707 |
| Tumor necrosis factor, α-induced protein 3 | 2.83 ± 0.32 | NM_009397 | 21929 |
| Chimerin (chimaerin) 2 | 2.15 ± 0.06 | NM_023543 | 69993 |
| Secretion | |||
| Synapsin III | 2.49 ± 0.18 | NM_013722 | 27204 |
| Immune response | |||
| Proteasome (prosome, macropain) 28 subunit, β, transcript variant 1 | 2.24 ± 0.08 | NM_011190 | 19188 |
| Proliferation/differentiation | |||
| Fibroblast growth factor 2 | 2.09 ± 0.04 | NM_008006 | 14173 |
| Unknown function | |||
| IBR domain containing 3 | 2.49 ± 0.01 | XM_991982 | 75234 |
The GenBank accession no. and the Gene cluster (GeneID) for each gene are indicated. The ratios ± sem were determined from three different experiments. The highlighted genes (in bold) correspond to TNF-α-stimulated genes in both 6- and 48-h microarray.
Table 2.
Named genes stimulated in 48 h TNF-α-treated chromaffin cells vs. control based on microarray analysis
| Gene | Ratio | Accession no. | GeneID |
|---|---|---|---|
| Signal transduction | |||
| Nuclear factor of κ light chain gene enhancer in B-cells inhibitor, α | 5.17 ± 0.38 | NM_010907 | 18035 |
| Tnf receptor associated factor 4 | 2.83 ± 0.08 | NM_009423 | 22032 |
| Guanine nucleotide binding protein (G protein), γ5 subunit | 2.66 ± 0.07 | NM_010318 | 14707 |
| Leucine-rich repeat kinase 2 | 2.66 ± 0.11 | NM_025730 | 66725 |
| Dual specificity phosphatase 10 | 2.57 ± 0.19 | NM_022019 | 63953 |
| Hypoxia inducible factor 1, α subunit | 2.49 ± 0.11 | NM_010431 | 15251 |
| Nucleotide-binding oligomerization domain containing 1 | 2.44 ± 0.06 | NM_172729 | 107607 |
| Forkhead box P4 | 2.44 ± 0.08 | NM_028767 | 74123 |
| CXXC finger 1 (PHD domain) | 2.36 ± 0.03 | NM_028868 | 74322 |
| Tumor necrosis factor, α-induced protein 3 | 2.33 ± 0.17 | NM_009397 | 21929 |
| PRP6 pre-mRNA splicing factor 6 homolog (yeast) | 2.25 ± 0.12 | NM_133701 | 68879 |
| Homeo box, msh-like 1 | 2.25 ± 0.12 | NM_010835 | 17701 |
| Bone morphogenetic protein receptor, type 1A | 2.14 ± 0.04 | BC042611 | 12166 |
| Zinc finger protein 239 (Zfp239), transcript variant 2 | 2.05 ± 0.04 | NM_008616 | 22685 |
| Immune response | |||
| Histocompatibility 2, Q region locus 10 | 3.33 ± 0.07 | BC011215 | 15007 |
| Histocompatibility 2, Q region locus 2 | 3.23 ± 0.18 | NM_010392 | 15013 |
| Histocompatibility 2, D region locus 1 | 3.10 ± 0.10 | NM_010380 | 14964 |
| Programmed cell death 1 ligand 2 | 2.38 ± 0.06 | NM_021396 | 58205 |
| Defensin β 7 | 2.21 ± 0.07 | NM_139220 | 246080 |
| Gene model 1960 (NCBI) | 2.08 ± 0.04 | NM_203320 | 330122 |
| Metabolism | |||
| Hexosaminidase B | 3.11 ± 0.02 | NM_010422 | 15212 |
| Ubiquitin specific peptidase 53 | 2.96 ± 0.05 | NM_133857 | 99526 |
| Phenylethanolamine-N-methyltransferase | 2.90 ± 0.27 | NM_008890 | 18948 |
| Cathepsin K | 2.74 ± 0.14 | NM_007802 | 13038 |
| Acetylcholinesterase | 2.66 ± 0.07 | NM_009599 | 11423 |
| Phosphoenolpyruvate carboxykinase 2 (mitochondrial) | 2.62 ± 0.13 | NM_028994 | 74551 |
| Enolase 1, α-nonneuron | 2.60 ± 0.17 | NM_023119 | 13806 |
| RAD52 homolog (S. cerevisiae) | 2.55 ± 0.12 | NM_011236 | 19365 |
| Proprotein convertase subtilisin/kexin type 7 | 2.52 ± 0.09 | NM_008794 | 18554 |
| F-box protein 17 | 2.40 ± 0.17 | NM_015796 | 50760 |
| Procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), α-polypeptide III | 2.38 ± 0.12 | NM_177161 | 320452 |
| arginase type II | 2.36 ± 0.08 | NM_009705 | 11847 |
| Protein-l-isoaspartate (d-aspartate) O-methyltransferase 1 | 2.30 ± 0.02 | NM_008786 | 18537 |
| Glycosyltransferase 25 domain containing 2 | 2.24 ± 0.01 | NM_177756 | 269132 |
| Glutamine fructose-6-phosphate transaminase 2 | 2.23 ± 0.12 | NM_013529 | 14584 |
| Traffic/secretion | |||
| Secretogranin II | 3.07 ± 0.13 | NM_009129 | 20254 |
| Hermansky-Pudlak syndrome 4 homolog (human) | 2.82 ± 0.12 | BC004668 | 192232 |
| Coatomer protein complex, subunit zeta 2 | 2.76 ± 0.18 | NM_019877 | 56358 |
| Uncoupling protein 2 (mitochondrial, proton carrier) | 2.57 ± 0.12 | NM_011671 | 22228 |
| Sorting nexin 2 | 2.40 ± 0.13 | NM_026386 | 67804 |
| Synuclein, α-interacting protein (synphilin) | 2.28 ± 0.12 | NM_026408 | 67847 |
| Proliferation/differentiation | |||
| Reelin | 2.93 ± 0.22 | NM_011261 | 19699 |
| Fibroblast growth factor 2 | 2.72 ± 0.09 | NM_008006 | 14173 |
| Myosin IB | 2.22 ± 0.09 | NM_010863 | 17912 |
| Vascular endothelial growth factor A, transcript variant 2 | 2.18 ± 0.05 | NM_009505 | 22339 |
| Unknown function | |||
| Moesin | 2.58 ± 0.05 | NM_010833 | 17698 |
| Zinc finger protein 291 | 2.55 ± 0.05 | NM_001081341 | 244891 |
| IBR domain containing 3 | 2.53 ± 0.04 | XM_991982 | 75234 |
| Hydrocephalus inducing | 2.47 ± 0.12 | NM_172916 | 244653 |
| Histidine triad nucleotide binding protein 2 | 2.42 ± 0.07 | NM_026871 | 68917 |
| Similar to l-lactate dehydrogenase A chain (LDH muscle subunit) | 2.42 ± 0.14 | XR_002509 | 385319 |
| Tripartite motif-containing 58 | 2.40 ± 0.12 | NM_001039047 | 216781 |
| Killer cell lectin-like receptor subfamily K, member 1, transcript variant 2 | 2.39 ± 0.05 | NM_001083322 | 27007 |
| Fibronectin type III domain containing 4 | 2.38 ± 0.22 | NM_022424 | 64339 |
| Hippocalcin-like 4 | 2.30 ± 0.14 | NM_174998 | 170638 |
| Spermine binding protein-like | 2.29 ± 0.16 | NM_001077421 | 638345 |
| Similar to ribosomal protein L18a | 2.26 ± 0.03 | XR_003475 | 625760 |
| Spermatogenic Zip 1 | 2.25 ± 0.09 | NM_030237 | 79401 |
| CUB and Sushi multiple domains 1 | 2.23 ± 0.10 | NM_053171 | 94109 |
| U1 small nuclear ribonucleoprotein polypeptide A | 2.19 ± 0.08 | NM_009224 | 20637 |
| Similar to Kruppel-like factor 5 | 2.13 ± 0.05 | XR_002503 | 546990 |
| Similar to solute carrier family 25, member A6 | 2.13 ± 0.02 | XR_003714 | 672115 |
| Seizure related 6 homolog like | 2.11 ± 0.05 | NM_019982 | 56747 |
| SET binding factor 1 | 2.09 ± 0.02 | NM_001081030 | 77980 |
The GenBank accession no. and the Gene cluster (GeneID) for each gene are indicated. The ratios ± sem were determined from three different experiments. The highlighted genes (in bold) correspond to TNF-α-stimulated genes in both 6- and 48-h microarray.
Table 3.
Named genes induced in 48-h TNF-α-treated chromaffin cells vs. control based on SSH analysis
| Gene | Ratio | Accession no. | Gene ID |
|---|---|---|---|
| Signal transduction | |||
| Nuclear factor of κ-light polypeptide gene enhancer in B-cells inhibitor, α | 2.47 ± 0.51 | NM_020529 | 4792 |
| Calcium modulating ligand | 1.83 ± 0.62 | NM_001745 | 819 |
| Mitogen-inducible gene 6 | 1.81 ± 0.38 | NM_018948 | 54206 |
| Stathmin-like 2 | 1.72 ± 0.26 | AK092187 | 11075 |
| Butyrate-induced transcript 1 | 1.52 ± 0.29 | BC058912 | 51495 |
| Transforming growth factor β receptor I | 1.51 ± 0.17 | BC071181 | 7046 |
| Intercellular signaling | |||
| Vasoactive intestinal polypeptide | 2.23 ± 0.28 | NM_173970 | 280956 |
| Proenkephalin | 1.88 ± 0.65 | NM_174141 | 281387 |
| Neuronal pentraxin | 1.69 ± 0.16 | NM_002522 | 4884 |
| Traffic/secretion | |||
| Secretogranin II (Chromogranin C) | 2.02 ± 0.75 | NM_174176 | 281477 |
| Sec61 γ subunit | 1.57 ± 0.28 | BC051840 | 23480 |
| Spastic paraplegia 3A (autosomal dominant) | 1.56 ± 0.21 | NM_181598 | 51062 |
| Metabolism | |||
| Similar to cytochrome P450 CYP3A24 | 2.40 ± 0.66 | XM_595414 | 517246 |
| Ubiquitin specific peptidase 12 | 1.65 ± 0.19 | AF022789 | 219333 |
| Phosphatidylinositol glycan, class S | 1.60 ± 0.15 | BC001319 | 94005 |
| Ferritin, heavy polypeptide 1 | 1.60 ± 0.41 | AF540563 | 281173 |
| Cellular adhesion/angiogenesis | |||
| Thrombospondin 1 | 1.69 ± 0.39 | NM_003246 | 7057 |
| Morphogenesis | |||
| Decorin | 1.61 ± 0.57 | NM_173906 | 280760 |
| Cell death | |||
| Tumor protein, translationally-controled 1 | 1.57 ± 0.24 | BC052333 | 7178 |
| Unknown function | |||
| VGF nerve growth factor inducible | 2.02 ± 0.37 | NM_003378 | 7425 |
| Selenoprotein K | 1.87 ± 0.22 | BC013162 | 58515 |
| Ribosomal protein L36a pseudogene | 1.59 ± 0.47 | NG_001571 | 207032 |
| LUC7-like 2 | 1.53 ± 0.23 | BC050708 | 51631 |
The GenBank accession no. and the Gene cluster (GeneID) for each gene are indicated. The ratios ± sem were determined from four different experiments.
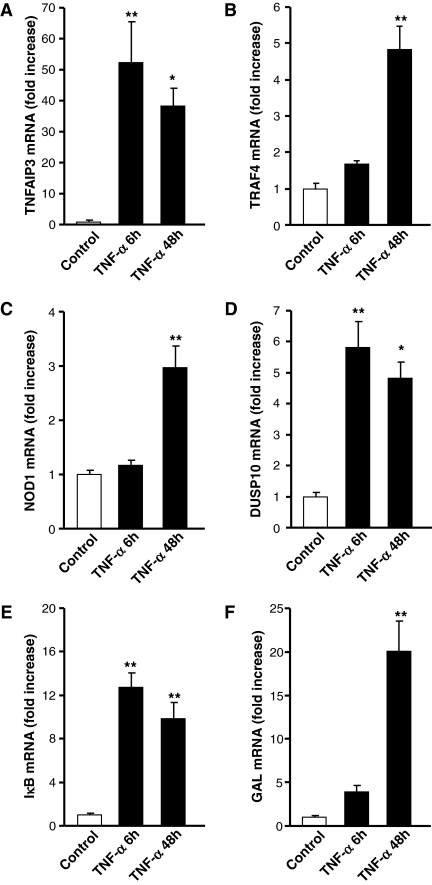
Because CSH is considered a nonstandard use of microarray technology (33), with a high potential for false-negative results, based on lack of hybridization due to input-to-target sequence mismatching, we assessed the validity of positive CSH results by measuring independently with Q-RT-PCR the effect of treatment with TNF-α at 6 and 48 h on five transcripts detected as up-regulated by bovine-mouse microarray hybridization (TRAF4, TNFAIP3, DUSP10, NOD1, and IκB), with GAL mRNA regulation determined as a positive control (Fig. 4, A–F). TNF-α (10 nm) induced a significant stimulation, ranging from 3- to 50-fold, of TNFAIP3 and IκB mRNA at 6 and 48 h (Fig. 4, A and E) and TRAF4 and NOD1 at 48 h (Fig. 4, B and C) as predicted by microarray. Only the results for DUSP10 mRNA levels as measured by Q-RT-PCR (5- to 6-fold at both 6 and 48 h; Fig. 4D) were slightly discordant from the microarray results (significant induction of DUSP10 at 48 but not 6 h after TNF-α), possibly attributable to cross-hybridization among the multiple DUSP targets present on the microarray, as reported for other large gene families (34).
Figure 4.
Time course of TNFAIP3, TRAF4, DUSP10, NOD1, and IκB gene stimulation by TNF-α determined by Q-RT-PCR. Cultured BCCs were incubated for 6 and 48 h in control conditions or with 10 nm TNF-α, and TNFAIP3 (A), TRAF4 (B), NOD1 (C), DUSP10 (D), IκB (E), and GAL (F) mRNA levels were measured by Q-RT-PCR as described in Materials and Methods. Values are the mean ± sem of four determinations for each condition. *, P < 0.05; **, P < 0.01 vs. the corresponding control (one-way ANOVA test, Bonferroni posttest).
SSH/macroarray analyses: expression of NF-κB-associated factors by TNF-α provides a potential mechanism for long-term NF-κB activation in BCCs
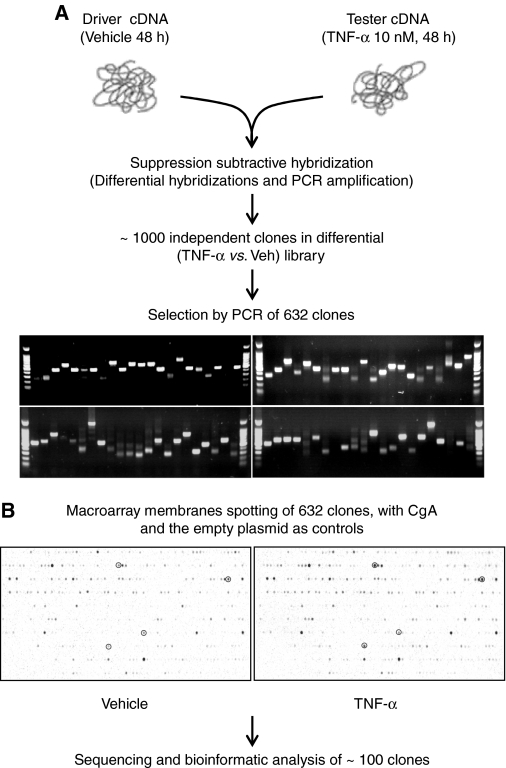
To examine further possible chromaffin cell-specific modulation of the NF-κB pathway by TNF-α, we next focused on the BCC transcriptome directly, to identify transcripts not represented through cross-hybridization to the mouse microarray (above). SSH, derived from differential display PCR (35) and cDNA representational difference analysis (36), makes it possible to isolate such chromaffin cell-specific genes. Using SSH, we were able to select more than 600 differentially expressed clones after 48 h of treatment of BCCs with TNF-α (Fig. 5A). To validate the results obtained with this technique, we spotted the cDNA isolated on nylon membranes and the macroarrays obtained were hybridized with reverse transcribed RNA extracted from untreated chromaffin cells or cells treated with TNF-α. Hybridization results from four independent cellular cultures allowed the identification of about 100 differentially expressed transcripts, with an induction threshold of 1.5-fold, which were then sequenced and identified bioinformatically (Fig. 5B). These 100 isolated clones corresponded to at least 30 unique transcripts, or TNF-α-target genes, classified in functional categories (Table 3) based on the consortium Gene Ontology criteria. Representation of VIP, SgII, and Pro-Enk A among these clones provided validation to the SSH technique because these three transcripts were previously shown by independent techniques to be up-regulated after TNF-α treatment of BCCs (3).
Figure 5.
Generation and validation by hybridization of a SSH library from TNF-α-treated BCCs. A, Schematic depiction of generation of SSH clone set. Chromaffin cells were incubated with TNF-α (10 nm) for 48 h, RNA was extracted, reverse transcribed, ligated to amplification primer-adapters (tester cDNA), denatured, and hybridized to excess control BCC cDNA (driver cDNA). Doubly primer-adapted (subtractive hybridization nonsuppressed) cDNA was amplified and cloned. The TNF-α vs. control differential library contained more than 1000 clones from which 632 inserts, ranging in size from about 500 to about 1500 bp were amplified. B, The 632 amplified inserts were spotted on nylon membranes, and the macroarrays obtained were hybridized with labeled cDNA from chromaffin cells treated with TNF-α (right) or vehicle (left) for 48 h. Four differentially expressed clones are circled, for purposes of illustration, from a total of 100 individual clones found to be differentially expressed based on densitometric analysis of hybridization results generated from four independent experiments similar to the one shown here. These clones were sequenced and identified through bioinformatics analysis and found to represent 23 distinct differentially expressed transcripts.
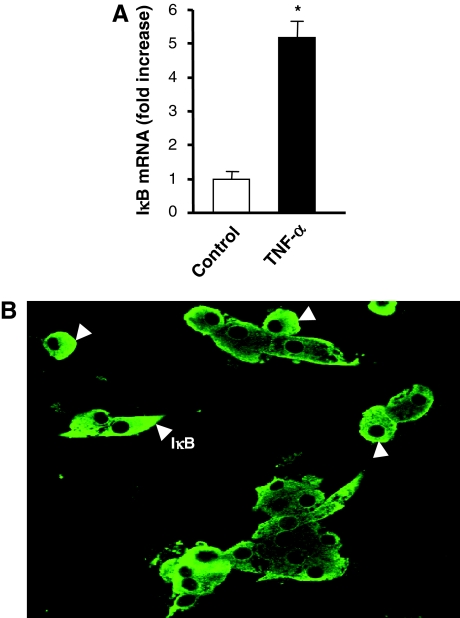
SSH confirms the persistent regulation of IκB mRNA induction by TNF-α in BCCs (Table 3). These results were further confirmed by Q-RT-PCR at the mRNA level (Fig. 6A), whereas immunocytochemistry showed that IκB protein was readily detected in cultured BCCs after 48 h of treatment by TNF-α (Fig. 6B). Together, these data indicate that NF-κB activation by TNF-α in BCCs persists over the entire 48-h treatment period, despite the ongoing translation and expression of IκB.
Figure 6.
Regulation of IκB by TNF-α in BCCs. A, Chromaffin cells were incubated for 48 h in control conditions or with 10 nm TNF-α, and IκB mRNA levels were measured by Q-RT-PCR as described in Materials and Methods. Values are the mean ± sem of four separate determinations for each condition from one experiment representative of three different experiments and are expressed as a fold increase over control untreated cells. *, P < 0.05 vs. control (Mann-Whitney U test). B, Chromaffin cells treated with 10 nm TNF-α for 48 h were fixed and processed for immunofluorescence. IκB was visualized by using an appropriate antibody and Alexa-488-conjugated goat antirabbit IgGs. IκB cytoplasmic staining is indicated by arrowheads in a representative field.
Several additional transcripts implicated in NF-κB signaling were revealed by SSH analysis. Thus, TNF-α stimulated expression of the gene encoding mitogen-inducible gene-6 (MIG-6), an adapter molecule induced by stress (37), which activates NF-κB by disrupting IκB/NF-κB binding (38), and also increased the level of mRNA encoding butyrate-induced transcript 1 (B-ind1), a factor shown to potentiate the activation of NF-κB by the GTPase Rac 1 (39).
The induction of calcium-modulating ligand by TNF-α at 48 h suggests a possible mechanism for stabilization of NF-κB activation in BCCs (40,41). Several studies have implicated Ca2+ as a second messenger for NF-κB activation in response to various stimuli in T lymphocytes and also in neurons (42,43,44). Activation of NF-κB triggered by glutamate in neurons (45,46), and by various stressors in other cell types (47,48,49,50), occurs via Ca2+ mobilization. Moreover, action of immunomodulatory factors such as epidermal growth factor, substance P, or TNF-α requires a Ca2+ accumulation to induce the activation of NF-κB (51,52,53). These data are consistent with the idea that Ca2+ mobilization, via the persistent activation and expression of calcium-modulating ligand, may play a role in supporting long-term activation of NF-κB by TNF-α in BCCs. Enhanced calcium mobilization might also play a role in activation of other transcription factors such as AP-1 induced by TNF-α in both monocytes (54) and BCCs (3).
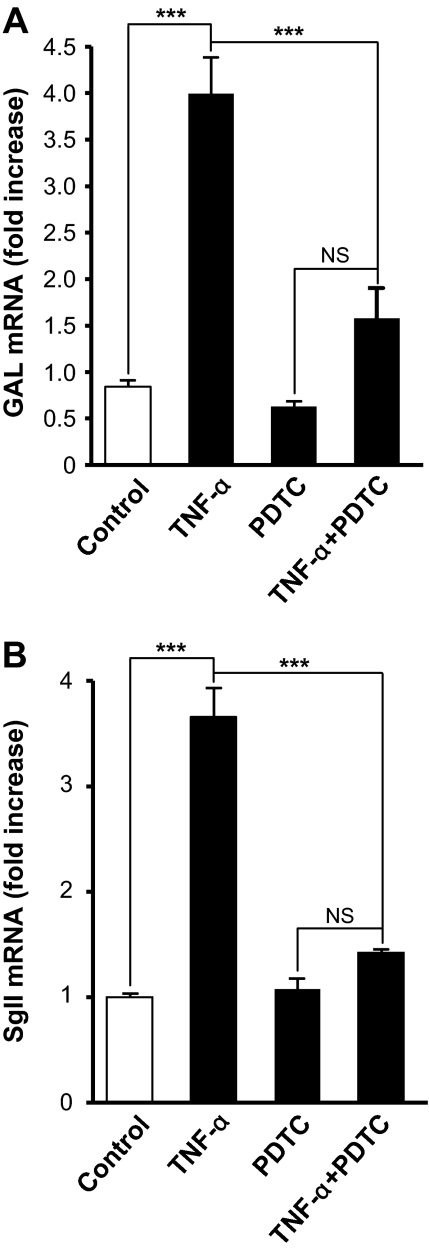
Role for NF-κB in TNF-α-regulated neuropeptide gene expression
Compared with the well documented role of AP-1 in neuropeptide gene regulation in adrenochromaffin cells (3,55,56), little is known about the involvement of NF-κB in neuropeptide gene transcription. To link NF-κB activation to the stimulatory effect of TNF-α on neuropeptide gene expression, BCCs were treated with TNF-α in the presence of PDTC, a specific inhibitor of NF-κB (57). PDTC (100 μm) suppressed the stimulatory effect of TNF-α on GAL and SgII mRNA levels at 48 h (Fig. 7, A and B). Taken together with our previous findings (3), these data suggest that the stimulatory effect of TNF-α on neuropeptide gene transcription in BCCs required the activation of both AP-1 and NF-κB. The effects of NF-κB on neuropeptide gene expression may be both direct and indirect. The GAL gene contains NF-κB binding sites (58) as does the Pro-Enk gene, whose transcription is activated by NF-κB in T cells (59). We have not yet examined the SgII promoter for the presence of a functional NF-κB response element.
Figure 7.
Implication of NF-κB in the effect of TNF-α on neuropeptide gene expression. Chromaffin cells were incubated for 48 h in control conditions or with 10 nm TNF-α, in the presence or absence of 100 mm PDTC as described in Materials and Methods. GAL (A) and SgII (B) mRNA levels, determined by Q-RT-PCR, are expressed as fold increase over corresponding control values and represents means ± sem of four determinations for each condition from one experiment representative of three different experiments. NS, Not significant (P > 0.05); ***, P < 0.001 vs. TNF-α alone (one-way ANOVA test, Bonferroni posttest).
TNF-α signaling in neuroendocrine cells: uniquely long-term regulation leading to neuropeptide gene expression
We have in the present report used two complementary approaches to analyze the transitional transcriptome created by TNF-α, underlying long-term regulation of neuropeptide gene expression in neuroendocrine cells: interspecies microarray (bovine on mouse), which allows the possibility of genome-wide, high-complexity screening for the regulation of highly conserved transcripts (interspecies microarray), albeit at a high threshold to eliminate potential false positives, and a technique of lower complexity (SSH) that is, however, not limited by loss of transcripts for which interspecies conservation is poor. The limitations of interspecies oligonucleotide microarray are apparent in the detection of only 25% of neuropeptide transcripts (SgII, but not Enk, VIP, or GAL at the stringency cutoff employed in Table 2), with detection of 75% of them using SSH (Table 3). The limitations of SSH, perhaps due to lack of enrichment of some classes of transcripts at the driver-tester discrimination step due to secondary structure, limited number of clones characterized by macroarray hybridization, or other unknown factors, are apparent in the lack of detection of several NF-κB-regulated transcripts (e.g. TRAF4, TNFAIP3, and NOD1) detected in the microarray screen and validated by Q-RT-PCR. However, despite the limitations of each approach, the combined use of interspecies microarray and SSH, in the absence of a bovine microarray, was sufficient to make the mechanistic connection between persistent NF-κB-dependent signaling and long-term regulation of neuropeptide gene expression described here.
The mechanism of TNF-α signaling in chromaffin cells is unique. It features immediate and persistent up-regulation of a group of NF-κB-dependent genes and results in NF-κB-dependent delayed transcription of a cohort of genes encoding neuropeptides that when released from the adrenal chromaffin cells have regulatory functions systemically and locally at the adrenal cortex. Most significantly, NF-κB-mediated signaling by TNF-α through the TNF-R2 in chromaffin cells provides a mechanism for regulating the expression of glucocorticoid biosynthesis-promoting neuropeptides based on long-term sampling by chromaffin cells of ΤΝF-α levels in blood and blood-borne TNF-α-expressing cells. Persistence of TNF-α signaling through NF-κB in chromaffin cells contrasts sharply with self-limiting signaling through NF-κB in immune/inflammatory cells mediated through IκB (30). The suppression of IκB inhibition in chromaffin cells is likely due to the up-regulation of factors such as mitogen-inducible gene-6 (MIG-6) and butyrate-induced transcript 1 (B-ind1), which could counterregulate the inhibitory effects of IκB induction by negating its inhibitory effects on NF-κB itself. Work is in progress to delineate the molecular basis for the neuroendocrine cell specificity of expression of these factors and the precise kinetics of their induction relative to IκB and neuropeptide gene induction in the chromaffin cell. The present results will also allow further determination of whether NF-κB acts directly on neuropeptide gene transcription or indirectly through persistent transcription, activation, or stabilization of other transcription factors that act directly on neuropeptide gene promoters.
Acknowledgments
We thank Drs. Abdel G. Elkahloun [National Human Genome Research Institute, National Institutes of Health (NIH)] and Babru Samal (National Institute of Mental Health, NIH) for assistance with microarray analysis and Huguette Lemonnier, Chang-Mei Hsu, Patrice Bizet, and David Huddleston for skillful technical assistance.
Footnotes
This work was supported by the INSERM Unité 413, the Conseil Regional de Haute-Normandie, the COMETE-3 Network (Grant PHRC AOM 06179), NIMH-IRP 1Z01-MH002386-20, and an NIMH Integrative Neuro-Immune Program (INIP) Post-Doctoral Fellowship (to D.A.-A.).
Disclosure Statement: The authors of this manuscript have nothing to disclose.
First Published Online February 21, 2008
Abbreviations: AP-1, Activator protein-1; BCC, bovine chromaffin cell; CgA, chromogranin A; CSH, cross-species hybridization; GAL, galanin; IκB, inhibitor κB; NF, nuclear factor; l-NMMA, NG-monomethyl-l-arginine; PDTC, pyrrolidine dithiocarbamate; PG, prostaglandin; Pro-Enk A, proenkephalin A; Q-RT-PCR, quantitative RT-PCR; SgII, secretogranin II; SSC, standard saline citrate; SSH, suppression subtractive hybridization; TNF-R1, type 1 TNF-α receptor; VIP, vasoactive intestinal polypeptide.
References
- Wrona D 2006 Neural-immune interactions: an integrative view of the bidirectional relationship between the brain and immune systems. J Neuroimmunol 172:38–58 [DOI] [PubMed] [Google Scholar]
- Andreis PG, Tortorella C, Ziolkowska A, Spinazzi R, Malendowicz LK, Neri G, Nussdorfer GG 2007 Evidence for a paracrine role of endogenous adrenomedullary galanin in the regulation of glucocorticoid secretion in the rat adrenal gland. Int J Mol Med 19:511–515 [PubMed] [Google Scholar]
- Ait-Ali D, Turquier V, Grumolato L, Yon L, Jourdain M, Alexandre D, Eiden LE, Vaudry H, Anouar Y 2004 The proinflammatory cytokines tumor necrosis factor-α and interleukin-1 stimulate neuropeptide gene transcription and secretion in adrenochromaffin cells via activation of extracellularly regulated kinase 1/2 and p38 protein kinases, and activator protein-1 transcription factors. Mol Endocrinol 18:1721–1739 [DOI] [PubMed] [Google Scholar]
- Bornstein SR, Haidan A, Ehrhart-Bornstein M 1996 Cellular communication in the neuro-adrenocortical axis: role of vasoactive intestinal polypeptide (VIP). Endocr Res 22:819–829 [DOI] [PubMed] [Google Scholar]
- Kruppa G, Thoma B, Machleidt T, Wiegmann K, Kronke M 1992 Inhibition of tumor necrosis factor (TNF)-mediated NF-κB activation by selective blockade of the human 55-kDa TNF receptor. J Immunol 148:3152–3157 [PubMed] [Google Scholar]
- Laegreid A, Medvedev A, Nonstad U, Bombara MP, Ranges G, Sundan A, Espevik T 1994 Tumor necrosis factor receptor p75 mediates cell-specific activation of nuclear factor kappa B and induction of human cytomegalovirus enhancer. J Biol Chem 269:7785–7791 [PubMed] [Google Scholar]
- McFarlane SM, Jupp OJ, Cobban HJ, Hunter I, Anderson HM, Vandenabeele P, Nixon GF, MacEwan DJ 2001 Stimulation of stress-activated but not mitogen-activated protein kinases by tumour necrosis factor receptor subtypes in airway smooth muscle. Biochem Pharmacol 61:749–759 [DOI] [PubMed] [Google Scholar]
- Thommesen L, Laegreid A 2005 Distinct differences between TNF receptor 1- and TNF receptor 2-mediated activation of NFκB. J Biochem Mol Biol 38:281–289 [DOI] [PubMed] [Google Scholar]
- Marchetti L, Klein M, Schlett K, Pfizenmaier K, Eisel UL 2004 Tumor necrosis factor (TNF)-mediated neuroprotection against glutamate-induced excitotoxicity is enhanced by N-methyl-d-aspartate receptor activation. Essential role of a TNF receptor 2-mediated phosphatidylinositol 3-kinase-dependent NF-κB pathway. J Biol Chem 279:32869–32881 [DOI] [PubMed] [Google Scholar]
- Anouar Y, MacArthur L, Cohen J, Iacangelo AL, Eiden LE 1994 Identification of a TPA-responsive element mediating preferential transactivation of the galanin gene promoter in chromaffin cells. J Biol Chem 269:6823–6831 [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N 1987 Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162:156–159 [DOI] [PubMed] [Google Scholar]
- Alexandre D, Vaudry H, Jegou S, Anouar Y 2000 Structure and distribution of the mRNAs encoding pituitary adenylate cyclase-activating polypeptide and growth hormone-releasing hormone-like peptide in the frog, Rana ridibunda. J Comp Neurol 421:234–246 [DOI] [PubMed] [Google Scholar]
- Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, Siebert PD 1996 Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci USA 93:6025–6030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais V, Rivest S 2004 Effects of TNF-α and IFN-γ on nitric oxide-induced neurotoxicity in the mouse brain. J Immunol 172:7043–7052 [DOI] [PubMed] [Google Scholar]
- Pascual RM, Carr EM, Seeds MC, Guo M, Panettieri Jr RA, Peters SP, Penn RB 2006 Regulatory features of interleukin-1β-mediated prostaglandin E2 synthesis in airway smooth muscle. Am J Physiol Lung Cell Mol Physiol 290:L501–L508 [DOI] [PubMed] [Google Scholar]
- Tammali R, Ramana KV, Srivastava SK 2007 Aldose reductase regulates TNF-α-induced PGE2 production in human colon cancer cells. Cancer Lett 252:299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdan C 2001 Nitric oxide and the immune response. Nat Immunol 2:907–916 [DOI] [PubMed] [Google Scholar]
- Rivest S 2001 How circulating cytokines trigger the neural circuits that control the hypothalamic-pituitary-adrenal axis. Psychoneuroendocrinology 26:761–788 [DOI] [PubMed] [Google Scholar]
- Salmenkivi K, Haglund C, Ristimaki A, Arola J, Heikkila P 2001 Increased expression of cyclooxygenase-2 in malignant pheochromocytomas. J Clin Endocrinol Metab 86:5615–5619 [DOI] [PubMed] [Google Scholar]
- Vicente S, Gonzalez MP, Oset-Gasque MJ 2002 Neuronal nitric oxide synthase modulates basal catecholamine secretion in bovine chromaffin cells. J Neurosci Res 69:327–340 [DOI] [PubMed] [Google Scholar]
- Zhang WY, Gotoh T, Oyadomari S, Mori M 2000 Coinduction of inducible nitric oxide synthase and arginine recycling enzymes in cytokine-stimulated PC12 cells and high output production of nitric oxide. Brain Res Mol Brain Res 83:1–8 [DOI] [PubMed] [Google Scholar]
- Suh HW, Hudson PM, Hong JS 1995 Expression of the proenkephalin A gene and [Met5]-enkephalin secretion induced by arachidonic acid in bovine adrenal medullary chromaffin cells: involvement of second messengers. J Neurochem 64:608–613 [DOI] [PubMed] [Google Scholar]
- Wolkersdorfer M, Egger C, Laslop A, Fischer-Colbrie R 1996 Nicotine and prostaglandin E induce secretogranin II levels in bovine chromaffin cells. Brain Res Mol Brain Res 38:260–266 [DOI] [PubMed] [Google Scholar]
- Ghosh S, Karin M 2002 Missing pieces in the NF-κB puzzle. Cell 109(Suppl):S81–S96 [DOI] [PubMed] [Google Scholar]
- Karin M, Cao Y, Greten FR, Li ZW 2002 NF-κB in cancer: from innocent bystander to major culprit. Nat Rev Cancer 2:301–310 [DOI] [PubMed] [Google Scholar]
- Li Q, Verma IM 2002 NF-κB regulation in the immune system. Nat Rev Immunol 2:725–734 [DOI] [PubMed] [Google Scholar]
- Beinke S, Ley SC 2004 Functions of NF-κB1 and NF-κB2 in immune cell biology. Biochem J 382:393–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek S, Huang DB, Huxford T, Ghosh S, Ghosh G 2003 X-ray crystal structure of an IκBβ × NF-κB p65 homodimer complex. J Biol Chem 278:23094–23100 [DOI] [PubMed] [Google Scholar]
- Rahman A, Anwar KN, True AL, Malik AB 1999 Thrombin-induced p65 homodimer binding to downstream NF-κB site of the promoter mediates endothelial ICAM-1 expression and neutrophil adhesion. J Immunol 162:5466–5476 [PubMed] [Google Scholar]
- Chiao PJ, Miyamoto S, Verma IM 1994 Autoregulation of IκBα activity. Proc Natl Acad Sci USA 91:28–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SC, Ganchi PA, Ballard DW, Greene WC 1993 NF-κB controls expression of inhibitor IκBα: evidence for an inducible autoregulatory pathway. Science 259:1912–1915 [DOI] [PubMed] [Google Scholar]
- Cooper JT, Stroka DM, Brostjan C, Palmetshofer A, Bach FH, Ferran C 1996 A20 blocks endothelial cell activation through a NF-κB-dependent mechanism. J Biol Chem 271:18068–18073 [DOI] [PubMed] [Google Scholar]
- Bar-Or C, Czosnek H, Koltai H 2007 Cross-species microarray hybridizations: a developing tool for studying species diversity. Trends Genet 23:200–207 [DOI] [PubMed] [Google Scholar]
- Hansen A, Chen Y, Inman JM, Phan QN, Qi ZQ, Xiang CC, Palkovits M, Cherman N, Kuznetsov SA, Robey PG, Mezey E, Brownstein MJ 2007 Sensitive and specific method for detecting G protein-coupled receptor mRNAs. Nat Methods 4:35–37 [DOI] [PubMed] [Google Scholar]
- Liang P, Pardee AB 1992 Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science 257:967–971 [DOI] [PubMed] [Google Scholar]
- Hubank M, Schatz DG 1994 Identifying differences in mRNA expression by representational difference analysis of cDNA. Nucleic Acids Res 22:5640–5648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makkinje A, Quinn DA, Chen A, Cadilla CL, Force T, Bonventre JV, Kyriakis JM 2000 Gene 33/Mig-6, a transcriptionally inducible adapter protein that binds GTP-Cdc42 and activates SAPK/JNK. A potential marker transcript for chronic pathologic conditions, such as diabetic nephropathy. Possible role in the response to persistent stress. J Biol Chem 275:17838–17847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunoda T, Inokuchi J, Baba I, Okumura K, Naito S, Sasazuki T, Shirasawa S 2002 A novel mechanism of nuclear factor κB activation through the binding between inhibitor of nuclear factor-κBα and the processed NH2-terminal region of Mig-6. Cancer Res 62:5668–5671 [PubMed] [Google Scholar]
- Courilleau D, Chastre E, Sabbah M, Redeuilh G, Atfi A, Mester J 2000 B-ind1, a novel mediator of Rac1 signaling cloned from sodium butyrate-treated fibroblasts. J Biol Chem 275:17344–17348 [DOI] [PubMed] [Google Scholar]
- Bram RJ, Crabtree GR 1994 Calcium signalling in T cells stimulated by a cyclophilin B-binding protein. Nature 371:355–358 [DOI] [PubMed] [Google Scholar]
- Holloway MP, Bram RJ 1998 Co-localization of calcium-modulating cyclophilin ligand with intracellular calcium pools. J Biol Chem 273:16346–16350 [DOI] [PubMed] [Google Scholar]
- Lilienbaum A, Israel A 2003 From calcium to NF-κB signaling pathways in neurons. Mol Cell Biol 23:2680–2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan NM, Bren GD, Frantz B, Tocci MJ, O'Neill EA, Paya CV 1995 Regulation of IκBα phosphorylation by PKC- and Ca2+-dependent signal transduction pathways. J Immunol 155:4685–4691 [PubMed] [Google Scholar]
- Trushin SA, Pennington KN, Algeciras-Schimnich A, Paya CV 1999 Protein kinase C and calcineurin synergize to activate IκB kinase and NF-κB in T lymphocytes. J Biol Chem 274:22923–22931 [DOI] [PubMed] [Google Scholar]
- Grilli M, Goffi F, Memo M, Spano P 1996 Interleukin-1β and glutamate activate the NF-κB/Rel binding site from the regulatory region of the amyloid precursor protein gene in primary neuronal cultures. J Biol Chem 271:15002–15007 [DOI] [PubMed] [Google Scholar]
- Guerrini L, Blasi F, Denis-Donini S 1995 Synaptic activation of NF-κB by glutamate in cerebellar granule neurons in vitro. Proc Natl Acad Sci USA 92:9077–9081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gewirtz AT, Rao AS, Simon Jr PO, Merlin D, Carnes D, Madara JL, Neish AS 2000 Salmonella typhimurium induces epithelial IL-8 expression via Ca2+-mediated activation of the NF-κB pathway. J Clin Invest 105:79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B, Logsdon CD 2000 CCK stimulates mob-1 expression and NF-kappaB activation via protein kinase C and intracellular Ca2+. Am J Physiol Cell Physiol 278:C344–C351 [DOI] [PubMed] [Google Scholar]
- Jefferson KK, Smith Jr MF, Bobak DA 1999 Roles of intracellular calcium and NF-κB in the Clostridium difficile toxin A-induced up-regulation and secretion of IL-8 from human monocytes. J Immunol 163:5183–5191 [PubMed] [Google Scholar]
- Pahl HL, Baeuerle PA 1996 Activation of NF-κB by ER stress requires both Ca2+ and reactive oxygen intermediates as messengers. FEBS Lett 392:129–136 [DOI] [PubMed] [Google Scholar]
- Quinlan KL, Naik SM, Cannon G, Armstrong CA, Bunnett NW, Ansel JC, Caughman SW 1999 Substance P activates coincident NF-AT- and NF-κB-dependent adhesion molecule gene expression in microvascular endothelial cells through intracellular calcium mobilization. J Immunol 163:5656–5665 [PubMed] [Google Scholar]
- Sun L, Carpenter G 1998 Epidermal growth factor activation of NF-κB is mediated through IκBα degradation and intracellular free calcium. Oncogene 16:2095–2102 [DOI] [PubMed] [Google Scholar]
- Tomsig JL, Sohma H, Creutz CE 2004 Calcium-dependent regulation of tumour necrosis factor-α receptor signalling by copine. Biochem J 378:1089–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra JP, Mishra S, Gee K, Kumar A 2005 Differential involvement of calmodulin-dependent protein kinase II-activated AP-1 and c-Jun N-terminal kinase-activated EGR-1 signaling pathways in tumor necrosis factor-α and lipopolysaccharide-induced CD44 expression in human monocytic cells. J Biol Chem 280:26825–26837 [DOI] [PubMed] [Google Scholar]
- Anouar Y, Lee HW, Eiden LE 1999 Both inducible and constitutive activator protein-1-like transcription factors are used for transcriptional activation of the galanin gene by different first and second messenger pathways. Mol Pharmacol 56:162–169 [DOI] [PubMed] [Google Scholar]
- Turquier V, Yon L, Grumolato L, Alexandre D, Fournier A, Vaudry H, Anouar Y 2001 Pituitary adenylate cyclase-activating polypeptide stimulates secretoneurin release and secretogranin II gene transcription in bovine adrenochromaffin cells through multiple signaling pathways and increased binding of pre-existing activator protein-1-like transcription factors. Mol Pharmacol 60:42–52 [DOI] [PubMed] [Google Scholar]
- Schreck R, Meier B, Mannel DN, Droge W, Baeuerle PA 1992 Dithiocarbamates as potent inhibitors of nuclear factor κB activation in intact cells. J Exp Med 175:1181–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler B, Evans HF, Liu ML, Falls V, Iismaa TP, Shine J, Herzog H 1995 Characterization of the 5′-flanking region of the human preprogalanin gene. DNA Cell Biol 14:321–329 [DOI] [PubMed] [Google Scholar]
- Rattner A, Korner M, Rosen H, Baeuerle PA, Citri Y 1991 Nuclear factor κB activates proenkephalin transcription in T lymphocytes. Mol Cell Biol 11:1017–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]