Summary
Accumulating evidence reveals hydrogen peroxide as a key player both as a damaging agent and, from emerging evidence over the last decade, as a second messenger in intracellular signaling. This rather mild oxidant acts upon downstream targets within signaling cascades to modulate the activity of a host of enzymes (e.g. phosphatases and kinases) and transcriptional regulators through chemoselective oxidation of cysteine residues. With the recent development of specific detection reagents for hydrogen peroxide and new chemical tools to detect the generation of the initial oxidation product, sulfenic acid, on reactive cysteines within target proteins, the scene is set to gain a better understanding of the mechanisms through which hydrogen peroxide acts as a second messenger in cell signaling.
Introduction
With the emerging understanding that hydrogen peroxide generation is a critical component of numerous receptor-mediated cell signaling events, investigators are seeking to identify, within given signaling networks, the particular oxidation-sensitive proteins, residues, and molecular events that lead to the modulation of cell signaling networks by hydrogen peroxide. We discuss herein some of the rapidly evolving chemical tools and experimental approaches being developed both for detecting oxidant generation and for identifying the molecular targets modified by the oxidants. We also summarize recent data supporting cysteine sulfenic acid as a key intermediate in the functional modulation of enzymes and transcription factors involved in peroxide-mediated cell signaling.
Localization and control of hydrogen peroxide for signaling
Reactive oxygen species (ROS) are produced in cells as a result of the partial reduction of oxygen by the electron transport chain in mitochondria, by lipoxygenases in the cytoplasm, and by flavoprotein oxidases in peroxisomes. It is increasingly appreciated that cytokines, growth factors and integrins also activate multicomponent NADPH oxidase enzymes (Nox) as part of the cell’s receptor-mediated signaling responses. Both superoxide and, hence, hydrogen peroxide are generated, with the latter serving a particularly significant role as a second messenger in signaling pathways. Targeting of Nox proteins to specific subcellular compartments, including lipid rafts within membranes, endosomal signaling vesicles and other cellular organelles, may serve to offset the highly diffusible nature of hydrogen peroxide and limit the oxidation signal to these microenvironments [1–3]. Consistent with this, evidence is accumulating that antioxidant enzymes that serve to control intracellular peroxide levels, including glutathione peroxidases, catalases and especially peroxiredoxins [4], may themselves be regulated by these oxidants, allowing a threshold concentration of peroxide to accumulate around the site of Nox localization while preventing damage to cellular components outside these regions (reviewed in [5]).
Transient generation of localized ROS is important in receptor-mediated cell signaling, yet the ability to detect ROS with a high degree of spatial and temporal resolution remains a challenge [6]. Oxidation of widely used 2′,7′-dichlorodihydrofluorescein (DCFH) and dihydrorhodamine 123 (DHR) reagents as detected by enhanced fluorescence gives a sense of the ROS burst, but these reagents exhibit a relative lack of specificity toward their oxidants, and are also subject to autoxidation and photo-oxidation. The recent design and synthesis of new, highly selective chemical sensors of hydrogen peroxide, particularly those which become fluorescent upon peroxide-mediated removal of a boronate-based protecting group, is a very exciting development in this regard [7]. These and other new ratiometric, protein-and/or nanoparticle-based reagents for peroxide sensing [8–10] are promising tools for continuously monitoring intracellular peroxide generation, yet will need to overcome limitations such as irreversibility and relatively slow rates of reaction to achieve the level of sensitivity required for “instantaneous” and nonperturbing detection of signaling-relevant hydrogen peroxide (for recent reviews, see [11,12]).
Cysteine redox chemistry
It is increasingly clear that a major component of the ROS-linked modulation of cell signaling pathways is the dynamic regulation of protein function by reversible thiol modification. For many proteins, the first product of cysteine oxidation by hydroperoxides, cysteine sulfenic acid (R-SOH) (Figure 1), undergoes rapid condensation either with another protein thiol (as an intra- or intermolecular interaction) or with a small molecule thiol like glutathione or cysteine to form a disulfide bond. Thiol-based reductants can then return this species to the fully reduced thiol state (Figure 1). Disulfide bonds can stabilize extracellular proteins, protect against irreversible inactivation, stabilize associations within protein complexes, modify structures to create, destroy or modulate functional sites, and ultimately regulate enzymatic or transcriptional activity of proteins. Under conditions where reactive nitrogen species (RNS) are generated, signaling through reversible S-nitrosation, perhaps in addition to the typical ROS-generated products, may also play an important role [13]. The features of the protein microenvironment that control thiol reactivity and stability of the resulting modifications are beginning to be analyzed [14].
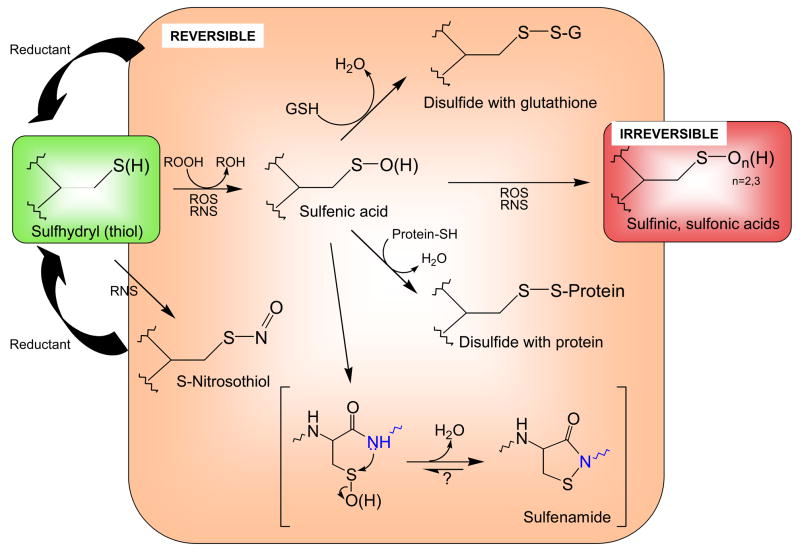
Figure 1. Biological modifications of cysteine thiols.
Reactive cysteine thiols (green), typically in their ionized, thiolate form (R-S−), are oxidized by such oxidants as hydrogen peroxide, organic hydroperoxides, hypochlorous acid and peroxynitrite to form sulfenic acids, which may be stabilized or go on to form other reversible (disulfides or sulfenamides, orange) or irreversible (sufinic and sulfonic acid, red) species. Both reactive oxygen species (ROS) and reactive nitrogen species (RNS) promote these oxidations. Note that the generation of sulfenamide (bottom) involves attack of a neighboring amino acid’s amide nitrogen (blue) on the sulfenic acid sulfur. Although sulfinic and sulfonic acids are shown here as irreversible modifications, recent discoveries show that some peroxiredoxins in this state can be recovered through action of specialized sulfinic acid reductases (sulfiredoxins).
In certain cases, the initial sulfenic acid may itself be stabilized within the protein microenvironment. This does, however, potentially leave the protein sulfenic acid vulnerable to hyperoxidation to the largely irreversible sulfinic or sulfonic acids (Figure 1), further modifications that could, in spite of their typically irreversible status, also provide a signaling role analogous to a “fire alarm” regarding cellular redox status [15].
Sulfenamide (sulfenyl-amide) has recently been recognized as another stabilized oxidation product generated through condensation of the sulfenic acid with a neighboring backbone amide nitrogen (Figure 1); this 5-membered ring structure appears to represent yet another form of oxidized cysteine resistant to hyperoxidation. The initial evidence for this species was crystallographic [16], leaving some doubt as to the relevance of this species in a biological setting. On the other hand, a functional role for the generation of such a species is suggested by the profound changes in structure imparted to the active site of protein tyrosine phosphatases with this modification, changes which may enhance access by reductants and/or modulate interactions with other domains ([17]; reviewed in [18]). Studies using model chemistry have also better defined the parameters which promote this cyclization reaction in the constrained setting of a protein active site [19]. Furthermore, recent studies of the peroxide sensitive transcriptional regulator, OhrR from Bacillus subtilis, support stable formation of the sulfenamide species in solution and possibly in vivo for this protein [20].
Redox regulated peroxiredoxin function
All of these examples of cysteine chemical states, with the possible exception of sulfenamides, are observed in the highly expressed peroxiredoxin enzymes (including recently-reported S-nitrosation [21]). These enzymes, which catalyze the reduction of hydroperoxide substrates via a catalytic cysteine at the active site, have been shown to exhibit high sensitivity (kcat/Km of ~107 M−1 s−1) and specificity toward H2O2 in at least some cases [22,23], and their activity is regulated by phosphorylation, oligomerization, and oxidation state [5]. During the normal catalytic cycle, the peroxidatic cysteine becomes oxidized to a sulfenic acid, then undergoes one or more steps of inter- or intrasubunit disulfide bond formation and thiol-disulfide exchange with other thiols in order to return these enzymes to their reduced, activated state [24,25]. In the mammalian peroxiredoxins, hyperoxidation competes successfully with disulfide bond formation under certain conditions, causing these enzymes to be “turned off” at their sulfenic acid redox switch when local peroxide levels rise (Figure 2). The physiological significance of this hyperoxidation is supported by several observations. (1) The susceptibility of eukaryotic peroxiredoxins toward hyperoxidation, a characteristic not observed in their prokaryotic counterparts, is linked to the presence of particular evolved structural features within these enzymes [4,25]. (2) For at least some peroxiredoxins, hyperoxidation to sulfinic acid is reversible as catalyzed, through highly specialized chemistry, by specific enzymes called sulfiredoxins (and perhaps sestrins), enzymes which appear to have co-evolved with this sensitivity [26,27]. (3) Hyperoxidation of Prxs has been detected as a result of tumor necrosis factor signaling, suggesting at least a role for this modification in apoptotic signaling processes [28]. While hyperoxidation of peroxiredoxin proteins on a global level has yet to be detected during growth factor signaling, such modifications, if generated in localized regions around signaling complexes, may be difficult to detect within the large pool of cytoplasmic peroxiredoxins. There is no doubt that it is early in our understanding of when and how this functional hyperoxidation switch contributes to biological processes. Still, there is a clear link between peroxide sensing and transcriptional activation through the modulation of the peroxiredoxin redox state in several yeast species (see [5] for a review), and both cell cycle arrest [15] and enhanced chaperone activity of the peroxiredoxin proteins [29–31] have been reported as functional outputs of peroxiredoxin hyperoxidation. Interestingly, recent studies in macrophages have demonstrated that nitric oxide affects expression of peroxiredoxins and sulfiredoxin, as well as sensitivity of peroxiredoxins toward hyperoxidation, highlighting the intricate interplay between RNS and ROS regulated signal transduction [32].
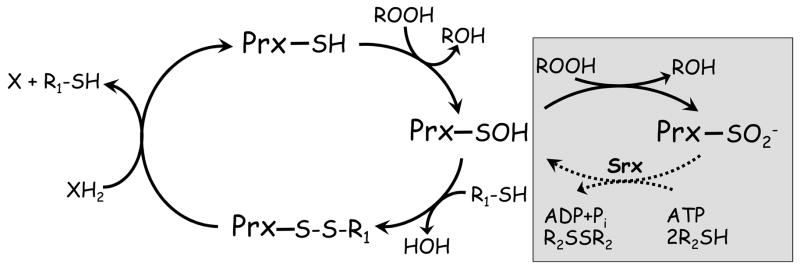
Figure 2. Peroxiredoxin catalytic and regulatory redox cycles.
Peroxiredoxins (Prx) have in common the first step of catalysis whereby the active site cysteine thiol (in its thiolate form) attacks the hydroperoxide substrate, releasing the corresponding alcohol and the enzyme in its sulfenic acid (SOH) form. For catalytic recycling, a resolving cysteine (R1-SH) in the same or another subunit typically forms a disulfide bond with the peroxidatic cysteine, and the enzyme is regenerated by small molecule or protein electron donors. The sulfenic acid can also act as a redox-sensitive switch, converting the enzyme to an inactive sulfinic acid form in the presence of excess hydroperoxide substrate (gray box). Enzymes called sulfiredoxins (and perhaps sestrins) can regenerate activity in some Prx through an ATP-dependent reduction (dotted line).
Detection of reversibly oxidized cysteine residues in proteins
As a category of regulatory posttranslational modifications (PTMs) acting as switches for modulating protein function, oxidation is a newcomer to the field relative to phosphorylation, glycosylation, ubiquitination and acetylation. Given the rapidly expanding interest in this field, a number of methods for the proteome-wide detection of reversible thiol oxidation have been developed, with most relying on the use of thiol modifying agents and detection of either thiols lost during oxidation (which would include those undergoing irreversible oxidation), or those regained (after blocking of free thiols initially present) upon reversal of specific or general oxidized species by reductants (for reviews, see [33–35]). Such approaches have been the basis for “biotin switch” methods to detect either S-nitrosation based on selective ascorbate reduction of these PTMs, or “sulfenation” (generation of sulfenic acid) based on selective arsenite reduction [36]. The successful use of such approaches requires not only comprehensive blocking of all free thiols at the beginning of the procedure, but also an absolute specificity of the reductant used for the targeted PTM, and a stability of that PTM through a multi-step procedure, all aspects which may limit the utility of such methods.
Alternative methods have been developed very recently which allow for the detection of sulfenic acid modifications on proteins based on the distinct chemical attributes of this species. Detection of sulfenic acids in proteins has the advantage of targeting the direct protein product of cysteine modification by hydrogen peroxide, lipid hydroperoxides and peroxynitrite; a potential disadvantage is that the lifetime of the sulfenic acid moiety may be quite short for many proteins, requiring a rapid chemical trapping procedure to detect such species. Another important advantage to the detection of the sites of sulfenic acid formation as one readout of protein oxidation is the identification via this method of the actual reactive site where the oxidation chemistry is initiated, whereas detection of downstream products may involve a great deal more complexity and would not necessarily identify the initial site of attack.
Although most of the new methods described here rely on small molecule probes for detecting sulfenic acid formation, one very recent article also reported the utility of using an engineered, sulfenic acid-sensing module of a yeast transcription factor, Yap-1, to detect protein sulfenic acid formation [37]. NBD-Cl (7-chloro-4-nitrobenzo-2-oxa-1,3-diazole) has been used in the past to distinguish sulfenic acids from thiols in pure proteins through UV-visible spectroscopy or mass spectrometry, but this labeling agent reacts with both thiols and sulfenic acids and is readily removed with dithiothreitol, thus limiting its utility for proteomics-based analyses [38]. On the other hand, dimedone irreversibly alkylates oxidized cysteine residues and has been used as a sulfenic acid specific modifying agent for years. Consequently, several newly developed methods have enabled the linkage of fluorescent [39,40] or affinity-based probes [39,41] to a dimedone-like core reagent to allow for the incorporation of detectable labels into proteins at their sulfenic acid sites (Figure 3). Initial results regarding the specificity of the reagents [40], as well as their detectability, rates of labeling and utility in cells and tissues [39,41,42], suggest that these will be highly useful chemical tools for addressing the biological relevance of cysteine oxidation in many systems in the future.
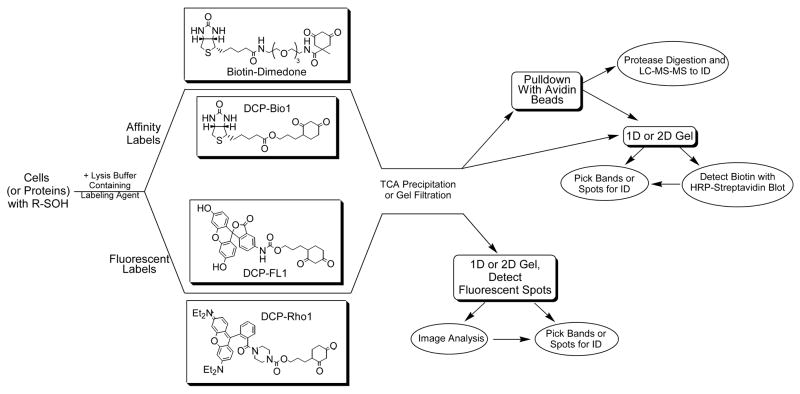
Figure 3. Strategies for detecting and isolating sulfenic acids in proteins.
Cells or proteins with cysteine sulfenic acid modifications are incubated with either affinity (biotin-dimedone or DCP-Bio1, or two other affinity probes [39,41]) or fluorescently-tagged reagents (DCP-FL1 or DCP-Rho1, or two other fluorescein- and rhodamine-based probes [39]). The chemically reactive probes based on dimedone include a nucleophilic carbon between the two carbonyls of the cyclohexane ring that exhibits specificity toward sulfenic acids. After incubation, unreacted probes are removed from the protein samples by trichloroacetic acid (TCA) precipitation or gel filtration chromatography. Subsequent analytical procedures can include one-dimensional or two-dimensional gels in both cases, from which bands or spots can be excised, digested and analyzed by mass spectrometry (MS) for identification of the labeled protein. For the biotinylated samples, “pulldown” of the affinity-labeled proteins with avidin-linked beads can be carried out either before or after proteolytic digestion of the proteins in the samples, enriching in either labeled proteins or labeled peptides. Subsequent LC-MS-MS analysis can then be used to identify labeled proteins/peptides.
Selected new examples of functional protein oxidation
The activities of many proteins in signaling pathways have been shown to be upregulated or downregulated in reponse to ROS, but many such reports lack clarity as to which protein, and especially which cysteine residue(s) within that protein, is/are actually sensitive to oxidants. We compile here a limited selection of recent reports describing new data for transcription factors and enzymes for which cysteine oxidation has been either proven or likely to occur through sulfenic acid formation and has recently been shown to have a functional effect on protein function (for other examples, see reference [43]).
Mechanistic details of the distinct molecular pathways for sensing organic hydroperoxides by a transcriptional repressor, OhrR, from two different organisms, Xanthomonas campestris (XcOhrR) and Bacillus subtilis (BcOhrR), have recently been elucidated. Although both proteins initially generate sulfenic acid at their reactive cysteine (Cys22 in XcOhrR and Cys15 in BsOhrR), XcOhrR quickly forms an intersubunit disulfide bond with Cys127 in that protein [44], whereas BsOhrR has no other cysteine [20]. Instead, formation in the latter protein of either a mixed disulfide with low molecular weight thiols (S-thiolation) or a protein sulfenamide (Figure 1) is required to abrogate DNA binding activity and allow transcription at the target sites.
Protein tyrosine phosphatases were the first signaling-relevant enzymatic activities clearly shown to undergo, and be inhibited by, sulfenic acid formation through oxidation at the nucleophilic cysteine that forms the phosphoenzyme intermediate of PTP1B during catalysis [45] (recent reviews include references [18,46]). Recently, Rinna et al. showed that PTP-1B becomes glutathionylated in a rat alveolar macrophage cell line (NR8383) after either exposure to 100 μM exogenous hydrogen peroxide or ADP-stimulation of the respiratory burst [47]. Recent strong evidence for sulfenic acid formation and subsequent disulfide bond formation has been presented for MAP kinase phosphatase 3, a dual-specficity phosphatase which dephosphorylates Jun N-terminal kinases (JNK) [48]. Finally, in naïve T-cells stimulated with anti-CD3 and anti-CD28 antibodies, both SHP-1 and SHP-2 were shown to form sulfenic acid by trapping oxidized cysteines with DCP-Bio1 (Figure 3), immunoprecipitation of the protein of interest, and blotting with streptavidin to detect incorporation of biotin [42].
Another category of oxidation targets important in signaling are regulatory protein kinases which are typically activated through phosphorylation cascades. While sulfenic acid formation has yet to be proven, the activity of many kinases appear to be regulated by hydrogen peroxide-mediated oxidation of non-catalytic cysteines (reviewed in [49]). New mechanistic details of the effects of oxidation are also emerging in this area. For example, when the catalytic subunit of cAMP-dependent protein kinase (PKA) was treated with hydrogen peroxide, Cys199 formed a disulfide bond with either glutathione or another cysteine residue within the protein, leading to dephosphorylation of a critical proximal Thr residue (Thr197) and inactivation of the protein [50]. cGMP-dependent protein kinase (PKGIα) was shown to form an interpro tein disulfide bond between Cys46 on two subunits after treatment of isolated perfused rat hearts with hydrogen peroxide that correlated with an increase in kinase activity and cellular relocalization of PKGIα; neither response was observed in the C46S mutant. On the other hand, the non-receptor tyrosine kinase c-Abl was shown to be inactivated by modification of cysteine residues by thiol alkylating agents and S-glutathionylation [51]. Other kinases that have been shown to be regulated by cysteine modification include IKKβ and MEKK1 (reviewed in [49,52]).
Other enzymes whose activity has recently been shown to be modified through the initial formation of sulfenic acid formation include Cathepsin B and L, two cysteine-based proteases that lose 50% to 75% of their activity after treatment with 10 μM hydrogen peroxide or protein/peptide hydroperoxides [53]. Rat phospholipase A2 (iPLA2β) exhibited a H2O2-dependent loss of activity correlated with loss of free thiol groups; cysteine sulfenic acid formation was proposed based on mass spectrometric evidence, dithiothreitol reversibility and a reduced sensitivity to oxidative inhibition upon mutagenesis of Cys651 [54]. Reversible redox regulation was also observed for SUMO proteases from human (SENP1) and yeast (Ulp1) through oxidation at the active site cysteine [55]. Cysteine sulfenic acid, as well as sulfinic and sulfonic acids, were identified at the active site of yeast Ulp1 through crystallographic studies. Finally, a comprehensive study of sulfenic acid formation and reactivity toward a panel of thiol-containing small molecules and potential reductants was conducted recently for human serum albumin, the highly abundant blood protein known to be reactive toward hydrogen peroxide [56].
Conclusions
Progress in and continued development of new chemical tools to evaluate, with high spatial and temporal resolution, the generation of hydrogen peroxide in response to cell signaling events, as well as to identify sites and in vivo relevance of peroxide-mediated oxidation of enzymes and transcription factors, bodes well for expanding our understanding of molecular events in ROS-dependent signal transduction. This will have important implications in better defining both normal physiological and pathological processes, and will allow for the identification of new therapeutic targets based on better defined oxidative mechanisms.
Acknowledgments
The authors thank Andy Karplus, Todd Lowther, Cristina Furdui, Bruce King and Jacque Fetrow for editorial suggestions. This work was supported by grants from the National Institutes of Health to L.B.P (RO1 GM50389 and R21 CA112145), K.J.N. (F32 GM074537), S. Bruce King (RO1 HL062198) and Jacquelyn S. Fetrow (GM 075304), and by a grant from the National Science Foundation to J.S.F. (MCB 0517343).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- ••1.Li Q, Harraz MM, Zhou W, Zhang LN, Ding W, Zhang Y, Eggleston T, Yeaman C, Banfi B, Engelhardt JF. Nox2 and Rac1 regulate H2O2-dependent recruitment of TRAF6 to endosomal interleukin-1 receptor complexes. Mol Cell Biol. 2006;26:140–154. doi: 10.1128/MCB.26.1.140-154.2006. Trafficking of both activated IL-1 receptor complexes and activated Nox complexes to endosomal compartments is demonstrated, along with evidence supporting the generation of superoxide and hydrogen peroxide within the lumen of the endosomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rhee SG. Cell signaling. H2O2, a necessary evil for cell signaling. Science. 2006;312:1882–1883. doi: 10.1126/science.1130481. [DOI] [PubMed] [Google Scholar]
- 3.Ushio-Fukai M. Localizing NADPH oxidase-derived ROS. Sci STKE. 2006;2006:re8. doi: 10.1126/stke.3492006re8. [DOI] [PubMed] [Google Scholar]
- 4.Wood ZA, Poole LB, Karplus PA. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science. 2003;300:650–653. doi: 10.1126/science.1080405. [DOI] [PubMed] [Google Scholar]
- 5.Veal EA, Day AM, Morgan BA. Hydrogen peroxide sensing and signaling. Mol Cell. 2007;26:1–14. doi: 10.1016/j.molcel.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 6.Rhee SG. Measuring H(2)O(2) produced in response to cell surface receptor activation. Nat Chem Biol. 2007;3:244–246. doi: 10.1038/nchembio0507-244. [DOI] [PubMed] [Google Scholar]
- ••7.Miller EW, Tulyathan O, Isacoff EY, Chang CJ. Molecular imaging of hydrogen peroxide produced for cell signaling. Nat Chem Biol. 2007;3:263–267. doi: 10.1038/nchembio871. This letter introduces the first chemoselective, boronate-based fluorescent probes able to image hydrogen peroxide in living cells at physiologically-meaningful levels, e.g. after growth factor stimulation. [DOI] [PubMed] [Google Scholar]
- 8.Albers AE, Okreglak VS, Chang CJ. A FRET-based approach to ratiometric fluorescence detection of hydrogen peroxide. J Am Chem Soc. 2006;128:9640–9641. doi: 10.1021/ja063308k. [DOI] [PubMed] [Google Scholar]
- 9.Belousov VV, Fradkov AF, Lukyanov KA, Staroverov DB, Shakhbazov KS, Terskikh AV, Lukyanov S. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat Methods. 2006;3:281–286. doi: 10.1038/nmeth866. [DOI] [PubMed] [Google Scholar]
- 10.Lee D, Khaja S, Velasquez-Castano JC, Dasari M, Sun C, Petros J, Taylor WR, Murthy N. In vivo imaging of hydrogen peroxide with chemiluminescent nanoparticles. Nat Mater. 2007;6:765–769. doi: 10.1038/nmat1983. [DOI] [PubMed] [Google Scholar]
- 11.Miller EW, Chang CJ. Fluorescent probes for nitric oxide and hydrogen peroxide in cell signaling. Curr Opin Chem Biol. 2007 doi: 10.1016/j.cbpa.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wardman P. Fluorescent and luminescent probes for measurement of oxidative and nitrosative species in cells and tissues: progress, pitfalls, and prospects. Free Radic Biol Med. 2007;43:995–1022. doi: 10.1016/j.freeradbiomed.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 13.Biswas S, Chida AS, Rahman I. Redox modifications of protein-thiols: emerging roles in cell signaling. Biochem Pharmacol. 2006;71:551–564. doi: 10.1016/j.bcp.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 14.Salsbury JFR, Knutson ST, Poole LB, Fetrow JS. Functional site profiling and electrostatic analysis of cysteines modifiable to cysteine sulfenic acid. Prot Sci. 2008 doi: 10.1110/ps.073096508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •15.Phalen TJ, Weirather K, Deming PB, Anathy V, Howe AK, van der Vliet A, Jönsson TJ, Poole LB, Heintz NH. Oxidation state governs structural transitions in peroxiredoxin II that correlate with cell cycle arrest and recovery. J Cell Biol. 2006;175:779–789. doi: 10.1083/jcb.200606005. Normal mitogenic signaling did not cause hyperoxidation of peroxiredoxins in epithelial cells, but hydrogen peroxide-dependent cell cycle arrest was correlated with the hyperoxidation of PrxII and its transition into large filamentous oligomers. Expression of cyclin D1 and cell proliferation did not resume until these oxidation and structural changes were reversed, suggesting PrxII as a sensor of perturbations in peroxide homeostasis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salmeen A, Barford D. Functions and mechanisms of redox regulation of cysteine-based phosphatases. Antioxid Redox Signal. 2005;7:560–577. doi: 10.1089/ars.2005.7.560. [DOI] [PubMed] [Google Scholar]
- 17.Yang J, Groen A, Lemeer S, Jans A, Slijper M, Roe SM, den Hertog J, Barford D. Reversible oxidation of the membrane distal domain of receptor PTPalpha is mediated by a cyclic sulfenamide. Biochemistry. 2007;46:709–719. doi: 10.1021/bi061546m. [DOI] [PubMed] [Google Scholar]
- 18.Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7:833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 19.Sarma BK, Mugesh G. Redox regulation of protein tyrosine phosphatase 1B (PTP1B): a biomimetic study on the unexpected formation of a sulfenyl amide intermediate. J Am Chem Soc. 2007;129:8872–8881. doi: 10.1021/ja070410o. [DOI] [PubMed] [Google Scholar]
- 20.Lee JW, Soonsanga S, Helmann JD. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc Natl Acad Sci U S A. 2007 doi: 10.1073/pnas.0702081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang J, Nakamura T, Cho DH, Gu Z, Lipton SA. S-nitrosylation of peroxiredoxin 2 promotes oxidative stress-induced neuronal cell death in Parkinson's disease. Proc Natl Acad Sci U S A. 2007;104:18742–18747. doi: 10.1073/pnas.0705904104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parsonage D, Karplus PA, Poole LB. Substrate specificity and redox potential of AhpC, a bacterial peroxiredoxin. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0708308105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peskin AV, Low FM, Paton LN, Maghzal GJ, Hampton MB, Winterbourn CC. The high reactivity of peroxiredoxin 2 with H(2)O(2) is not reflected in its reaction with other oxidants and thiol reagents. J Biol Chem. 2007;282:11885–11892. doi: 10.1074/jbc.M700339200. [DOI] [PubMed] [Google Scholar]
- 24.Poole LB. The Catalytic Mechanism of Peroxiredoxins. In: Flohé L, Harris JR, editors. Peroxiredoxin Systems. Springer; 2007. pp. 61–81. [DOI] [PubMed] [Google Scholar]
- 25.Karplus PA, Hall A. Structural Survey of the Peroxiredoxins. In: Flohé L, Harris JR, editors. Peroxiredoxin Systems. Springer; 2007. pp. 41–60. [DOI] [PubMed] [Google Scholar]
- 26.Jeong W, Park SJ, Chang TS, Lee DY, Rhee SG. Molecular mechanism of the reduction of cysteine sulfinic acid of peroxiredoxin to cysteine by mammalian sulfiredoxin. J Biol Chem. 2006;281:14400–14407. doi: 10.1074/jbc.M511082200. [DOI] [PubMed] [Google Scholar]
- 27.Jönsson TJ, Johnson LC, Lowther WT. Structure of the sulphiredoxin-peroxiredoxin complex reveals an essential repair embrace. Nature. 2007 doi: 10.1038/nature06415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rabilloud T, Heller M, Gasnier F, Luche S, Rey C, Aebersold R, Benahmed M, Louisot P, Lunardi J. Proteomics analysis of cellular response to oxidative stress. Evidence for in vivo overoxidation of peroxiredoxins at their active site. J Biol Chem. 2002;277:19396–19401. doi: 10.1074/jbc.M106585200. [DOI] [PubMed] [Google Scholar]
- 29.Lee W, Choi KS, Riddell J, Ip C, Ghosh D, Park JH, Park YM. Human peroxiredoxin 1 and 2 are not duplicate proteins: the unique presence of CYS83 in Prx1 underscores the structural and functional differences between Prx1 and Prx2. J Biol Chem. 2007;282:22011–22022. doi: 10.1074/jbc.M610330200. [DOI] [PubMed] [Google Scholar]
- 30.Jang HH, Kim SY, Park SK, Jeon HS, Lee YM, Jung JH, Lee SY, Chae HB, Jung YJ, Lee KO, et al. Phosphorylation and concomitant structural changes in human 2-Cys peroxiredoxin isotype I differentially regulate its peroxidase and molecular chaperone functions. FEBS Lett. 2006;580:351–355. doi: 10.1016/j.febslet.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 31.Chuang MH, Wu MS, Lo WL, Lin JT, Wong CH, Chiou SH. The antioxidant protein alkylhydroperoxide reductase of Helicobacter pylori switches from a peroxide reductase to a molecular chaperone function. Proc Natl Acad Sci U S A. 2006;103:2552–2557. doi: 10.1073/pnas.0510770103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •32.Diet A, Abbas K, Bouton C, Guillon B, Tomasello F, Fourquet S, Toledano MB, Drapier JC. Regulation of peroxiredoxins by nitric oxide in immunostimulated macrophages. J Biol Chem. 2007;282:36199–36205. doi: 10.1074/jbc.M706420200. This paper reports that the upregulation of PrxI and VI, as well as sulfiredoxin, is mediated by NO in macrophages stimulated with interferon γ or lipopolysaccharide. NO was also shown to protect against hyperoxidation of peroxiredoxins and hasten their recovery, highlighting the intricate interplay between NO- and ROS-dependent signaling. [DOI] [PubMed] [Google Scholar]
- 33.Leichert LI, Jakob U. Global methods to monitor the thiol-disulfide state of proteins in vivo. Antioxid Redox Signal. 2006;8:763–772. doi: 10.1089/ars.2006.8.763. [DOI] [PubMed] [Google Scholar]
- 34.Spickett CM, Pitt AR, Morrice N, Kolch W. Proteomic analysis of phosphorylation, oxidation and nitrosylation in signal transduction. Biochim Biophys Acta. 2006;1764:1823–1841. doi: 10.1016/j.bbapap.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 35.Ying J, Clavreul N, Sethuraman M, Adachi T, Cohen RA. Thiol oxidation in signaling and response to stress: detection and quantification of physiological and pathophysiological thiol modifications. Free Radic Biol Med. 2007;43:1099–1108. doi: 10.1016/j.freeradbiomed.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eaton P. Protein thiol oxidation in health and disease: techniques for measuring disulfides and related modifications in complex protein mixtures. Free Radic Biol Med. 2006;40:1889–1899. doi: 10.1016/j.freeradbiomed.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 37.Takanishi CL, Ma LH, Wood MJ. A Genetically Encoded Probe for Cysteine Sulfenic Acid Protein Modification in Vivo. Biochemistry. 2007 doi: 10.1021/bi701625s. [DOI] [PubMed] [Google Scholar]
- 38.Poole LB, Ellis HR. Identification of cysteine sulfenic acid in AhpC of alkyl hydroperoxide reductase. Methods Enzymol. 2002;348:122–136. doi: 10.1016/s0076-6879(02)48632-6. [DOI] [PubMed] [Google Scholar]
- ••39.Poole LB, Klomsiri C, Knaggs SA, Furdui CM, Nelson KJ, Thomas MJ, Fetrow JS, Daniel LW, King SB. Fluorescent and affinity-based tools to detect cysteine sulfenic acid formation in proteins. Bioconjug Chem. 2007;18:2004–2017. doi: 10.1021/bc700257a. Seven new affinity-based or fluorescent tagging reagents for detecting sulfenic acid formation in proteins are presented and tested in vitro with pure proteins known to form stable sulfenic acids. At least some rhodamine and biotin probes are cell permeable for use in in vivo studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poole LB, Zeng BB, Knaggs SA, Yakubu M, King SB. Synthesis of chemical probes to map sulfenic acid modifications on proteins. Bioconjug Chem. 2005;16:1624–1628. doi: 10.1021/bc050257s. [DOI] [PubMed] [Google Scholar]
- ••41.Charles RL, Schröder E, May G, Free P, Gaffney PR, Wait R, Begum S, Heads RJ, Eaton P. Protein sulfenation as a redox sensor: proteomics studies using a novel biotinylated dimedone analogue. Mol Cell Proteomics. 2007;6:1473–1484. doi: 10.1074/mcp.M700065-MCP200. Synthesis and use of a biotin-dimedone conjugate for labeling protein sulfenic acids is reported. Labeled proteins were identified from hydrogen peroxide treated rat ventricular myocytes and perfused hearts, and detected by fluorescence microscopy throughout fixed cells probed with avidin-FITC. [DOI] [PubMed] [Google Scholar]
- ••42.Michalek RD, Nelson KJ, Holbrook BC, Yi JS, Stridiron D, Daniel LW, Fetrow JS, King SB, Poole LB, Grayson JM. The requirement of reversible cysteine sulfenic acid formation for T cell activation and function. J Immunol. 2007;179:6456–6467. doi: 10.4049/jimmunol.179.10.6456. This paper represents one of the first uses of sulfenic acid directed probes in detecting signaling relevant protein oxidation, and concludes that reversible cysteine sulfenic acid formation is an important regulatory mechanism by which naïve CD8(+) T cells undergo activation and proliferation. [DOI] [PubMed] [Google Scholar]
- 43.Poole LB, Karplus PA, Claiborne A. Protein sulfenic acids in redox signaling. Annu Rev Pharmacol Toxicol. 2004;44:325–347. doi: 10.1146/annurev.pharmtox.44.101802.121735. [DOI] [PubMed] [Google Scholar]
- 44.Panmanee W, Vattanaviboon P, Poole LB, Mongkolsuk S. Novel organic hydroperoxide-sensing and responding mechanisms for OhrR, a major bacterial sensor and regulator of organic hydroperoxide stress. J Bacteriol. 2006;188:1389–1395. doi: 10.1128/JB.188.4.1389-1395.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Denu JM, Tanner KG. Specific and reversible inactivation of protein tyrosine phosphatases by hydrogen peroxide: Evidence for a sulfenic acid intermediate and implications for redox regulation. Biochemistry. 1998;37:5633–5642. doi: 10.1021/bi973035t. [DOI] [PubMed] [Google Scholar]
- 46.Chiarugi P, Buricchi F. Protein Tyrosine Phosphorylation and Reversible Oxidation: Two Cross-Talking Posttranslation Modifications. Antioxid Redox Signal. 2007;9:1–24. doi: 10.1089/ars.2007.9.1. [DOI] [PubMed] [Google Scholar]
- 47.Rinna A, Torres M, Forman HJ. Stimulation of the alveolar macrophage respiratory burst by ADP causes selective glutathionylation of protein tyrosine phosphatase 1B. Free Radic Biol Med. 2006;41:86–91. doi: 10.1016/j.freeradbiomed.2006.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seth D, Rudolph J. Redox regulation of MAP kinase phosphatase 3. Biochemistry. 2006;45:8476–8487. doi: 10.1021/bi060157p. [DOI] [PubMed] [Google Scholar]
- 49.Cross JV, Templeton DJ. Regulation of signal transduction through protein cysteine oxidation. Antioxid Redox Signal. 2006;8:1819–1827. doi: 10.1089/ars.2006.8.1819. [DOI] [PubMed] [Google Scholar]
- 50.Humphries KM, Deal MS, Taylor SS. Enhanced dephosphorylation of cAMP-dependent protein kinase by oxidation and thiol modification. J Biol Chem. 2005;280:2750–2758. doi: 10.1074/jbc.M410242200. [DOI] [PubMed] [Google Scholar]
- 51.Leonberg AK, Chai YC. The functional role of cysteine residues for c-Abl kinase activity. Mol Cell Biochem. 2007;304:207–212. doi: 10.1007/s11010-007-9501-y. [DOI] [PubMed] [Google Scholar]
- 52.Pantano C, Reynaert NL, van der Vliet A, Janssen-Heininger YM. Redox-sensitive kinases of the nuclear factor-kappaB signaling pathway. Antioxid Redox Signal. 2006;8:1791–1806. doi: 10.1089/ars.2006.8.1791. [DOI] [PubMed] [Google Scholar]
- 53.Headlam HA, Gracanin M, Rodgers KJ, Davies MJ. Inhibition of cathepsins and related proteases by amino acid, peptide, and protein hydroperoxides. Free Radic Biol Med. 2006;40:1539–1548. doi: 10.1016/j.freeradbiomed.2005.12.036. [DOI] [PubMed] [Google Scholar]
- 54.Song H, Bao S, Ramanadham S, Turk J. Effects of biological oxidants on the catalytic activity and structure of group VIA phospholipase A2. Biochemistry. 2006;45:6392–6406. doi: 10.1021/bi060502a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu Z, Mi Lam LS, Lam LH, Chau SF, Ng TB, Au SW. Molecular basis of the redox regulation of SUMO proteases: a protective mechanism of intermolecular disulfide linkage against irreversible sulfhydryl oxidation. Faseb J. 2007 doi: 10.1096/fj.06-7871com. [DOI] [PubMed] [Google Scholar]
- 56.Turell L, Botti H, Carballal S, Ferrer-Sueta G, Souza JM, Duran R, Freeman BA, Radi R, Alvarez B. Reactivity of Sulfenic Acid in Human Serum Albumin. Biochemistry. 2007 doi: 10.1021/bi701520y. [DOI] [PubMed] [Google Scholar]