Abstract
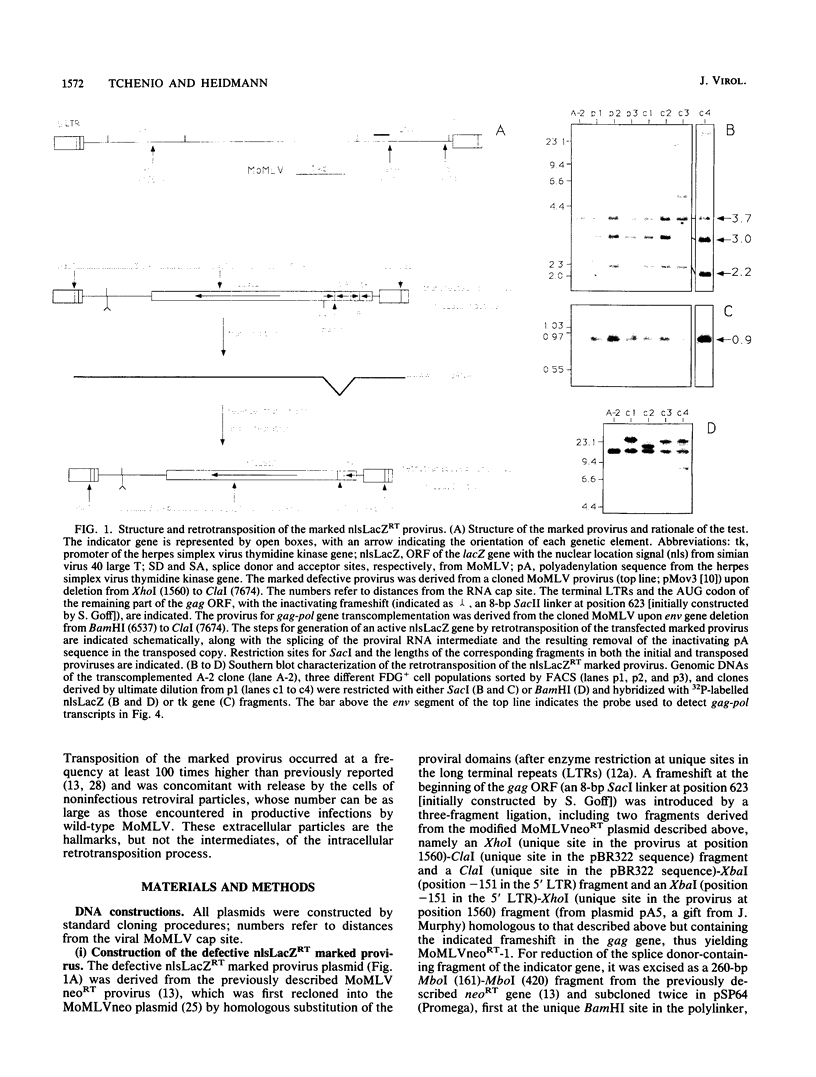
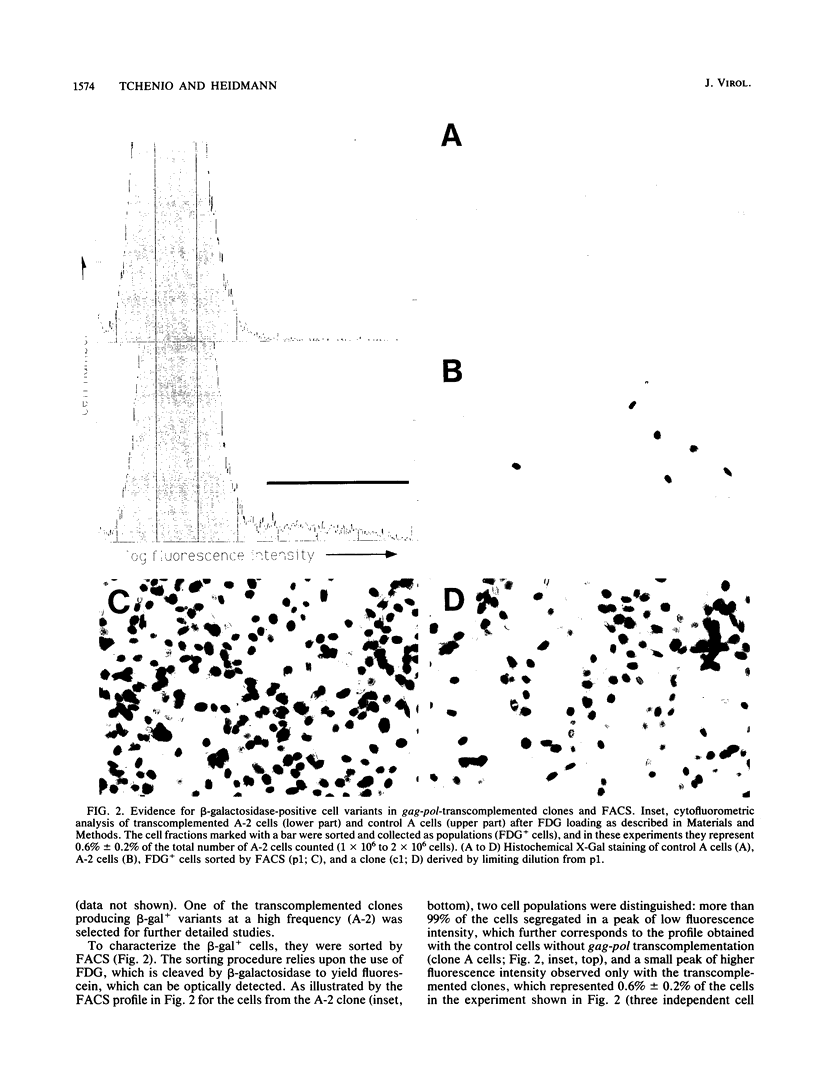
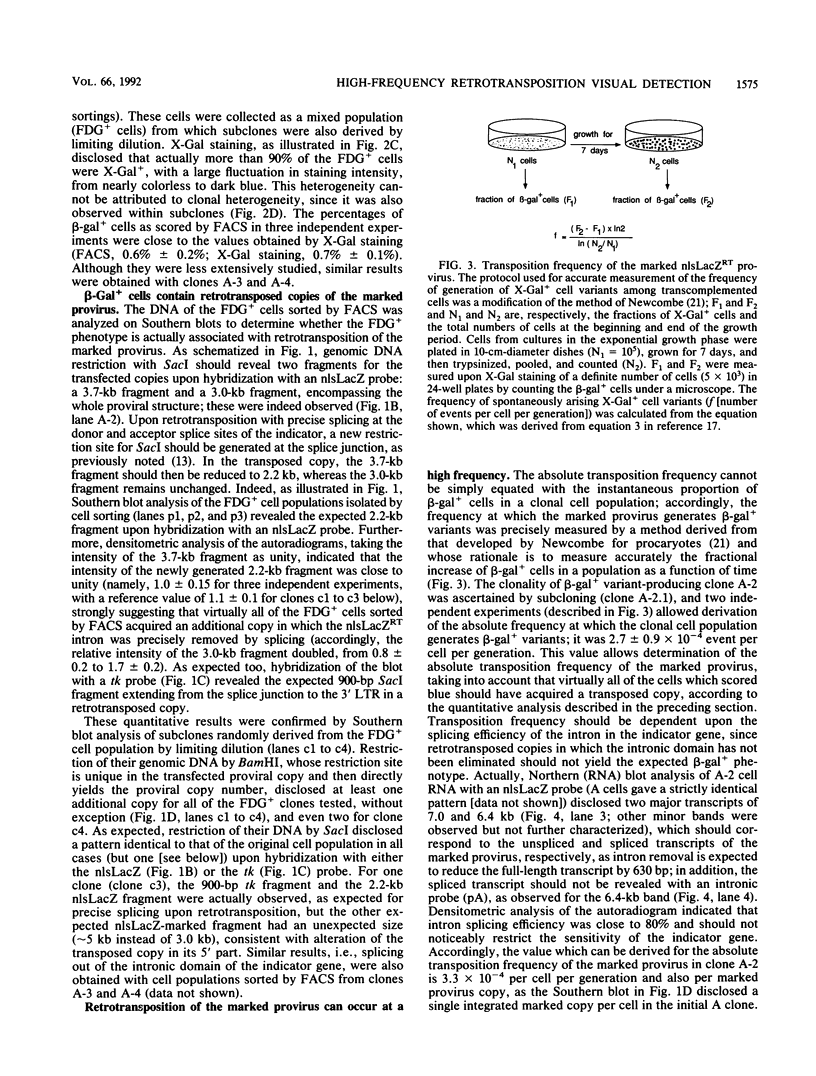
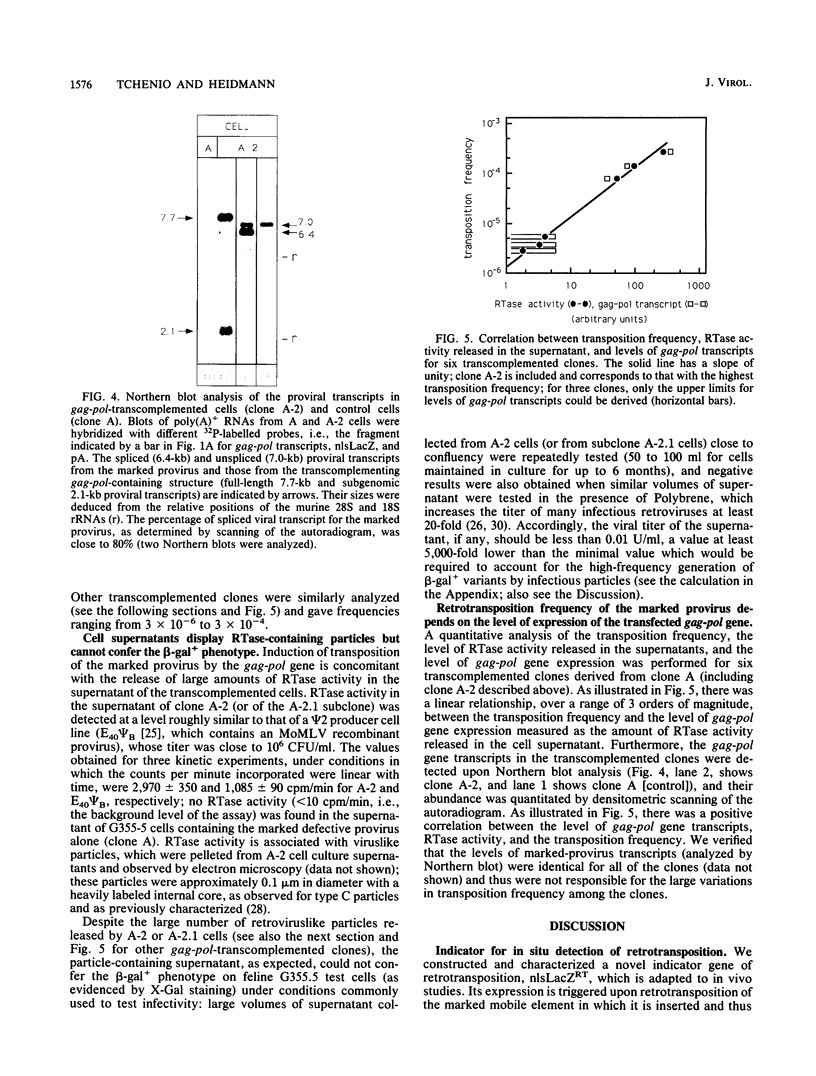
We devised an indicator gene for retrotransposition, nlsLacZRT, which contains the Escherichia coli lacZ gene fused to a nuclear location signal (nlsLacZ), engineered in such a way that the gene is expressed only if the structure in which it has been inserted transposes itself through an RNA intermediate. A cloned murine leukemia retrovirus with an ecotropic host range (Moloney murine leukemia virus), rendered defective by a large deletion encompassing the three viral gag, pol, and env open reading frames, was marked with this indicator gene and introduced by transfection into heterologous feline cells. No beta-galactosidase activity could be detected among the clonal cell population, unless the defective provirus was complemented in trans by the gag-pol gene products. Under these conditions, cell variants which disclosed an easily detectable nuclear blue coloration upon in situ 5-bromo-4-chloro-3-indolyl-beta-D-galactopyranoside staining were observed. Fluorescence-activated cell sorting of the beta-galactosidase-positive cells, followed by Southern blot analysis, demonstrated an unambiguous correlation between nlsLacZRT activation and retrotransposition of the marked provirus. Transposition occurs at a high frequency (up to 10(-4) events per cell per generation), which is dependent on the level of expression of the gag-pol gene and is concomitant with the release of noninfectious retroviruslike particles which are the hallmarks, but not the intermediates, of the intracellular transposition process.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armentano D., Yu S. F., Kantoff P. W., von Ruden T., Anderson W. F., Gilboa E. Effect of internal viral sequences on the utility of retroviral vectors. J Virol. 1987 May;61(5):1647–1650. doi: 10.1128/jvi.61.5.1647-1650.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender M. A., Palmer T. D., Gelinas R. E., Miller A. D. Evidence that the packaging signal of Moloney murine leukemia virus extends into the gag region. J Virol. 1987 May;61(5):1639–1646. doi: 10.1128/jvi.61.5.1639-1646.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnerot C., Rocancourt D., Briand P., Grimber G., Nicolas J. F. A beta-galactosidase hybrid protein targeted to nuclei as a marker for developmental studies. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6795–6799. doi: 10.1073/pnas.84.19.6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dührsen U., Stahl J., Gough N. M. In vivo transformation of factor-dependent hemopoietic cells: role of intracisternal A-particle transposition for growth factor gene activation. EMBO J. 1990 Apr;9(4):1087–1096. doi: 10.1002/j.1460-2075.1990.tb08214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echalier G. Drosophila retrotransposons: interactions with genome. Adv Virus Res. 1989;36:33–105. doi: 10.1016/s0065-3527(08)60582-5. [DOI] [PubMed] [Google Scholar]
- Gallagher R. E., Gallo R. C. Type C RNA tumor virus isolated from cultured human acute myelogenous leukemia cells. Science. 1975 Jan 31;187(4174):350–353. doi: 10.1126/science.46123. [DOI] [PubMed] [Google Scholar]
- Harbers K., Schnieke A., Stuhlmann H., Jähner D., Jaenisch R. DNA methylation and gene expression: endogenous retroviral genome becomes infectious after molecular cloning. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7609–7613. doi: 10.1073/pnas.78.12.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberlein C., Kawai M., Franz M. J., Beck-Engeser G., Daniel C. P., Ostertag W., Stocking C. Retrotransposons as mutagens in the induction of growth autonomy in hematopoietic cells. Oncogene. 1990 Dec;5(12):1799–1807. [PubMed] [Google Scholar]
- Heidmann O., Heidmann T. Retrotransposition of a mouse IAP sequence tagged with an indicator gene. Cell. 1991 Jan 11;64(1):159–170. doi: 10.1016/0092-8674(91)90217-m. [DOI] [PubMed] [Google Scholar]
- Heidmann T., Heidmann O., Nicolas J. F. An indicator gene to demonstrate intracellular transposition of defective retroviruses. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2219–2223. doi: 10.1073/pnas.85.7.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon D., Roberts B. L., Richardson W. D., Smith A. E. A short amino acid sequence able to specify nuclear location. Cell. 1984 Dec;39(3 Pt 2):499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Kuff E. L. Intracisternal A particles in mouse neoplasia. Cancer Cells. 1990 Dec;2(12):398–400. [PubMed] [Google Scholar]
- Kuff E. L., Lueders K. K. The intracisternal A-particle gene family: structure and functional aspects. Adv Cancer Res. 1988;51:183–276. doi: 10.1016/s0065-230x(08)60223-7. [DOI] [PubMed] [Google Scholar]
- Luria S. E., Delbrück M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics. 1943 Nov;28(6):491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooslehner K., Karls U., Harbers K. Retroviral integration sites in transgenic Mov mice frequently map in the vicinity of transcribed DNA regions. J Virol. 1990 Jun;64(6):3056–3058. doi: 10.1128/jvi.64.6.3056-3058.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan R. C., Berg P. Selection for animal cells that express the Escherichia coli gene coding for xanthine-guanine phosphoribosyltransferase. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2072–2076. doi: 10.1073/pnas.78.4.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mével-Ninio M., Mariol M. C., Gans M. Mobilization of the gypsy and copia retrotransposons in Drosophila melanogaster induces reversion of the ovo dominant female-sterile mutations: molecular analysis of revertant alleles. EMBO J. 1989 May;8(5):1549–1558. doi: 10.1002/j.1460-2075.1989.tb03539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan G. P., Fiering S., Nicolas J. F., Herzenberg L. A. Fluorescence-activated cell analysis and sorting of viable mammalian cells based on beta-D-galactosidase activity after transduction of Escherichia coli lacZ. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2603–2607. doi: 10.1073/pnas.85.8.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein J. L., Nicolas J. F., Jacob F. Construction of a retrovirus capable of transducing and expressing genes in multipotential embryonic cells. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7137–7140. doi: 10.1073/pnas.81.22.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes J. R., Rubenstein J. L., Nicolas J. F. Use of a recombinant retrovirus to study post-implantation cell lineage in mouse embryos. EMBO J. 1986 Dec 1;5(12):3133–3142. doi: 10.1002/j.1460-2075.1986.tb04620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherdin U., Rhodes K., Breindl M. Transcriptionally active genome regions are preferred targets for retrovirus integration. J Virol. 1990 Feb;64(2):907–912. doi: 10.1128/jvi.64.2.907-912.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchenio T., Heidmann T. Defective retroviruses can disperse in the human genome by intracellular transposition. J Virol. 1991 Apr;65(4):2113–2118. doi: 10.1128/jvi.65.4.2113-2118.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima K., Vogt P. K. Enhancement and inhibition of avian sarcoma viruses by polycations and polyanions. Virology. 1969 Jul;38(3):414–426. doi: 10.1016/0042-6822(69)90154-8. [DOI] [PubMed] [Google Scholar]
- de la Luna S., Soria I., Pulido D., Ortín J., Jiménez A. Efficient transformation of mammalian cells with constructs containing a puromycin-resistance marker. Gene. 1988;62(1):121–126. doi: 10.1016/0378-1119(88)90585-9. [DOI] [PubMed] [Google Scholar]