Summary
Targeting protein for Xklp2 (TPX2) activates the Ser/Thr-kinase Aurora A in mitosis and targets it to the mitotic spindle [1, 2]. These effects on Aurora A are mediated by the N-terminal domain of TPX2, whereas a C-terminal fragment has been reported to affect microtubule nucleation [3]. Using the Xenopus system, we identified a novel role of TPX2 during mitosis. Injection of TPX2 or its C-terminus (TPX2-CT) into blastomeres of two-cell embryos led to potent cleavage arrest. Despite cleavage arrest, TPX2-injected embryos biochemically undergo multiple rounds of DNA synthesis and mitosis, and arrested blastomeres have abnormal spindles, clustered centrosomes, and an apparent failure of cytokinesis. In Xenopus S3 cells, transfection of TPX2-FL causes spindle collapse, whereas TPX2-CT blocks pole segregation, resulting in apposing spindle poles with no evident displacement of Aurora A. Analysis of TPX2-CT deletion peptides revealed that only constructs able to interact with the class 5 kinesin-like motor protein Eg5 induce the spindle phenotypes. Importantly, injection of Eg5 into TPX2-CT arrested blastomeres causes resumption of cleavage. These results define a discrete domain within the C-terminus of TPX2 that exerts a novel, Eg5 dependent function in spindle pole segregation.
Keywords: TPX2, Eg5, mitotic spindle, cytokinesis, Xenopus embryos
Results and Discussion
Injection of the TPX2 carboxy-terminus induces cleavage arrest in Xenopus embryos
We injected bacterially expressed, His-tagged TPX2 proteins into one blastomere of a two-cell embryo. Full length TPX2 (TPX2-FL) at about 4-fold over the endogenous level (Fig. S1A) induced cleavage arrest in the injected blastomere, whereas uninjected or BSA-injected blastomeres underwent multiple rounds of cell division and cleavage furrow ingression (Fig. 1A). To identify the domain(s) in TPX2 responsible for cleavage arrest, we generated constructs containing only the TPX2 N-terminal half (TPX2-NT: aa 1–364) or the C-terminal half (TPX2-CT: aa 365–715). Injection of TPX2-NT, which is sufficient for targeting and activating the mitotic Ser/Thr protein kinase Aurora A [1, 4], had no effect on early embryonic divisions. In contrast, TPX2-CT induced potent cleavage arrest in the injected blastomere (Fig. 1A). Importantly, deletion of the last 35 aa in TPX2-CT (TPX2-365–680, herein termed TPX2-CTΔ35) completely abolished this phenotype, and these embryos cleaved normally. Morphologically, blastomeres injected with TPX2-CT looked similar to those injected with Emi1-CT, a mitotic inhibitor [5] (Fig. 1A). To define the minimal region in TPX2 inducing cleavage arrest, we analyzed a panel of TPX2-CT deletion constructs. TPX2 (423–715) and TPX2 (475–715) led to cleavage arrest in all of the injected blastomeres. Shorter fragments, e.g., TPX2 (500–680) and TPX2 (515–715) did not induce any visible defects (Fig. 1A and Fig. S1C), and TPX2 (500–715) was greatly impaired in inducing cleavage arrest (Fig. 1A and S1B). Thus, the minimal fragment inducing the phenotype in all of the injected embryos at a final intracellular concentration of 0.5 μM encompassed amino acids 475–715.
Figure 1. TPX2 induces cleavage arrest in Xenopus embryos and disrupts spindles.
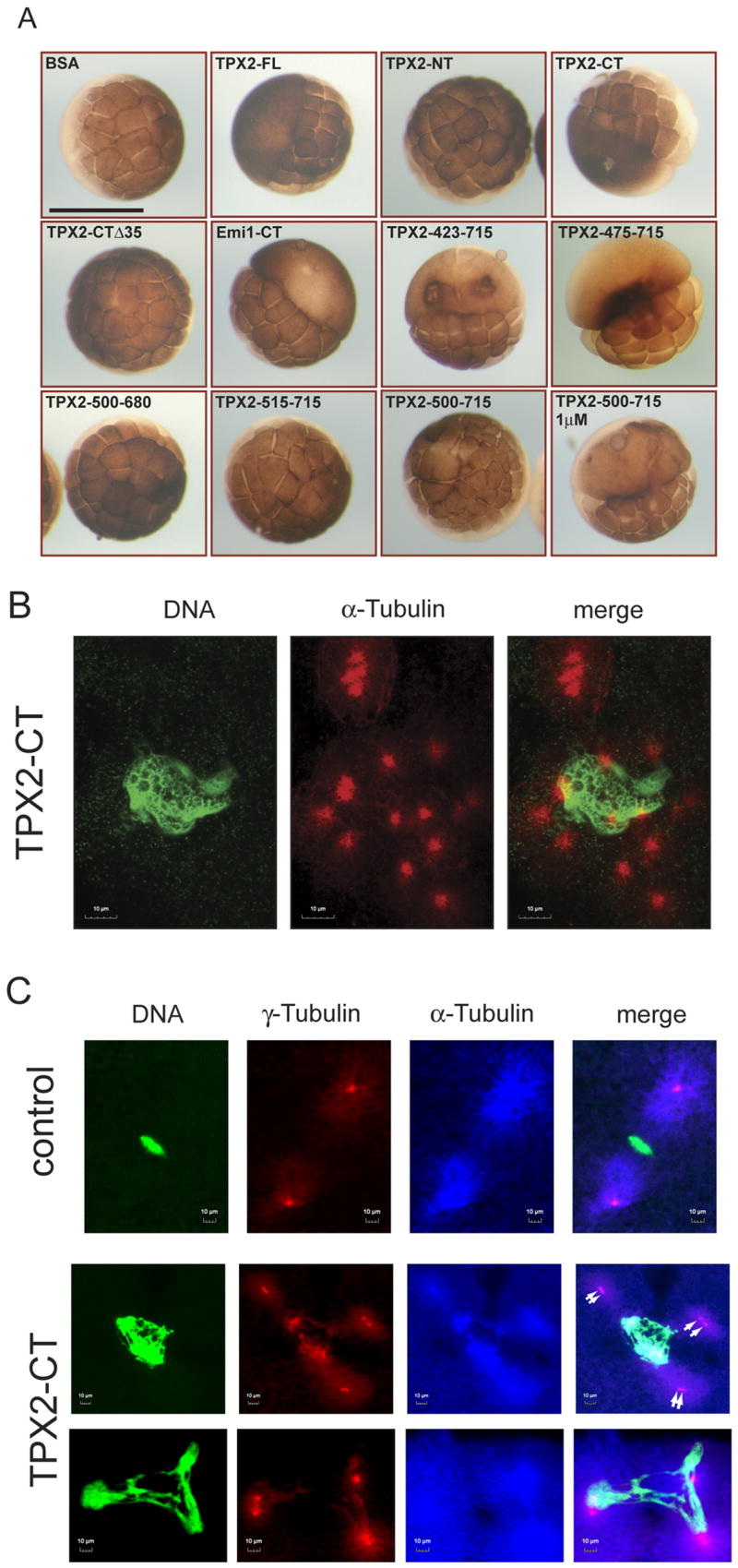
(A) Mapping of cleavage arrest-inducing activity in TPX2 in living Xenopus embryos. One blastomere of a two-cell embryo was injected with the indicated proteins at a final intracellular concentration of 0.5 μM, unless otherwise indicated. For TPX2-FL, TPX2-CT, TPX2 (423–715), TPX2 (475–715) and Emi1-CT, all of the injected blastomeres were arrested. Only TPX2 (500–715) failed to inhibit cleavage in all embryos. Other constructs i.e. TPX2-NT, TPX2-CTΔ35, TPX2 (500–680), TPX2 (515–715) did not have any visible effect on embryos. Three independent experiments were performed, and for each injected protein the number of embryos injected was >25 per experiment. Bar, 1 mm.
(B) Clustered centrosomes in TPX2-CT injected embryos. Confocal laserscan microscopy (CLSM) of Xenopus embryos injected with TPX2-CT. Embryos were fixed ~3 hrs after fertilization and stained for DNA (Sytox Green) and α-tubulin (red). Embryos were analyzed with the 100x oil Plan Apo objective.
(C) Morphology of abnormal mitotic spindles in TPX2-CT injected embryos. Embryos were fixed ~ 2 hrs after fertilization and triple stained for DNA (green), γ-tubulin (red) and α-tubulin (far red, shown in blue) and analyzed using the 100x oil Plan Apo objective. Control embryos exhibit bipolar metaphase spindles containing one centrosome per spindle pole (upper panel). Injection of TPX2-CT leads to formation of abnormal spindles, often with two centrosomes (white arrows) per spindle pole (middle panel) and chromosomes stretched between poles (lower panel).
Embryos injected with TPX2-CT fail to establish a bipolar spindle
Biochemical analyses revealed that embryos injected with Emi1-CT are in mitosis, whereas those injected with TPX2-CT continue to cycle through DNA synthesis and mitosis while cleavage furrow ingression is inhibited (see Fig. S2 and Supplementary Results and Discussion). To investigate the mechanistic events leading to cytokinesis failure, Xenopus embryos injected with various purified TPX2 proteins were analyzed by confocal laserscan microscopy (CLSM). Embryos injected with BSA or TPX2-CTΔ35, a C-terminal construct that does not induce cleavage arrest (see Fig. 1A), exhibited metaphase spindles with chromosomes aligned at the equatorial plate (data not shown). Control embryos contained multiple nuclei associated with one or two centrosomes/spindle poles (Fig. S3). By contrast, embryos injected with TPX2-CT displayed obvious spindle- and chromatin abnormalities and also numerous nuclei, but most of these were not associated with α-tubulin staining or centrosomes/spindle poles (Fig. S3, D-F, white arrows). TPX2-CT arrested embryos formed apparent centrosomes/spindle poles, but these were present as pair-wise structures lacking DNA (Fig. S3, E and G). Strikingly, all TPX2-CT injected embryos analyzed also had one or more nuclei that were larger than other nuclei within the same blastomere and were associated with multiple centrosomes/spindle poles (Fig. S3F, yellow arrow). Higher magnification objectives revealed that these oversized interphase nuclei contain a large mass of DNA surrounded by multiple, pair-wise arranged centrosomes loosely attached to the genetic material (Fig. 1B). Some of these pair-wise arranged centrosomes appeared detached from chromatin, leading to formation of free spindle poles that fail to segregate within the embryo. These pair-wise arranged centrosomes are a predicted result of failure to establish pushing forces by interpolar microtubules that are required for segregation of spindle poles and formation of a bipolar spindle.
To further analyze these spindle poles, we triple-stained TPX2-CT injected embryos for DNA, α- and γ-tubulin. Figure 1C depicts blastomeres in mitosis as judged by condensed DNA and spindle staining. Control embryos in mitosis exhibited chromosomes aligned on the metaphase plate with a bipolar spindle and one centrosome at each spindle pole (Fig. 1C, upper panel). By contrast, embryos injected with TPX2-CT exhibited three or more spindle poles surrounding a large mass of condensed chromatin (Fig. 1C, lower panels). Furthermore, we often observed two neighboring γ-tubulin dots within one spindle pole (Fig. 1C, middle panel, white arrows), most likely a result of two separated centrosomes that have failed to move apart. Moreover, in some cases the condensed chromosomes were stretched between three or more spindle poles, each consisting of two centrosomes (Fig. 1C, lower panel). The stretched chromosome morphology suggests that pulling forces emanate from the poles, and hence poles must be attached to chromatin. We conclude that overexpression of TPX2-CT inhibits forces that push centrosomes apart but does not block microtubule-kinetochore attachments.
Xenopus S3 cells expressing TPX2 or its C-terminus fail to segregate spindle poles and arrest in mitosis
Given that spindle poles do not move apart in Xenopus embryos expressing TPX2-CT, we investigated whether TPX2 would produce a similar phenotype in somatic cells. We transfected Xenopus S3 cells stably expressing GFP-α-tubulin with TPX2 constructs cloned into the pDsRed2-C1 vector that produces proteins fused to a red fluorescent tag. The protein expressed from the parental vector itself did not localize to any mitotic structures (Fig. S4A). In interphase cells, both TPX2-FL and TPX2-NT were exclusively nuclear and as cells approached mitosis, these proteins colocalized with condensed chromosomes (Fig. S4B). By contrast, TPX2-CT was localized mostly in the cytoplasm and concentrated at the centrosome (Fig. S4B, lower panel). This observation supports previous evidence that a functional NLS in TPX2 is centered at aa 284 [6]. Western Blot analysis indicates that the level of expressed TPX2 is about 4–6 times greater than endogenous TPX2 in asynchronous cells, and each expressed protein is at a similar level (Fig. S4C). Strikingly, almost all cells expressing TPX2-FL or TPX2-CT arrested in mitosis with two neighboring spindle poles (Fig. 2). In some cases spindle poles disintegrated (Fig. 2, third panel), and cells unable to enter anaphase or overcome the arrest eventually died (data not shown). Similar results were observed for TPX2 (423–715) and TPX2 (475–715) (Fig. 2A, lower panels).
Figure 2. Xenopus S3 cells, expressing TPX2 proteins, arrest in mitosis with apposed spindle poles.
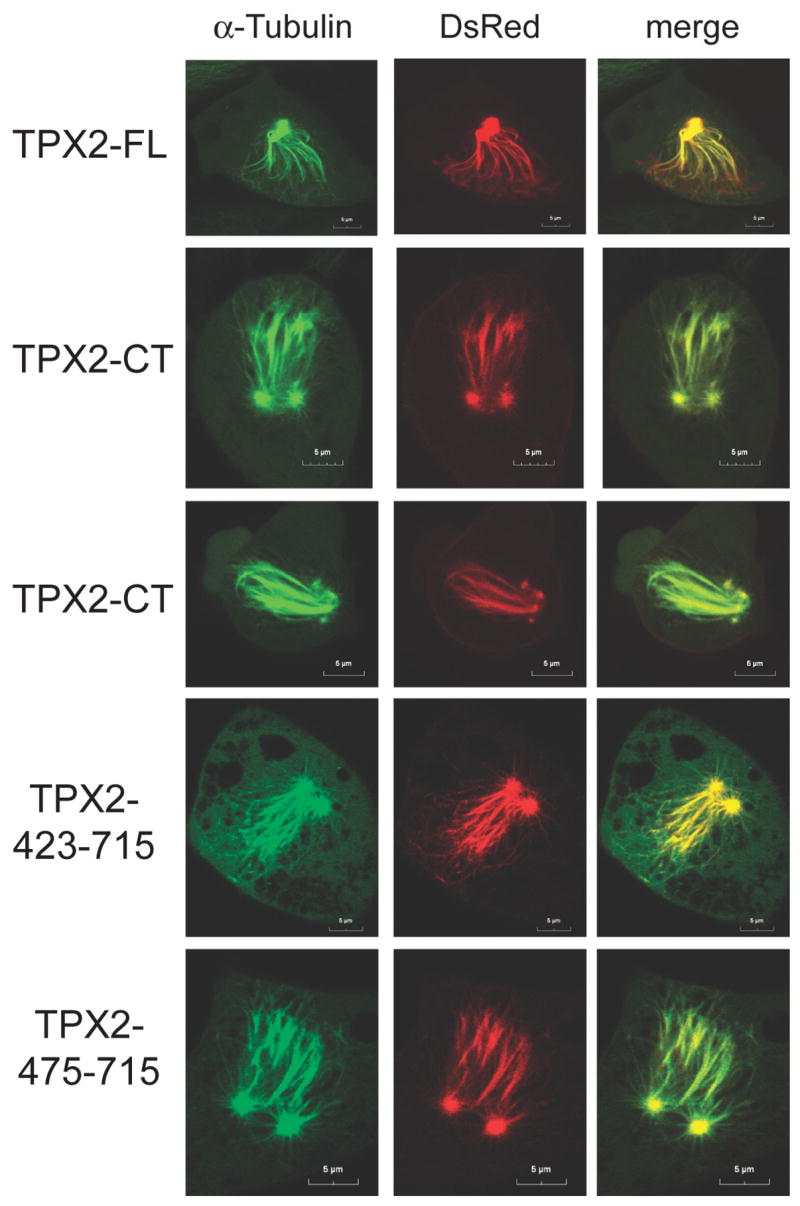
Effect of TPX2 expression in X S3 cells. Live-cell imaging of X S3 cells stably expressing GFP-α-tubulin (green) arrested in mitosis after transfection of DsRed-TPX2 or DsRed-TPX2-CT (red), as indicated. Cells were seeded onto glass bottom wells. Images were taken by CLSM 8 hrs after release from the second thymidine block as described in Experimental Procedures.
These apposing spindle poles show striking similarity to the pair-wise arranged centrosomes within one spindle pole observed in embryos, suggesting that expression of TPX2 or its C-terminal fragments hamper bipolar spindle formation in both systems. Recent studies have shown that both centrosomes and chromosomes are dispensable for inducing cleavage furrow ingression, as this event is mainly triggered by signals emanating from the spindle midzone, including components of the centralspindlin complex [7, 8]. Embryos injected with TPX2-FL or TPX2-CT do not form a functional bipolar spindle, do not exhibit a spindle midzone, and therefore cannot produce a functional centralspindlin complex. We speculate that this accounts for their failure to induce cleavage furrow ingression. As spindle checkpoint signaling is absent in early embryonic cell divisions [9], these embryos continuously undergo multiple rounds of DNA synthesis and mitosis without cleaving. By contrast, mitotic progression in somatic cells is under the control of the spindle assembly checkpoint. Accordingly, S3 cells overexpressing TPX2-FL or TPX2-CT fail to establish or maintain a bipolar spindle and arrest in mitosis as a result of spindle checkpoint signaling.
However, under these conditions microtubule polymerization is not inhibited per se because dense microtubule fibers emanating from the paired spindle poles are evident in the arrested cells. In TPX2-FL transfected cells, TPX2 was still colocalized with mitotic chromosomes and kinetochore microtubules (Fig. 2A, upper panel), indicating that microtubules are connected to chromatin. Transfection of TPX2-FL or TPX2-CT caused cells to arrest in mitosis with two apposing spindle poles from which microtubules point out towards the condensed chromatin, and all these microtubule plus ends are decorated with Aurora B (data not shown), which localizes at kinetochores lacking tension [10]. Bub1 staining revealed that kinetochores in S3 cells expressing TPX2-CT are extensively loaded with Bub1 as compared to TPX2-NT expressing prometaphase cells (Fig. S5A). This suggests that TPX2-CT expressing cells fail to form a functional bipolar spindle and activate the spindle checkpoint, leading to mitotic arrest. Taken together, our results indicate that both embryos and somatic cells expressing the C-terminus of TPX2 lack pushing forces generated by interpolar microtubule connections and fail to move spindle poles apart, whereas microtubule nucleation and capture at chromosomes seems unperturbed because microtubule-kinetochore connections are established.
This distinction is important because it has been reported that a major function of TPX2 is the induction of microtubule nucleation around chromosomes [11]. However, most nucleation studies have been in vitro using pure tubulin solutions or Xenopus egg extracts. A detailed study performed by Brunet and coworkers [3] reported an Aurora A independent role for the C-terminus of TPX2 in microtubule nucleation around chromosomes in vitro. In that study, TPX2 (319–715) was able to restore microtubule nucleation and spindle formation in egg extracts, whereas smaller constructs were deficient in these functions. In contrast, we show here that much smaller constructs, e.g., TPX2 (475–715) are still able to induce cleavage arrest in embryos and failure of spindle pole segregation in S3 cells. Moreover, microtubule-kinetochore connections do not appear to be inhibited in embryos or S3 cells, suggesting that the novel role of TPX2-CT in spindle pole segregation in vivo is distinct from its function in microtubule nucleation around chromosomes.
Given that mutation of Aurora A leads to decreased length of astral microtubules [12], we examined whether the spindle phenotype observed with TPX2-FL or TPX2-CT expression is a consequence of displacing Aurora A from spindle poles. However, Aurora A staining was evident on spindle poles in TPX2-FL or TPX2-CT expressing cells that are arrested in mitosis with the characteristic apposing spindle poles (Fig. S5B). Remarkably, even as spindle poles started to disintegrate, Aurora A staining was still present (Fig. S5B lower middle panel). In addition, expression of TPX2-NT, which targets and activates Aurora A [1, 2], had no effect in either S3 cells or Xenopus embryos. Taken together, the spindle defects observed in TPX2-FL and TPX2-CT overexpressing cells are unlikely to be a consequence of Aurora A mislocalization.
TPX2 induces bipolar spindle collapse and TPX2-CT inhibits spindle pole segregation
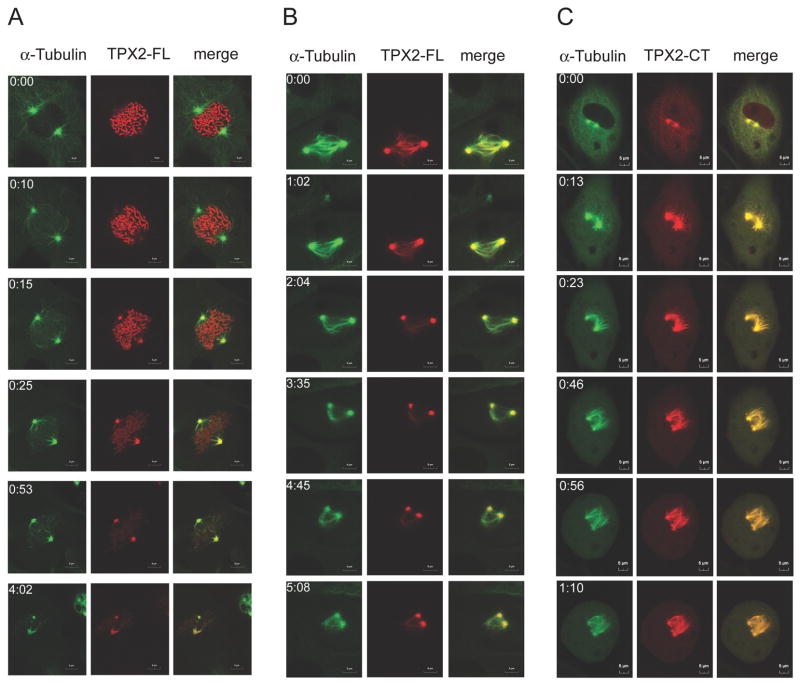
It was important to determine how apposing spindle poles are generated by TPX2-FL and TPX2-CT expression. To investigate spindle pole separation and segregation in real time, we subjected Xenopus S3 cells expressing GFP-α-tubulin to time-lapse, live-cell-imaging by CLSM after transfection of red fluorescent protein-tagged TPX2-proteins. After nuclear envelope breakdown (NEBD), most TPX2-NT relocalizes from chromosomes to spindle poles, and mitotic division is unperturbed (Fig. S6A). TPX2-FL shows a subcellular distribution throughout mitosis similar to that of TPX2-NT, but its overexpression has a severe effect, causing mitotic arrest. Like TPX2-NT, TPX2-FL co-localizes exclusively with condensed chromatin as cells enter mitosis (Fig. 3A). After NEBD, TPX2-FL relocates from chromosomes to spindle poles, which separate and initially move apart but then arrest, often for more than four hours. After long periods of time the morphology of the mitotic spindle changes and spindle poles approach each other, indicating a “collapsing spindle” [13] (Fig. 3A and B and Movie S1).
Figure 3. TPX2-FL induces spindle pole collapse, whereas TPX2-CT inhibits spindle pole segregation.
Live cell imaging by CLSM of GFP-α-tubulin (green) expressing X S3 cells transiently transfected with the indicated DsRed-TPX2 (red) constructs. Cells were seeded onto glass bottom wells, transfected with the indicated pDsRed2-TPX2 constructs, and subjected to double thymidine treatment as described in Experimental Procedures. Live-cell monitoring began ~5 hrs after release from the second thymidine block. Time (hr:min) after imaging began is indicated in the left panels.
(A) TPX2-FL in early mitosis. Cells expressing full length TPX2 enter mitosis, form a bipolar spindle, but then arrest in mitosis for more than 4 hr. Before NEBD, TPX2 protein is nuclear and stains chromosomes. After NEBD, the protein relocates from the condensed chromosomes to the spindle poles. Over time spindle poles approach each other.
(B) TPX2-FL in mitotically arrested cells. Full length DsRed-TPX2 induces collapsing spindle poles in mitotic X S3 cells.
(C) TPX2-CT in mitosis. Spindle poles in cells expressing TPX2-CT separate, but fail to move apart, resulting in apposing spindle poles.
The effect of TPX2-CT is even more severe than TPX2-FL. As mentioned above, TPX2-CT localizes in the cytoplasm in interphase with the protein concentrating at centrosomes even before NEBD. In this case, spindle poles separate but do not move apart, suggesting that the establishment of pushing forces generated by interpolar microtubules might be impaired (Fig. 3C and Movie S2). These cells are not able to proceed into anaphase or cytokinesis and eventually die. However, in all cases the formation of other spindle microtubules appears to be unperturbed, as microtubules rapidly emanate in a unidirectional manner from both spindle poles, which eventually disintegrate. Transfection of DsRed-tagged TPX2 (423–715) and TPX2 (475–715) leads to the same phenotype (data not shown). Notably, transfection of TPX2 (500–715), a 25 aa shorter fragment that only induces cleavage arrest in embryos at very high concentrations, did not induce failure of spindle pole segregation in S3 cells but instead led to a prolonged metaphase arrest of ~ 1 hr (Fig. S6B). This construct localized at spindle poles, spindle microtubules and finally the midzone and midbody. These cells eventually entered anaphase and completed cytokinesis. Thus, TPX2 (500–715), still localizes to spindle structures and interferes with formation of a bipolar spindle, resulting in activation of the spindle checkpoint but not spindle collapse.
TPX2-CTΔ35 in S3 cells showed the same subcellular localization as TPX2-CT, concentrated at centrosomes and the spindle, but this construct did not cause any spindle defects (Fig. S6C and Movie S3). Noteworthy, these cells established interpolar forces pushing spindle poles apart, leading to the formation of a spindle midzone. Importantly, both TPX2-CT and TPX2-CT-Δ35 localized at centrosomes in interphase and at the spindle throughout mitosis, but only TPX2-CT induced spindle defects and arrested cells in mitosis. Thus, besides localization at the mitotic spindle, an activity residing within the last 35 aa is required for inducing spindle pole collapse in S3 cells and cleavage arrest in embryos (Fig. 1A and S1C).
Studies using HeLa cells have reported that knockdown of TPX2 leads to multipolar spindles [14]. Conversely, overexpression of TPX2 in HeLa cells or in egg extracts results in the formation of monopolar spindles with abnormally enlarged spindle poles [15, 16]. Our study adds a new dimension to these reports because time-lapse microscopy reveals that TPX2 overexpression does not induce the formation of monopolar spindles, but rather induces collapse of spindle poles. Therefore, the ‘abnormally enlarged spindle poles’ reported previously [16] are likely the result of collapsed spindles – a conclusion only evident with live-cell imaging.
Eg5 binds TPX2-CT and restores cleavage in embryos
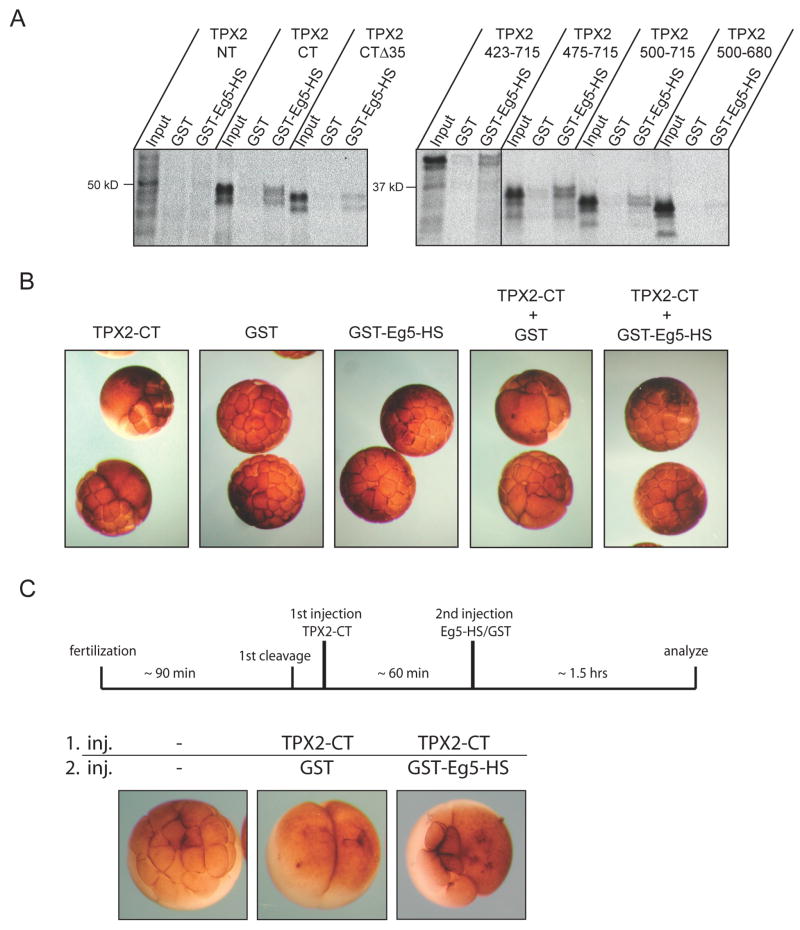
It was important to determine the mechanism by which TPX2-CT induces failure of spindle pole segregation. In early prophase, the subcellular localization of TPX2-CT and its derivatives strikingly correlates with the localization of the motor protein Eg5 [17], which was reported to ‘appear very prominently along microtubules close to the centrosome’ [18] (see Fig. S4B). Based on the observation that TPX2 and Eg5 coexist in a large HURP complex [19], we investigated a possible role for direct binding of TPX2 to Eg5 during cleavage arrest in embryos and spindle collapse in S3 cells. As full-length Eg5 is difficult to express and purify from bacteria, we utilized an Eg5 construct with a GST-tag encompassing the head and stalk domain (Eg5-HS), corresponding to aa 1–685. Pull-down assays demonstrated GST-Eg5-HS interacts with radiolabeled TPX2 (data not shown). Furthermore, TPX2-CT, but not TPX2-NT, was able to bind Eg5 (Fig. 4A). Coomassie staining confirmed equal amounts of GST-proteins (Fig. S7A). Strikingly, TPX2-CTΔ35 was greatly impaired in its ability to bind Eg5, and only those constructs able to induce cleavage arrest in embryos and collapsing or apposing spindle poles in S3 cells (i.e. 423–715, 475–715) interact with Eg5. Furthermore, the amount of TPX2 (500–715) bound to Eg5 was reduced compared to the amount of TPX2 (475–715). This reduced binding is significant because this construct only induced cleavage arrest in embryos at very high concentrations (Fig. S1B) and had only a weak phenotype in S3 cells (Fig. S6C). Finally, TPX2 (500–680) did not interact at all with Eg5, consistent with no phenotype after expression in embryos or S3 cells. We confirmed this interaction is direct by utilizing only bacterially expressed proteins (Fig. S7C).
Figure 4. Eg5 directly interacts with TPX2 and rescues its effects in embryos.
(A) Pull-down analysis of Eg5 and TPX2. Pull-down assays using radiolabeled TPX2 proteins and GST or GST-Eg5-HS were performed as described in Experimental Procedures and define a discrete domain within the C-terminus of TPX2 that binds to Eg5. For input control, see Fig. S7A.
(B) Effect of co-injection of Eg5 and TPX2-CT in embryos. Eg5 blocks cleavage arrest in TPX2-CT injected embryos. One blastomere of a two-cell embryo was injected with similar amounts of the indicated proteins and cleavage was monitored with a dissecting microscope for 4 hrs. For input control see Fig. S7B.
(C) Eg5 expression rescues TPX2-CT-induced cleavage arrest. Upper panel: Scheme of the experimental design. Lower panel: After cleavage of both blastomeres of two-cell embryos was arrested for ~1hr by injection of TPX2-CT, one of the arrested blastomeres was injected with GST or GST-Eg5-HS and cleavage monitored for 90 minutes with a dissecting microscope.
This report is the first to demonstrate a direct interaction between TPX2 and Eg5. Both proteins have been reported to be part of a large HURP complex that also contains Aurora A [19]. However, Aurora A localization is not altered after overexpressing TPX2-CT (Fig. S5B), and knockdown of HURP itself results in defects in chromosome capture and alignment as well as in spindle assembly but not in spindle pole segregation [19]. Therefore, TPX2-dependent regulation of spindle pole segregation via Eg5 is unlikely to involve HURP or Aurora A. If Eg5 is the target of TPX2-CT, then Eg5 itself should block the effects of TPX2-CT. Single injection of GST alone or GST-Eg5-HS had no effect on embryos as they cleaved at normal rates (Fig. 4B). Embryos coinjected with GST and TPX2-CT still failed to induce cleavage furrow ingression, indicating that GST does not interfere with the ability of TPX2-CT to inhibit cleavage. However, coinjection of Eg5 and TPX2-CT at similar concentrations (see Fig. S7B) blocked the induction of cleavage arrest (Fig. 4B). Importantly, we also performed a specific rescue experiment with Eg5. Both blastomeres of a two-cell embryo were injected with TPX2-CT and incubated ~ 1hr to establish TPX2-CT induced arrest. Then one of the arrested blastomeres was injected with GST or GST-Eg5-HS. The arrested blastomere injected with GST-Eg5-HS, but not GST, resumed cleavage, largely on one side of the blastomere (Fig. 4C). This cleavage pattern may be explained by the observation that centrosomes in TPX2-CT injected embryos duplicate and separate in each round of mitosis but fail to migrate away from each other, resulting in clustered centrosomes (see Fig. 1B). After injecting Eg5, these centrosomes may regain their ability to establish pushing forces and bipolar spindles, but they induce cleavage only near the clustered area. The fact that coinjection of Eg5 rescues the TPX2-CT cleavage arrest in embryos supports the idea that Eg5 is a downstream target of TPX2 in the context of spindle pole separation. Taken together, these results indicate that the C-terminus of TPX2 regulates spindle pole movement by a novel Eg5-dependent mechanism in vivo.
Supplementary Material
Acknowledgments
We thank Dr. Gary Gorbsky (Oklahoma Medical Research Foundation) for the Xenopus S3 cell lines, Dr. Bryn Grimison (University of Colorado Health Sciences Center) for providing Emi1-CT protein, and Dr. Thierry Lorca (CNRS, Montpellier) for Xenopus securin cDNA. We also thank Dr. Johné Liu (Ottawa Health Research Institute) for providing Aurora B antibodies. CP is financed by the CNRS, the UICC and the LNCC Equipe Labellisée. FE is a Research Associate and JLM an Investigator of the Howard Hughes Medical Institute.
Abbreviations
- APC/C
anaphase-promoting complex cyclosome
- APD
aphidicolin
- BSA
bovine serum albumin
- CLSM
confocal laser scanning microscopy
- CSF
cytostatic factor
- CT
Carboxy-terminus
- Emi1
early mitotic inhibitor 1
- HURP
hepatoma up-regulated protein
- NEBD
nuclear envelope breakdown
- NT
Amino-terminus
- TPX2
targeting protein for Xklp2
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bayliss R, Sardon T, Vernos I, Conti E. Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol Cell. 2003;12:851–862. doi: 10.1016/s1097-2765(03)00392-7. [DOI] [PubMed] [Google Scholar]
- 2.Eyers PA, Erikson E, Chen LG, Maller JL. A novel mechanism for activation of the protein kinase Aurora A. Curr Biol. 2003;13:691–697. doi: 10.1016/s0960-9822(03)00166-0. [DOI] [PubMed] [Google Scholar]
- 3.Brunet S, Sardon T, Zimmerman T, Wittmann T, Pepperkok R, Karsenti E, Vernos I. Characterization of the TPX2 domains involved in microtubule nucleation and spindle assembly in Xenopus egg extracts. Mol Biol Cell. 2004;15:5318–5328. doi: 10.1091/mbc.E04-05-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eyers PA, Maller JL. Regulation of Xenopus Aurora A activation by TPX2. J Biol Chem. 2004;279:9008–9015. doi: 10.1074/jbc.M312424200. [DOI] [PubMed] [Google Scholar]
- 5.Reimann JD, Freed E, Hsu JY, Kramer ER, Peters JM, Jackson PK. Emi1 is a mitotic regulator that interacts with Cdc20 and inhibits the anaphase promoting complex. Cell. 2001;105:645–655. doi: 10.1016/s0092-8674(01)00361-0. [DOI] [PubMed] [Google Scholar]
- 6.Schatz CA, Santarella R, Hoenger A, Karsenti E, Mattaj IW, Gruss OJ, Carazo-Salas RE. Importin alpha-regulated nucleation of microtubules by TPX2. Embo J. 2003;22:2060–2070. doi: 10.1093/emboj/cdg195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bringmann H, Hyman AA. A cytokinesis furrow is positioned by two consecutive signals. Nature. 2005;436:731–734. doi: 10.1038/nature03823. [DOI] [PubMed] [Google Scholar]
- 8.D’Avino PP, Savoian MS, Glover DM. Cleavage furrow formation and ingression during animal cytokinesis: a microtubule legacy. J Cell Sci. 2005;118:1549–1558. doi: 10.1242/jcs.02335. [DOI] [PubMed] [Google Scholar]
- 9.Minshull J, Sun H, Tonks NK, Murray AW. A MAP kinase-dependent spindle assembly checkpoint in Xenopus egg extracts. Cell. 1994;79:475–486. doi: 10.1016/0092-8674(94)90256-9. [DOI] [PubMed] [Google Scholar]
- 10.Carmena M, Earnshaw WC. The cellular geography of aurora kinases. Nat Rev Mol Cell Biol. 2003;4:842–854. doi: 10.1038/nrm1245. [DOI] [PubMed] [Google Scholar]
- 11.Gruss OJ, Vernos I. The mechanism of spindle assembly: functions of Ran and its target TPX2. J Cell Biol. 2004;166:949–955. doi: 10.1083/jcb.200312112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giet R, McLean D, Descamps S, Lee MJ, Raff JW, Prigent C, Glover DM. Drosophila Aurora A kinase is required to localize D-TACC to centrosomes and to regulate astral microtubules. J Cell Biol. 2002;156:437–451. doi: 10.1083/jcb.200108135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barbosa V, Gatt M, Rebollo E, Gonzalez C, Glover DM. Drosophila dd4 mutants reveal that gammaTuRC is required to maintain juxtaposed half spindles in spermatocytes. J Cell Sci. 2003;116:929–941. doi: 10.1242/jcs.00295. [DOI] [PubMed] [Google Scholar]
- 14.Garrett S, Auer K, Compton DA, Kapoor TM. hTPX2 is required for normal spindle morphology and centrosome integrity during vertebrate cell division. Curr Biol. 2002;12:2055–2059. doi: 10.1016/s0960-9822(02)01277-0. [DOI] [PubMed] [Google Scholar]
- 15.Stewart S, Fang G. Anaphase-promoting complex/cyclosome controls the stability of TPX2 during mitotic exit. Mol Cell Biol. 2005;25:10516–10527. doi: 10.1128/MCB.25.23.10516-10527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wittmann T, Wilm M, Karsenti E, Vernos I. TPX2, A novel xenopus MAP involved in spindle pole organization. J Cell Biol. 2000;149:1405–1418. doi: 10.1083/jcb.149.7.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valentine MT, Fordyce PM, Block SM. Eg5 steps it up! Cell Div. 2006;1:31. doi: 10.1186/1747-1028-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sawin KE, Mitchison TJ. Mutations in the kinesin-like protein Eg5 disrupting localization to the mitotic spindle. Proc Natl Acad Sci U S A. 1995;92:4289–4293. doi: 10.1073/pnas.92.10.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koffa MD, Casanova CM, Santarella R, Kocher T, Wilm M, Mattaj IW. HURP is part of a Ran-dependent complex involved in spindle formation. Curr Biol. 2006;16:743–754. doi: 10.1016/j.cub.2006.03.056. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.