Abstract
Certain proteins can undergo polyglycylation and polyglutamylation. Polyglutamylases have recently been identified in a family of tubulin tyrosine ligase-like (TTLL) proteins. However, no polyglycylase (glycine ligase) has yet been reported. Here we identify a polyglycylase in the TTLL proteins by using an anti-poly-glycine antibody. The antibody reacted with a cytoplasmic 60-kDa protein that accumulated in elongating spermatids. Using tandem mass spectrometry of trypsinized samples, immunoprecipitated by the antibody from the TTLL10-expressing cells, we identified protein as nucleosome assembly protein 1 (NAP1). Recombinant TTLL10 incorporated glycine into recombinant NAP1 in vitro. Mutational analyses indicated that Glu residues at 359 and 360 in the C-terminal part of NAP1 are putative sites for the modification. Thus, TTLL10 is a polyglycylase for NAP1.
Keywords: glycylation, glycylase, nucleosome assembly protein, tubulin tyrosine ligase, histone, spermiogenesis
1. Introduction
Post-translational modification (PTM) facilitates dynamic regulation of a variety of protein properties. Many PTM involve the reversible addition of a range of molecules of variable size that range from methyl, phosphate, and acetyl groups, to lipids and small proteins. Glutamylation [1] and glycylation [2] are unusual PTMs, as the amino acids are added as homopolymeric chains of differing length that are covalently attached to a side-chain carboxyl group of individual glutamic acid residues near the carboxyl terminus of specific proteins. These modifications were originally discovered in tubulins. Proteins other than tubulin can also undergo these modifications. Nucleosome-assembly proteins (NAPs) are subjected to glutamylation [3], and 14-3-3 proteins [4] and Hsp70/Grp170-related protein [5] can be glycylated.
Recent strategies to identify enzymes performing the modifications have proven successful. Genetic and biochemical approaches enabled identification of tubulin polyglutamylases (i.e. tubulin glutamate ligases), as well as functions for this activity in vivo [6,7]. More recently, tubulin polyglutamylases have been identified as members of the tubulin tyrosine ligase-like (TTLL) family [8–11]. Knockdown of TTLL7, a polyglutamylase for β-tubulin, demonstrated the essential role of the β-tubulin glutamylation in neurite growth [9]. In addition, analysis of ROSA22 mice [6] that lack PGs1, a non-catalytic subunit of a polyglutamylase protein complex with specificity for α-tubulin [8], demonstrated that α-tubulin glutamylation is required for intracellular targeting of KIF1 kinesin motor and its cargo synaptic vesicles, as well as for continuous synaptic transmission [12]. In addition, knockdown of TTLL6 in zebrafish eliminates cilia formation in olfactory placodes [13]. Despite these advances in our understanding of the mechanism and function of tubulin glutamylation, there remains little information about protein polyglycylases (i.e. glycine ligases). Here we identify TTLL10 as a glycylase for NAP1 using an experimental approach of immunoprecipitation of proteins with an anti-poly glycine antibody, followed by two-dimensional gel electrophoresis and mass spectrometry.
2. Materials and Methods
2.1. Antibodies
The anti-poly-glycine polyclonal (R-polygly) antibody, which was generated by using Cys-(Gly)9 as a immunogen [14], was the generous gift of Dr. M. Gorovsky (U. Rochester, NY). The monoclonal antibody (mAb) 4A8 for NAP1 was the kind gift of Dr. Y. Ishimi (Ibaraki Univ., Mito, Japan) [15]. Additional, commercially available, antibodies used in this study and their targets were as follows; -tubulin (mAb DM1A), FLAG tag (mAb M2, or polyclonal), Sigma-Aldrich (St Louis, MO); GAPDH (mAb 6C5, Chemicon International, Temecula, CA); Anti-GFP polyclonal antibody (Medical & Biological Laboratories, Nagoya, Japan); Alexa fluorophore-conjugated secondary antibodies (Invitrogen, Carlsbad, CA); HRP-conjugated secondary antibodies for western blot analysis (Jackson ImmunoResearch Laboratories, West Grove, PA).
2.2. Immunoblotting and immunohistochemistry
Immunoblotting and immunohistochemistry were performed as described [9]. Total testicular protein was isolated from C57BL/6J mice of the indicated age using standard methods. For electrophoresis, Precision Plus Protein Standards, all blue prestained, (Bio-Rad Laboratories, Hercules, CA) were used as molecular weight markers. For immunohistochemistry, the stage of spermatogenesis was determined using criteria as described [16]. In the absence of acrosomal staining, staging was based on the relative size of pachytene spermatocytes, the size of round spermatids, the location and shape of elongate spermatid nuclei, the size of elongate spermatid cytoplasm, and the distance between elongate spermatids and lumen of the tubules.
2.3. Plasmids
Complementary DNA (cDNA) containing whole coding sequences of each mouse TTLL protein and NAP1 were cloned by polymerase chain reaction (PCR). Primers were designed using reference sequences deposited in the NCBI database. Amplified coding regions were inserted in pCMV-Tag 4A vector (Stratagene, La Jolla, CA). For expression in insect cells, the full-length coding sequence of mouse TTLL10 was cloned into a pENTR vector (Invitrogen). The insert was moved into baculovirus-mediated expression vector with GATEWAY system (Invitrogen). Point mutant and deletion mutants were constructed with Site-Directed Mutagenesis kit (Stratagene). Plasmids were transfected using Lipofectamine2000 (Invitrogen) according to the manufacturer’s instructions.
2.4. Immunoprecipitation
Tissues or cells were lysed with 20 mM Tris, pH 7.6, 140 mM NaCl, 0.1% Triton X-100. To immunoprecipitate R-polygly-reactive proteins, the antibody was covalently linked to protein G-sepharose beads (GE Healthcare) with dimethyl pimelimidate. For immunoprecipitation of FLAG-tagged proteins, M2 mAb covalently bound to agarose beads (Sigma) was used. The beads were added to centrifuged tissue or cell lysates then incubated at 4 °C for a few hours. Bound proteins were directly eluted in electrophoresis sample buffer.
2.5. Two-dimensional electrophoresis
Immunoprecipitates were resolved in lysis buffer (7 M urea, 2 M thiourea, 4% CHAPS, 40 mM DTT, 2% IPG buffer (pH range 3–11), containing Complete™ EDTA-free protease inhibitor cocktail (Roche, Indianapolis, IN). First and second-dimension electrophoresis was performed according to the manufacturer’s instructions. Proteins on the gel slabs were stained with SyproRuby (Invitrogen). Signals were visualized with a UV transilluminator.
2.6. Mass spectrometry
Protein spots were excised. Preparation of samples was performed as described [12]. ZipTip (Millipore) was used to desalt the sample. Eluted peptides were applied to Prespotted AnchorChip™ (Bruker Daltonics, Bremen, Germany), which was prespotted with an α-cyano-4-hydroxycinnamic acid matrix. Mass spectrometry was performed with a MALDI-TOF MS instrument, ULTRA FLEX II TOF/TOF (Bruker). Data acquisition was performed in a positive ion mode using an external calibration method with a peptide mixture of bradykinin 1–7 ([M+H]+: 757.40), angiotensin II ([M+H]+: 1046.54), angiotensin I ([M+H]+: 1296.68), neurotensin ([M+H]+: 1672.92), renin substrate ([M+H]+: 1758.93), ACTH clip 1–17 ([M+H]+: 2093.09), ACTH clip 18–39 ([M+H]+: 2465.20), and ACTH clip 1–24 ([M+H]+: 2932.59).
2.6. In vitro enzyme activity assay for polyglycylase
Glutathione-S-transferase-fused mouse TTLL10 and mouse NAP1 synthesized in insect Sf9 cells were purified by chromatography on glutathione-sepharose 4B beads (GE Healthcare). Enzyme activities were quantified by measuring incorporation of 3H-labeled glycine into substrates. After reaction at 30 °C overnight, radioactivity incorporated into NAP1 was quantified using a Winspectral 1414 liquid scintillation counter (Wallac, Turku, Finland) for 1 min.
3. Results
3.1. R-polygly antibody detects cytoplasmic proteins in mouse spermatids
As no report has used the R-polygly antibody [14] in mammalian tissues and cells, we first examined what the antibody detected in extracts of mammalian (mouse) tissues. The strongest signal was observed in testis with ~60 kDa migrating position in SDS-PAGE (Fig. 1A). We next investigated if the antibody was capable of immunoprecipitating the protein detected. Under the conditions used, the 60-kDa protein band detected in testis was immunoprecipitated by the R-polygly antibody (Fig. 1B).
Figure 1. Detection of polyglycylated proteins in mouse tissues.
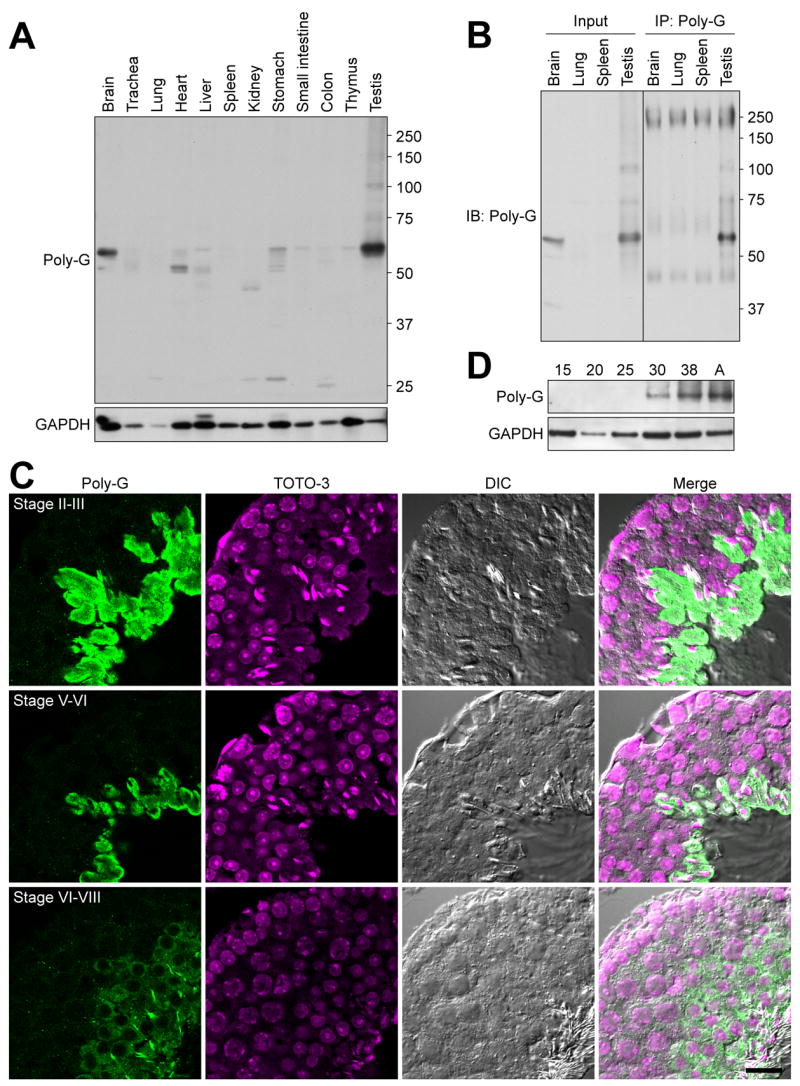
(A) Western blot analyses of adult mouse tissues with R-polygly antisera. Testis showed strongest signal at approximately 60 kDa. (B) Immunoprecipitation of the 60-kDa protein from testis. The 60-kDa protein detected in testis lysates was immunoprecipitated. (C) Cytoimmunofluorescence analyses of R-polygly-reactive proteins in testis. The immunoreactivity was localized in cytoplasm of spermatids. Elongating spermatids showed strongest signals, with weaker signals in round spermatids. TOTO-3; a DNA dye for staining nuclei. DIC; differential interference contrast microscopy. Scale bar, 20 μm. (D) Western analysis of testis protein isolated from mice at 15, 20, 25, 30 38-day and adult (12-week) age. Accumulation of the polyglycylated 60-kDa protein in mice older than 25 days is consistent with the localization of the same protein detected in panel C.
To identify where the protein identified by R-polygly antibody was expressed in testis, we performed immunohistochemistry. The antibody cross-reacted with protein localized in spermatids (Fig. 1C). During spermiogenesis, signals were first observed in the cytoplasm of round spermatids between steps 6–8 (Fig 1C, lowest row). The intensity of the signal increased within the cytoplasm of elongate step 14–15 spermatids (Fig 1C, top row), before becoming reduced in later stage elongate spermatids (Fig 1C, middle row). Results of western analysis of total protein isolated from testis of prepubertal mice of different ages supports the conclusion that the protein accumulating in elongating spermatids is the 60-kDa protein (Fig. 1D). These results indicate that glycylation of the ~60-kDa protein accumulates in cytoplasm of spermatids during spermiogenesis.
3.2. TTLL10 performs polyglycylation of NAP1 in vitro and in vivo
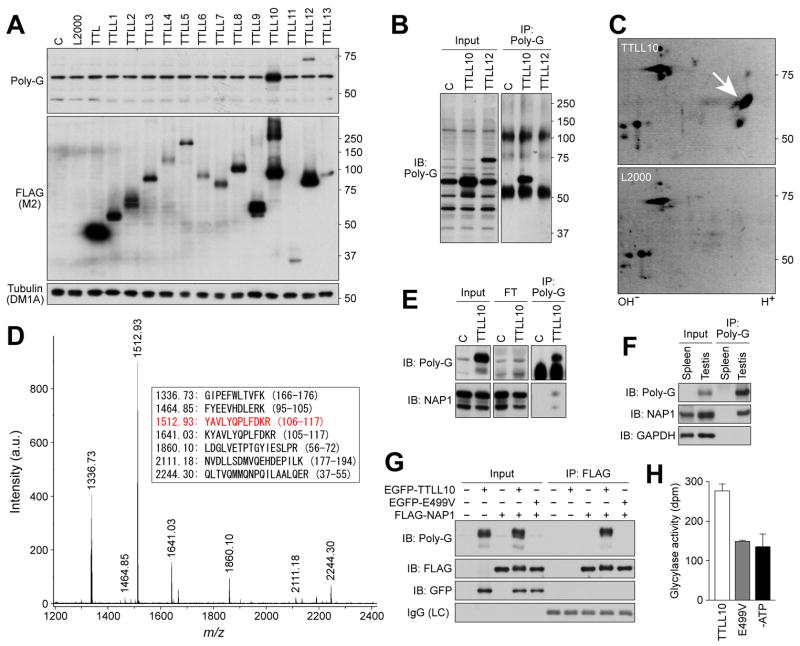
Both we, and others [10], have predicted that TTLL family members other than those that show glutamylase activity on tubulin or NAP would display glycylase activity. To identify protein glycylase(s), we expressed cDNA clones containing all currently identified TTLL proteins in HEK293T cells and screened total cell protein extracts with the R-polygly antibody. Ectopic-expression of TTLL10 generated a significant increase in an R-polygly-reactive protein migrating at 60 kDa (Fig. 2A).
Figure 2. Identification of TTLL10 as a glycine ligase for NAP1 in vitro and in vivo.
(A) Screening of HEK293T cells expressing thirteen TTLL proteins with the R-polygly antibody. In TTLL10-expressing cell lysates, there was a significant increase in a 60-kDa protein detected by the antibody. (B) Immunoprecipitation of the 60-kDa protein. Note that non-specific R-polygly reactive proteins present in control samples and TTLL12-generated proteins were not precipitated with the antibody. (C) Two-dimensional gel electrophoresis resolution of the immunoprecipitated protein. Arrow indicates spots observed only in TTLL10 expressing sample. (D) Tandem mass spectrometry of trypsin digests of the immunoprecipitated protein. The sequences corresponding to the peaks are listed in the box. The peptide sequence in red was further identified by tandem mass spectrometry. (E-G) Immunoprecipitation-western blot analyses. Protein immunoprecipitated from TTLL10-expressing cell lysates or testis lysates with the R-polygly antibody was detected by an anti-NAP1 antibody (E,F). Almost no glycylated proteins remained in flow through (FT) in panel E. FLAG-NAP1 immunoprecipitated from lysates of cells co-expressing TTLL10 was detected by the R-polygly antibody (G). Note that FLAG-NAP1 co-expressed with wild-type TTLL10 displayed both slightly slower mobility and blurred tailing in electrophoresis, indicative of change of molecular weight by addition of multiple glycines (G). (H) In vitro enzyme activity of TTLL10. The activity was measured by [3H]-glycine incorporation into recombinant NAP1.
To identify the substrate of TTLL10, we immunoprecipitated the 60-kDa protein using R-polygly antibody (Fig. 2B). Two-dimensional electrophoresis was used to separate the immunoprecipitated protein from immunoglobulin heavy chains and the protein was concentrated as a few spots at a position of pI ~4 and 60 kDa (Fig. 2C). Tandem mass spectrometry of trypsin digests of the protein eluted from the spots revealed that the protein was NAP1 (Fig. 2D). This observation was verified by western blot analysis with a monoclonal antibody against NAP1 (Fig 2E). The majority of NAP1 remained in flow-through of immunoprecipitation whereas almost all glycylated proteins were adsorbed to the R-polygly antibody and eluted into precipitated fraction (Fig. 2E), suggesting that only small proportion of endogenous NAP1 was glycylated by TTLL10. The testicular ~60-kDa protein precipitated with the R-polygly antibody was also detected by the anti-NAP1 mAb (Fig. 2F). FLAG-tagged NAP1 immunoprecipitated with an anti-FLAG antibody from cell lysates co-expressing EGFP-TTLL10 was strongly detected by the R-polygly antibody (Fig. 2G).
To investigate whether TTLL10 displays glycylase activity on NAP1, we performed in vitro enzyme activity assay with recombinant TTLL10. The recombinant TTLL10 was capable of incorporating glycine onto the recombinant NAP1 in vitro (Fig. 2H). Recombinant TTLL10 enzyme activity was abolished when the reaction mixture lacked ATP or when a mutant of TTLL10 was used, in which the glutamate within the active site was replaced by valine (E499V) [8,9] (Fig. 2H). Taken together, these findings indicate that TTLL10 can function as a protein glycylase for NAP1 in vivo and in vitro.
3.3. Glycylation of NAP1 by TTLL10 requires Glu residues at 359 and 360 in the C-terminus of NAP1
NAP1 can be glutamylated at Glu-356 and -357 in the C-terminal Glu-rich regions [3]. To investigate which Glu residues are required for polyglycine ligation to NAP1, we generated various mutants of NAP1 (Fig. 3A), and co-expressed each of them with TTLL10. To separate ectopic NAP1 from co-immunoprecipitated endogenous NAP1, we used Δ371-NAP1, which retained the ability to be glycylated by TTLL10 (Fig. 3B). Substitution of either Glu-359 or -360 with Asp caused a loss of the reactivity to the R-polygly antibody (Fig. 3D), whereas either multi Glu repeats in 363–368 (Fig. 3C) or glutamylation sites (Fig. 3D) were dispensable. These indicate that both Glu-359 and -360 are potential sites for TTLL10-mediated glycylation.
Figure 3. Amino acids of NAP1 required for NAP1 to be glycylated.
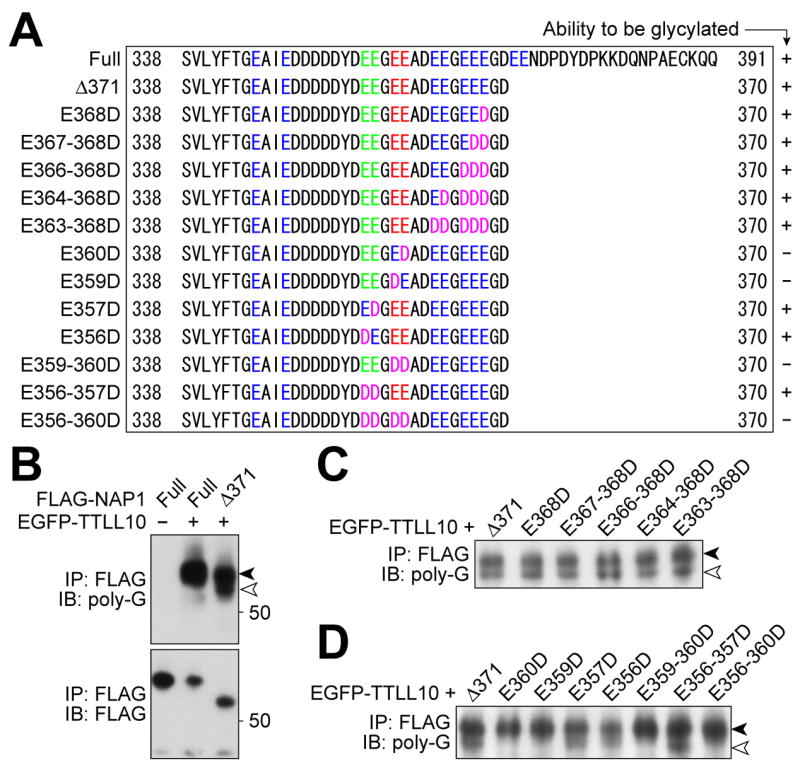
(A) Alignment of C-termini of mouse NAP1 mutants used. Glu (“E”) residues in C-terminal acidic domain are colored. Green text indicates the sites for glutamylation (Regnard et al, 2000). Glu residues colored red were required for glycylation. Asp (“D”) residues replacing Glu are colored magenta. A summary of the relative ability of each protein to be glycylated is shown on the right side. (B-D) Western blot analyses of NAP1 mutants with the R-polygly antibody. The Δ371-NAP1 (white arrowhead) is distinguishable from co-precipitated endogenous NAP1 (black arrowhead), with both species detected by the R-polygly antibody (B). Substitution of all Glu residues between 363 and 368 with Asp did not affect the ability to be a substrate for TTLL10 (C). Replacement of either Glu359 or 360 by Asp is sufficient to cause a loss of reactivity to the R-polygly antibody, while substituting the glutamylation sites with Asp residues did not impair the NAP1 ability to be glycylated by TTLL10 (D).
4. Discussion
We have identified TTLL10 as a glycylase (i.e. glycine ligase) for NAP1. The TTLL10 enzyme activity detected was extremely low. In vivo, TTLL10 glycylated only a small portion of total NAP1, and in vitro, it required more than 20 hours to make glycylated NAP1. Given that an oligo-peptide composed of nine glycines was used as the antigen for generating the antibody used, we should detect “poly”-glycine side chains on NAP1. A plausible explanation for the relatively low protein glycylase activity observed is that TTLL10 might serve as an elongase that can only add glycines to the carboxy terminus of a glycine previously attached by an initiase to a γ-carboxyl group of glutamic acids in NAP1 backbone. If cells lack initiase activity, addition of poly-glycine chains would be difficult to detect. There is precedence for this mechanism involving tubulin polyglutamylation [10]. Other TTLL proteins might perform initiation.
TTLL proteins were initially identified as a family harboring tubulin glutamylases. Available evidence and phylogenetic relationship support that TTLLs 1, 2, 4, 5, 6, 7, 9, 11, and 13 constitute a glutamylase subfamily[8–11]. Based on the similarity in the PTMs, both we and others predicted that some of the remaining TTLL proteins (TTLLs 3, 8, 10 and 12) are glycylase(s). Surprisingly, none of the individually expressed constructs generated tubulin polyglycylation (Ikegami and Setou, unpublished observation). Some TTLLs may have only initiase activity, and thus we simply failed to detect such “mono”-glycylated tubulins with the R-polygly antibody. Alternatively, it is possible that co-expression of such initiases with TTLLs having elongase activity is required for generating “poly”-glycylated tubulins up to detectable level. Additional studies involving co-expression of different combinations of TTLLs are required to discriminate between these possibilities.
NAP1 was originally identified as a histone chaperone [15,17–19]. This function of NAP1 relies on its ability to bind histones. Inspection of the three-dimensional structure of NAP1 suggests that the acidic surface of the NAP1 dimer contributes to the interaction between NAP1 and histones [20]. The C-terminal acidic domain participates in the interaction [21], Addition of poly-glycine chains to the C-terminus could block the acidic effect, either by steric hindrance of the electrostatic charge, or even simpler by preventing the acidification caused by polyglutamylation as the modification sites were close to each other. Thus, the blockade of acidic C-terminus could reduce NAP1’s ability to interact with basic histones. Alternatively, glycylation might affect nuclear import of NAP1, as the import of NAP1 into nucleus depends on the C-terminal acidic domain [22].
Glycylation occurred during spermiogenesis. The timing of appearance of polyglycylation during mouse spermatogenesis is coincident with cessation of transcription that begins in step 8 spermatids. An interesting hypothesis is that glycylation of a proportion of NAP1 modulates NAP1-histone interactions, including their ability to mediate H2A/B exchange and nucleosomal sliding required for transcription. Hence, glycylation of NAP1 could play a role in mediating transcriptional repression during spermiogenesis. Second, the spermatid nucleus alters shape dramatically during spermiogenesis, becoming highly compact through replacement of histones by protamines. NAP1 can also facilitate decondensation of highly condensed sperm nuclei [23]. Hence, glycylation might counteract the NAP1 function, thereby promoting initial replacement of histone octamers with the transition proteins, followed by protamines. One possible reason why NAP1 polyglycylation was detected exclusively in elongating spermatids may lie in the fact that replacement of histones with transition proteins, then protamine occurs exclusively in these cells.
In summary, we have demonstrated that TTLL10 has polyglycylase (protein glycine ligase) activity, that this enzyme can modify NAP1 and that a proportion of NAP1 is polyglycylated during spermiogenesis in mice. Ttll10 is expressed in mouse testis [10] and analysis of NCBI EST database identifies four independent Ttll10 EST cDNA clones (accession numbers CF105733.1, CF105131.1, BF021739.1 and BF319334.1) produced from RNA isolated from purified round spermatids. These data indicate that NAP1 is capable of being modified by polyglycylation via TTLL10 in cells in culture and that this modification could also be performed by TTLL10 in mouse spermatids. Based on those findings, we speculate that glycylation of nucleosome assembly protein contributes to two processes during spermiogenesis; i.e. regulation of transcriptional activity, as well as histone replacement and chromatin remodeling.
Acknowledgments
We thank Drs. M. Gorovsky and Y. Ishimi for generous gifts of antibodies. We thank members of MITILS, especially Dr. Noce for valuable discussion, and other members of Setou Lab., especially Dr. S. Sato, K. Yasutake, M. Takamatsu, Y. Sugiura for technical assistance and advice. We thank Dr. Sekiya for generous support and constructive discussion. This research was supported by a grant from the NICHD, NIH (GM), and by PRESTO and SENTAN grant from the JST, and WAKATE-A grant from the JSPS (MS)
Abbreviations
- PTM
post-translational modification
- NAP
nucleosome assembly protein
- TTLL
tubulin tyrosine ligase-like
- R-polygly
anti-poly-glycine polyclonal antibody
- mAb
monoclonal antibody
- cDNA
complementary DNA
- PCR
polymerase chain reaction
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Eddé B, Rossier J, Le Caer JP, Desbruyères E, Gros F, Denoulet P. Posttranslational glutamylation of alpha-tubulin. Science. 1990;247:83–85. doi: 10.1126/science.1967194. [DOI] [PubMed] [Google Scholar]
- 2.Redeker V, Levilliers N, Schmitter JM, Le Caer JP, Rossier J, Adoutte A, Bré MH. Polyglycylation of tubulin: a posttranslational modification in axonemal microtubules. Science. 1994;266:1688–1691. doi: 10.1126/science.7992051. [DOI] [PubMed] [Google Scholar]
- 3.Regnard C, Desbruyères E, Huet JC, Beauvallet C, Pernollet JC, Eddé B. Polyglutamylation of nucleosome assembly proteins. J Biol Chem. 2000;275:15969–15976. doi: 10.1074/jbc.M000045200. [DOI] [PubMed] [Google Scholar]
- 4.Lalle M, Salzano AM, Crescenzi M, Pozio E. The Giardia duodenalis 14-3-3 protein is post-translationally modified by phosphorylation and polyglycylation of the C-terminal tail. J Biol Chem. 2006;281:5137–5148. doi: 10.1074/jbc.M509673200. [DOI] [PubMed] [Google Scholar]
- 5.Xie R, Clark KM, Gorovsky MA. Endoplasmic reticulum retention signal-dependent glycylation of the Hsp70/Grp170-related Pgp1p in Tetrahymena. Eukaryotic Cell. 2007;6:388–397. doi: 10.1128/EC.00366-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell PK, Waymire KG, Heier RL, Sharer C, Day DE, Reimann H, Jaje JM, Friedrich GA, Burmeister M, Bartness TJ, Russell LD, Young LJ, Zimmer M, Jenne DE, MacGregor GR. Mutation of a novel gene results in abnormal development of spermatid flagella, loss of intermale aggression and reduced body fat in mice. Genetics. 2002;162:307–320. doi: 10.1093/genetics/162.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Regnard C, Fesquet D, Janke C, Boucher D, Desbruyéres E, Koulakoff A, Insina C, Travo P, Eddé B. Characterisation of PGs1, a subunit of a protein complex co-purifying with tubulin polyglutamylase. J Cell Sci. 2003;116:4181–4190. doi: 10.1242/jcs.00743. [DOI] [PubMed] [Google Scholar]
- 8.Janke C, Rogowski K, Wloga D, Regnard C, Kajava AV, Strub JM, Temurak N, van Dijk J, Boucher D, van Dorsselaer A, Suryavanshi S, Gaertig J, Eddé B. Tubulin polyglutamylase enzymes are members of the TTL domain protein family. Science. 2005;308:1758–1762. doi: 10.1126/science.1113010. [DOI] [PubMed] [Google Scholar]
- 9.Ikegami K, Mukai M, Tsuchida JI, Heier RL, MacGregor GR, Setou M. TTLL7 is a mammalian beta-tubulin polyglutamylase required for growth of MAP2-positive neurites. J Biol Chem. 2006;281:30707–30716. doi: 10.1074/jbc.M603984200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Dijk J, Rogowski K, Miro J, Lacroix B, Eddé B, Janke C. A targeted multienzyme mechanism for selective microtubule polyglutamylation. Mol Cell. 2007;26:437–448. doi: 10.1016/j.molcel.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 11.van Dijk J, Miro J, Strub JM, Lacroix B, van Dorsselaer A, Eddé B, Janke C. Polyglutamylation is a post-translational modification with a broad range of substrates. J Biol Chem. 2007;283:3915–3922. doi: 10.1074/jbc.M705813200. [DOI] [PubMed] [Google Scholar]
- 12.Ikegami K, Heier RL, Taruishi M, Takagi H, Mukai M, Shimma S, Taira S, Hatanaka K, Morone N, Yao I, Campbell PK, Yuasa S, Janke C, MacGregor GR, Setou M. Loss of alpha-tubulin polyglutamylation in ROSA22 mice is associated with abnormal targeting of KIF1A and modulated synaptic function. Proc Natl Acad Sci USA. 2007;104:3213–3218. doi: 10.1073/pnas.0611547104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pathak N, Obara T, Mangos S, Liu Y, Drummond IA. The zebrafish fleer gene encodes an essential regulator of cilia tubulin polyglutamylation. Mol Biol Cell. 2007;18:4353–4364. doi: 10.1091/mbc.E07-06-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duan J, Gorovsky MA. Both carboxy-terminal tails of alpha- and beta-tubulin are essential, but either one will suffice. Curr Biol. 2002;12:313–316. doi: 10.1016/s0960-9822(02)00651-6. [DOI] [PubMed] [Google Scholar]
- 15.Ishimi Y, Kikuchi A. Identification and molecular cloning of yeast homolog of nucleosome assembly protein I which facilitates nucleosome assembly in vitro. J Biol Chem. 1991;266:7025–7029. [PubMed] [Google Scholar]
- 16.Russell LD, Ren HP, Sinha Hikim I, Schulze W, Sinha Hikim AP. A comparative study in twelve mammalian species of volume densities, volumes, and numerical densities of selected testis components, emphasizing those related to the Sertoli cell. Am J Anat. 1990;188:21–30. doi: 10.1002/aja.1001880104. [DOI] [PubMed] [Google Scholar]
- 17.Ishimi Y, Yasuda H, Hirosumi J, Hanaoka F, Yamada M. A protein which facilitates assembly of nucleosome-like structures in vitro in mammalian cells. J Biochem (Tokyo) 1983;94:735–744. doi: 10.1093/oxfordjournals.jbchem.a134414. [DOI] [PubMed] [Google Scholar]
- 18.Ishimi Y, Hirosumi J, Sato W, Sugasawa K, Yokota S, Hanaoka F, Yamada M. Purification and initial characterization of a protein which facilitates assembly of nucleosome-like structure from mammalian cells. Eur J Biochem. 1984;142:431–439. doi: 10.1111/j.1432-1033.1984.tb08305.x. [DOI] [PubMed] [Google Scholar]
- 19.Ishimi Y, Kojima M, Yamada M, Hanaoka F. Binding mode of nucleosome-assembly protein (AP-I) and histones. Eur J Biochem. 1987;162:19–24. doi: 10.1111/j.1432-1033.1987.tb10535.x. [DOI] [PubMed] [Google Scholar]
- 20.Park YJ, Luger K. The structure of nucleosome assembly protein 1. Proc Natl Acad Sci USA. 2006;103:1248–1253. doi: 10.1073/pnas.0508002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McBryant SJ, Park YJ, Abernathy SM, Laybourn PJ, Nyborg JK, Luger K. Preferential binding of the histone (H3-H4)2 tetramer by NAP1 is mediated by the amino-terminal histone tails. J Biol Chem. 2003;278:44574–44583. doi: 10.1074/jbc.M305636200. [DOI] [PubMed] [Google Scholar]
- 22.Miyaji-Yamaguchi M, Kato K, Nakano R, Akashi T, Kikuchi A, Nagata K. Involvement of nucleocytoplasmic shuttling of yeast Nap1 in mitotic progression. Mol Cell Biol. 2003;23:6672–6684. doi: 10.1128/MCB.23.18.6672-6684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito T, Tyler JK, Bulger M, Kobayashi R, Kadonaga JT. ATP-facilitated chromatin assembly with a nucleoplasmin-like protein from Drosophila melanogaster. J Biol Chem. 1996;271:25041–25048. doi: 10.1074/jbc.271.40.25041. [DOI] [PubMed] [Google Scholar]