Abstract
Virus assembly occurs in a complex environment and is dependent upon viral and cellular components being properly correlated in time and space. The simplicity of the Flock House virus (FHV) capsid and the extensive structural, biochemical, and genetic characterization of the virus make it an excellent system for studying in vivo virus assembly. The tetracysteine motif (CCPGCC), that induces fluorescence in bound biarsenical compounds (FlAsH and ReAsH), was genetically inserted in the coat protein, to visualize this gene product during virus infection. The small size of this modification when compared to those made by traditional fluorescent proteins minimizes disruption of the coat proteins numerous functions. ReAsH not only fluoresces when bound to the tetracysteine motif but also allows correlated electron microscopy (EM) of the same cell following photoconversion and osmium staining. These studies demonstrated that the coat protein was concentrated in discrete patches in the cell. High pressure freezing (HPF) followed by freeze substitution (FS) of infected cells showed that these patches were formed by virus particles in crystalline arrays. EM tomography (EMT) of the HPF/FS prepared samples showed that these arrays were proximal to highly modified mitochondria previously established to be the site of RNA replication. Two features of the mitochondrial modification are ~60 nm spherules that line the outer membrane and the large chamber created by the convolution induced in the entire organelle.
1. Introduction
Modern cell biology has revealed that all cellular functions require their components to be correlated in space and time. It is clear that viral infection and replication share this organizational hierarchy. Often the infection alters cellular chemistry and organelle morphology to enhance virus production. Genetic, biochemical, and structural studies of the Flock House virus (FHV) have provided valuable insights into its assembly, making it an excellent system for the study of virus production in the context of the cell. Replication of the viral RNA genome causes drastic changes in the mitochondria (Miller et al. 2001). Expression of just the RNA dependent RNA polymerase (RdRp) results in invaginations on the outer membrane called spherules in which RNA replication occurs (Miller et al. 2001). Following viral RNA replication, RNA2 is translated to produce the viral coat protein α (407 amino acids), and 180 copies of α assemble into a T=3 icosahedral shell containing one copy of RNA1 and RNA2 (Gallagher and Rueckert 1988; Krishna and Schneemann 1999). Infectivity depends on a subsequent autocatalytic maturation in which α is cleaved into the β protein (1–363) and γ peptide (364–407) (Gallagher and Rueckert 1988). Only coat protein synthesized from replicating RNA2 properly incorporates the genome (Venter et al. 2005). This indicates that RNA replication and virion formation are coupled, suggesting that assembly occurs in close proximity to the mitochondria, the site of RNA synthesis.
The location of protein expression, protein age, and the spatial relationship of the coat protein to cellular structures is crucial to understanding virus assembly. Standard genetic tags such as green fluorescent protein or dsRed are often larger than the protein being studied. In order to obtain a good fluorescent signal the protein is appended to coat proteins that repeat in a pattern reflecting its icosahedral symmetry. For FHV, the addition of 180 fluorescent proteins (FPs) is only possible on the outside of the virus because the FP’s would be too bulky to be accommodated inside the virus. However, these large knob-like features on the outside of the virus eliminate infectivity since the relatively large size of GFP sterically hinders receptor binding sites. Published studies on Ebola virus (Panchal et al. 2003) and HIV (Rudner et al. 2005) have shown that tetracysteine tags provide closer to native results than fluorescent protein tags.
Questions associated with protein age and location have been addressed in protein trafficking studies with the development of the tetracysteine motif (CCPGCC) that induces fluorescence when it binds specifically to the small biarsenical molecules FlAsH and ReAsH (Gaietta et al. 2002). The tetracysteine technology has obvious advantages over other methods for labeling proteins in cells. It is relatively small (~6 residues), reducing the chance that it will interfere with the biological activity of the protein (Hoffmann et al. 2005), older protein can be distinguished from newer protein by sequential labeling with ReAsH-EDT2 (red emission) followed by FlAsH-EDT2 (green emission) in live cells (Gaietta et al. 2002), and ReAsH can be subjected to photoconversion to generate electron dense material localized at the ReAsH labeled sites providing selective staining for EM specimens (Gaietta et al. 2002).
A FHV infection can also be established in Drosophila cells by simultaneously transfecting them with RNA1 and RNA2, allowing assembly to be studied in the absence of virus entry. This allowed coat protein containing genetically encoded tetracysteine motif (FHVtetracysteine) to be expressed in Drosophila cells and the protein was observed with fluorescence microscopy. Then higher resolution EM images of photoconverted FHVtetracysteine were collected with the same cell. Electron dense aggregates were observed in Drosophila cells transfected with the FHV mutants containing the tetracysteine motifs and these appeared to displace cellular components. Additionally, cells infected with wild type FHV were prepared for EM using high pressure freezing (HPF) followed by freeze substitution (FS) and examined with electron microscope tomography (EMT), which revealed that large chambers formed in the mitochondria.
2. Materials and methods
2.1. Cell culture and site-directed mutagenesis of FHV coat protein
Drosophila melanogaster cells, Schneider’s line 1 and Schneider line 2 cells (DL1 and S2 respectively), were maintained in Schneider’s insect medium supplemented with 15% heat inactivated fetal bovine serum and penicillin/streptomycin (Gibco BRL) (Friesen and Rueckert 1981) and Schneider’s Drosophila medium (Gibco BRL) supplemented with 10% heat inactivated fetal bovine serum and penicillin/streptomycin, respectively.
The tetracysteine motif was inserted into the FHV coat protein in two locations, substituting residues 4–9 (CCPGCC) or between residues 268–269 (FLNCCPGCCMEP). Using PCR, the tetracysteine motif was inserted into a plasmid containing a cDNA copy of wild-type (wt) FHV coat protein (p2BS(+)-wt) (Schneemann et al. 1992) to produce p2BS(+)-tetracysteine 4–9 and p2BS(+)-tetracysteine 268–269. These plasmids were then used to generate RNA2 by in vitro transcription (Ambion) (Schneemann and Marshall 1998). RNA1 was produced by in vitro transcription (Ambion), transfected into DL1 cells using transfectin (BioRad), and isolated from cells (Schneemann and Marshall 1998).
2.2. ReAsH labeling
For light microscopy 1.5 × 106 S2 cells were plated in Schneider’s Drosophila medium (Gibco BRL) onto 35 mm glass bottom dishes (MatTek), which had previously been covered with a solution of 0.5 mg/ml concanavalin A (Sigma-Aldrich) in water and allowed to dry (Rogers et al. 2002). After allowing the cells to attach for 1 h they were transfected simultaneously with RNA1 and RNA2tetracysteine268–269 using methods similar to those previously described (Schneemann and Marshall 1998). After 3 h the medium was replaced with Schneider’s Drosophila medium supplemented with 10% fetal bovine serum and penicillin/streptomycin. At 21 or 27 h post transfection the media was removed, rinsed one time with Schneider’s Drosophila medium, placed in 1 ml Schneider’s Drosophila medium containing 1.8 µM ReAsH and 25 µM EDT for 1 hour. The cells were then rinsed with Schneider’s Drosophila medium containing 500 µM EDT2 and incubated in Schneider’s Drosophila medium containing 500 µM EDT2 for 10 minutes, and fixed in 2% formaldehyde (EM sciences).
2.3. Photoconversion of ReAsH labeled Drosophila cells
Following labeling with ReAsH-EDT2 the cells were fixed with 2% glutaraldehyde (EM sciences) in 100 mM sodium cacodylate buffer (pH 7.4) for 30 min, rinsed in buffer, and treated in 10 mM KCN, 10 mM aminotriazole, 0.01% hydrogen peroxide, and 50 mM glycine in 100 mM cacodylate buffer (pH 7.4) to reduce background staining (Gaietta et al. 2002). The cells were rinsed in buffer and the area of interest was identified using fluorescence microscopy (MRC-1024 confocal microscope, BioRad). A solution of 1 mg/ml diaminobenzidine in oxygenated 100 mM sodium cacodylate buffer (pH 7.4) was added to the cells, and photoconversion was performed by intense illumination with 585 nm light from a xenon lamp until a brownish reaction product appeared (~8 min). The cells were then washed in buffer, post-fixed in 1% osmium tetroxide for 30 min, rinsed in distilled water, dehydrated in an ethanol gradient, embedded in Durcupan (Fluka), and polymerized at 60° C for 48 hours.
2.4. Sample preparation for electron microscopy
Drosophila cells were plated onto a 35 mm tissue culture plates at a total cell count of 1 × 106 cells. After adherence, cells were infected with FHV at a multiplicity of infection (MOI) of 5. At 18 h post infection the cells were fixed in 2% glutaraldehyde (EM Sciences) in 100 mM sodium cacodylate buffer (pH 7.4) for 30 min, washed in buffer, post-fixed in 1% osmium tetroxide for 30 min, rinsed in distilled water, stained in 2% uranyl acetate in water for 1 h, rinsed in distilled water, dehydrated in an ethanol gradient, embedded in Durcupan, and polymerized at 60° C for 48 hours.
For samples prepared by cryofixation, Drosophila cells were infected with FHV at a MOI of 5. At 12 h post infection the cells were gently removed from the plates and centrifuged at 500g for 5 min. A small sample from the resulting pellet was placed into a brass planchet (Ted Pella) and rapidly frozen in a high pressure freezer (BAL-TEC HPM 010). The frozen planchets were placed into a freeze substitution device (Leica EM-AFS) at −90 °C then treated with 1% OsO4 and 0.1% uranyl acetate for 72 hours at −60 °C. Alternatively the samples treated with 0.1% tannic acid in acetone for 24 h, washed three times with cold acetone, and then treated with 1% OsO4 and 0.1% uranyl acetate for 72 hours at −60 °C. The sample were then warmed to 4° C and were infiltrated with 50:50 Durcupan:acetone for 1 hour, 100% Durcupan for 24 hours, and 100% Durcupan for 24 hours, then polymerized at 60 °C for 48 hours.
2.5. Electron tomography and data analysis
Sections of 500 nm thickness were cut and stained with Sato’s lead and 2% uranyl acetate. A cell of interest was located and a series of micrographs were collected on a JEOL 4000EX at 400 keV while the sample was tilted from −60° to + 60° degrees in 2 degree increments. The grid was then rotated in plane 90° and another tilt series was collected. The micrographs were then digitized (0.8 nm/pixel) and aligned using IMOD software (Kremer et al. 1996). The TxBR (transform-based back projection) software package was then used to create the final alignment and back projection resulting in a three-dimensional volume (Lawrence et al. 2006).
3. Results
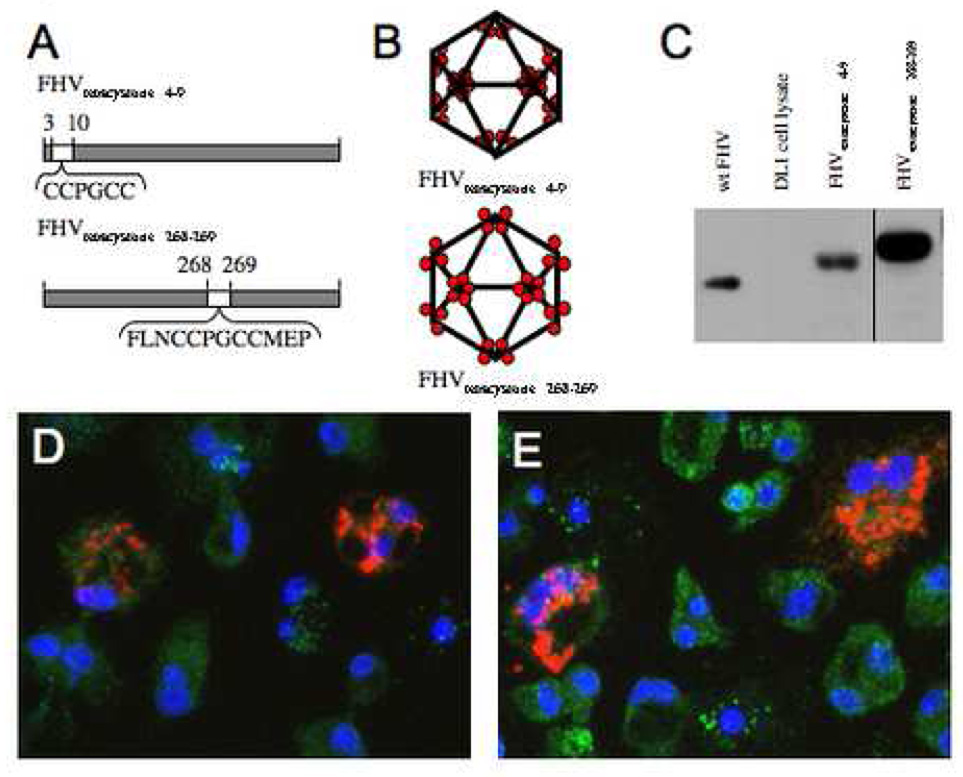
The FHV coat protein displays remarkable functional diversity, including genome recognition and packaging, self-assembly to form a protective capsid, autocatalytic cleavage, binding to the surface receptor, and permeabilization of the membrane for genome delivery. Due to this maximization of functionality it is difficult to alter the coat protein without reducing the efficiency of viral infectivity. The small size of the inserted tetracystiene motif reduces the likelihood of interference with the coat protein’s biological functions when compared to larger fluorescent proteins. Previous structural work showed that the N-terminus of the coat protein is located in the interior of the virus capsid (Fisher and Johnson 1993). Based on this information, the first construct substituted N-terminal residues 4 to 9 with the CCPGCC (FHVtetracysteine 4–9). The surface of the FHVtetracysteine 4–9 virus particles was expected to be identical to wild type, and therefore capable of binding the receptor (Fig. 1A and B). Genetic analysis in combination with structural information showed that the loops on the surface of the virus capsid are responsible for binding the receptor. The second construct optimized the tetracysteine motif for binding efficiency (FLNCCPGCCMEV) (Adams et al. 2002) and was inserted between coat protein residues 268 and 269 (FHVtetracystein 268–269), altering the particle surface and potentially inhibiting binding to the receptor (Fig. 1A and B).
Fig. 1.
Insertion of the tetracysteine motif into the FHV coat protein and expression in Drosophila cells. (A and B) A schematic representation of where the tetracysteine motif was inserted into coat protein for FHVtetracysteine 4–9 and FHVtetracysteine 268–269. (C) DL1 cells were transfected with RNA1 and either RNAtetracysteine 4–9 or RNA2tetracysteine 268–269, and 27 h post infection the cellular lysate was subjected to SDS-PAGE, which was probed with an anti FHV antibody. S2 cells were transfected with both RNA1 and RNA2tetracysteine 268–269 and labeled with ReAsH (red) at 21 h (D) and 27 h (E) post transfection. The mitochondria were labeled with an anti-biotin antibody (green) and the nucleus was labeled with DAPI (blue).
Drosophila cells (S2) were transfected with RNA1 (encoding the viral polymerase) and RNA2 encoding either FHVtetracysteine 4–9 or FHVtetracystein 268–269 (denoted as RNA2tetracysteine 4–9 and RNA2tetracysteine 268–269 respectively). Western blot analysis of cell lysates expressing RNA2tetracysteine 4–9 or RNA2tetracysteine 268–269 had one band, which migrated slightly slower then wt coat protein, consistent with the incorporation of the tetracysteine motif (Fig. 1C). The RNA2tetracysteine 268–269 migrates slowest because it has the larger size tetracysteine domain insertion.
The appearance of fluorescent signal in transfected Drosophila cells confirms that the cells were expressing coat protein capable of binding ReAsH. At 21 and 27 hours post-transfection the cells were placed in serum free media containing ReAsH-EDT2 for one hour. The cells were fixed and observed with the fluorescence microscope. Cells expressing FHVtetracystein 268–269 showed fluorescent patches indicating that the tetracysteine motif in these virus particles was able to bind ReAsH (Figure 1D and E). Fluorescent patches were only observed in ~5% of the cells, a typical transfection efficiency (Schneemann and Marshall 1998). The non-transfected cells serve as an internal control since no fluorescence was observed in those cells. The coat protein is located throughout the cytoplasm in dense patches. Studies with FHVtetracsyteine 4–9 showed a similar fluorescence pattern but the signal was less intense (data not shown).
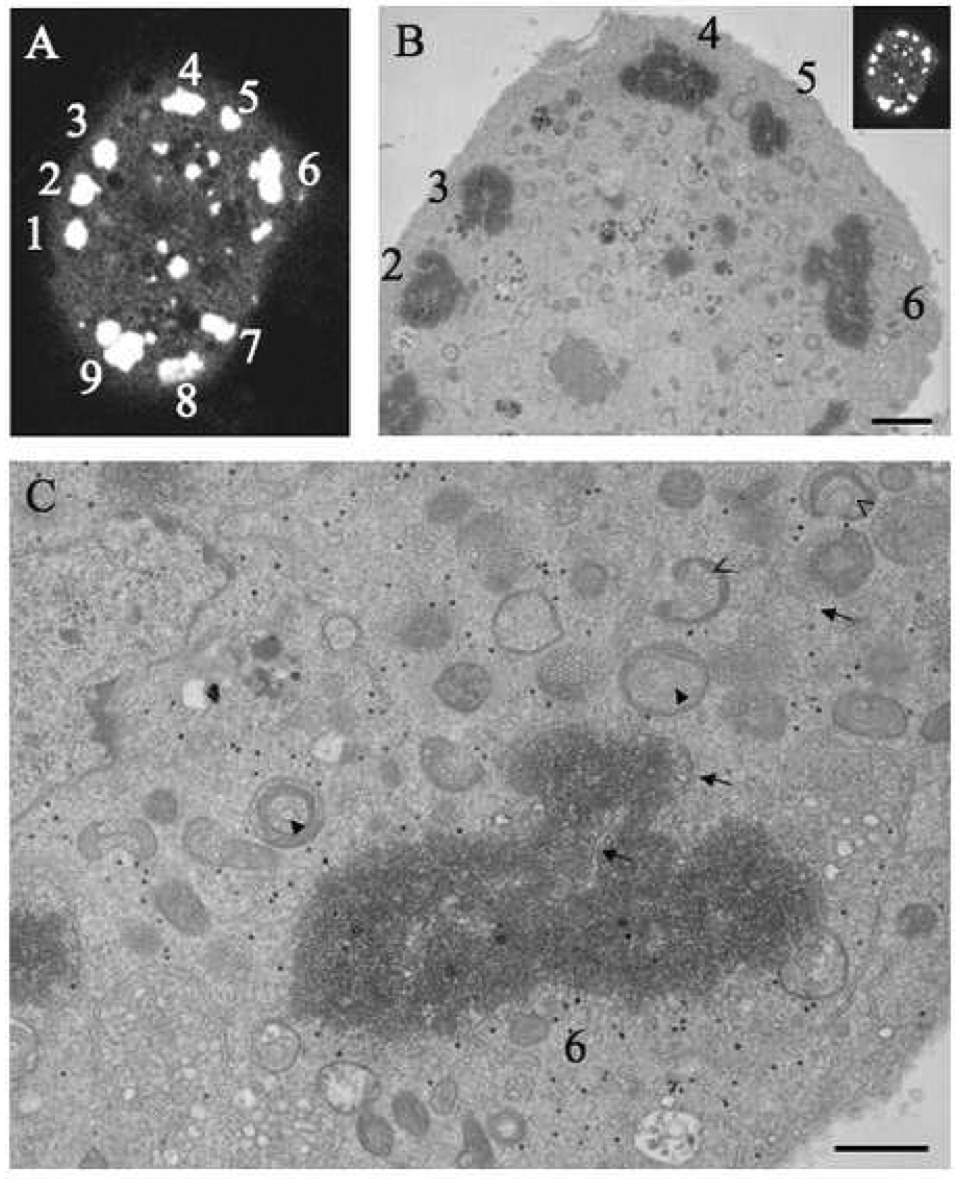
Drosophila cells transfected with RNA1 and RNA2tetracysteine 268–269 and labeled with ReAsH at 27 hours were viewed by fluorescence microscopy and chosen for photo-conversion (Fig. 2A). The cells were dehydrated, embedded in resin, thin sectioned and viewed with the electron microscope. There was striking correlation between the images obtained by fluorescence and EM methods (Fig. 2B).
Fig. 2.
EM of ReAsH labeled FHVtetracysteine 268–269. S2 cells expressing FHVtetracysteine were labeled with ReAsH and a cell of interest was imaged with fluorescence microscopy (A). The cell was then subjected to photoconversion, which created an electron dense-precipitate around the FHV coat protein and was imaged with EM (B and C). The corresponding coat protein patches are numbered. The donut shape mitochondria are indicated by closed arrow while open arrows indicate the horseshoe shaped mitochondria. Scale bars: B – 5 µM, and C – 2 µM.
The EM images showed that the coat protein was confined in large patches observed throughout the cytoplasm (Fig. 2B). These patches were surrounded by mitochondria that have the characteristic spherule structures that are generated by the viral RNA polymerase. The observed spherules were approximately 60 nm, which is in agreement with the previous reported dimensions (Miller et al. 2001; Kopek et al. 2007). Some of the mitochondria have a donut appearance where the spherules form rings on the inside of the mitochondria as well as lining the outside surface (Fig. 2B and C, closed arrow heads). Other mitochondria form horseshoe shapes with spherules lining the inside and outside surfaces of the horseshoe (Fig. 2B and C, open arrow heads). These mitochondria probably have similar structures but the observed shape is dependent upon the direction in which they are sliced during sectioning of the block. This suggests that many of the mitochondria form small chambers lined with spherules that are open to the cytoplasm. It was difficult to determine the overall morphology of these mitochondria since only a thin section was observed. The smooth ER was neighboring the mitochondria and was associated with all the coat protein patches observed (Fig 2C, arrows).
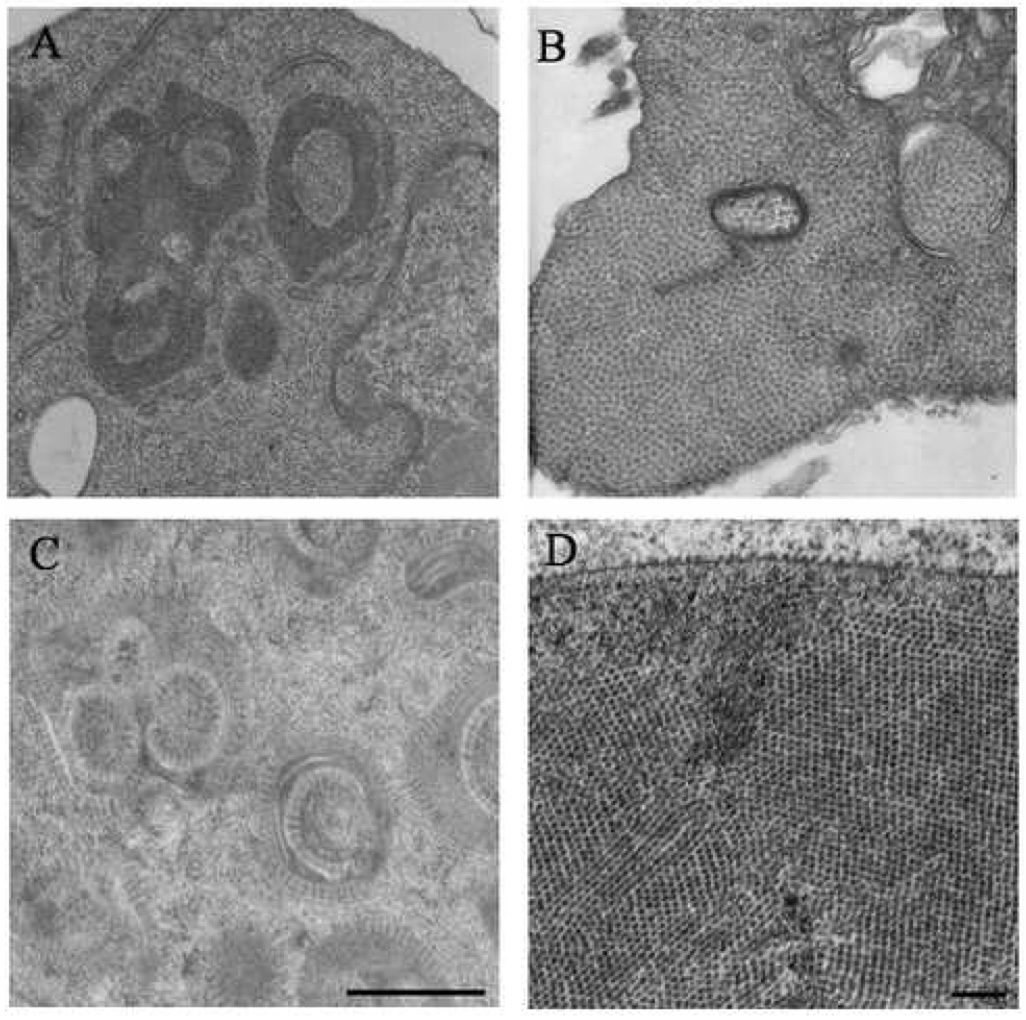
The photo-converted images allowed the location of the coat protein to be determined but it was not possible to establish its assembly state within these patches. The photoconversion process deposits electron dense material around the labeled protein causing it to obscure structural definition. More detailed information on the state of the coat protein was obtained with samples prepared by traditional chemical fixation or HPF and FS. Drosophila cells were infected with wild type FHV and 18 h post infection the cells were prepared for EM analysis. The chemically fixed and HPF/FS prepared samples both showed that the patches were formed by arrays of virus particles (Fig. 3B and D), however the arrays in cells prepared with chemical fixation were not well-ordered and the particles appeared to lie in a matrix, while arrays in the HPF/FS prepared cells were well-ordered, which is more evident when the center of the virus particles were plotted (Sosinsky et. al., in preparation). The cellular membrane is better preserved in HPF/FS cells allowing virus particles to be easily observed bound to the outer surface of the cell membrane. The increased sample quality is most likely due to the minimization of leaching and denaturation of cellular material during steps necessary for staining and embedding.
Fig. 3.
Comparison of ultrastructure using chemical fixation versus high pressure freezing and freeze substitution. Drosophila cell were infected with FHV and at 12 h post infection the cells were prepared for EM using traditional chemical fixation (A and B) or HPF and FS (C and D). The samples prepared by HPF/FS had significant improvement in preservation compared to the samples prepared by traditional chemical fixation. With this improved quality of preservation the paracrystaline nature of the arrays is apparent and the virus particles are observed bound to the cell membrane (D). Scale bars: A and C – 1 µM, and B and D – 250 nm.
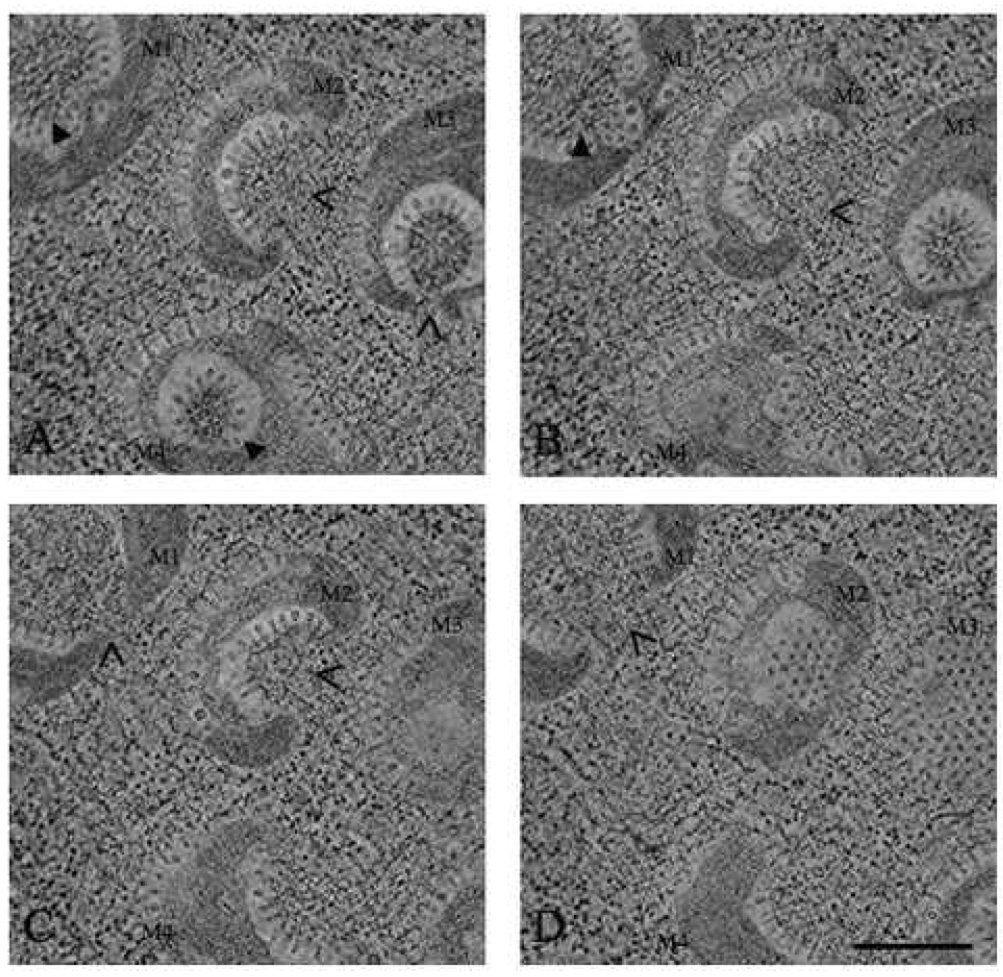
Although HPF allowed better preservation of the cellular features, the overall structure of the mitochondria was still ambiguous due to the two-dimensional nature of these images. A three-dimensional tomographic reconstruction of the mitochondria and their surroundings was computed from 120 images taken in two-degree increments. The thicker sections (500 nm) used for the tomogram allowed the morphology of four mitochondria to be examined in planes of multiple heights. Sections displaying the inner most regions of the mitochondria show that each chamber is open to the cytoplasm at one location (M2 and M3 in Fig. 4A, M2 in Fig. 4B, M1 and M2 in Fig. 4C, and M1 in Fig. 4D). In regions further from the center of the mitochondria the chambers are closed (M1 and M4 in Fig. 4A, and M1 and M3 in Fig. 4B). Sections displaying the outer most portions of the mitochondria display spherules arranged on well-ordered lattices (M2 and M3 in Fig. 4D). Some of the mitochondria have a larger opening in the chamber that appears to be closing (M2 in Fig. 4A, B, and C). These tomographic studies demonstratedthe mitochondria do not have multiple forms but are in fact close to uniform with a shape resembling a small cement mixer. All the spherules observed are formed on the outer mitochondrial membrane, which is convoluted in to form the observed chamber. The formation of these large chambers appear to displace the mitochondrial cristae, packing them into adjacent regions in the mitochondria (M3 in Fig. 4A).
Fig. 4.
EMT of FHV in Drosophila cells. Drosophila cells at 12 h post infection were subjected to HPF and FS, embedded, and cut into ~500 nm sections. Two orthogonal −60 to +60 degree tilt series in 2 degree increments were collected. Four sections approximately 120 nm apart and about 12 nm thick were extracted from the reconstructed volume (A, B, C, and D). Four mitochondria were observed in all the sections and are labeled M1, M2, M3, and M4. A closed arrow indicates the sections where the mitochondrial chambers are closed to the cytoplasm. Open arrows indicate the openings in the mitochondrial chambers. Scale bar: 500 nm.
4. Discussion
Inserting the tetracysteine motif into the viral coat protein allowed its location to be determined in both the fluorescence and electron microscope. The virus particles were primarily located within large paracrystalline arrays, which neighbored some of the mitochondria. However, many polymerase-altered mitochondria were distant from the arrays. One model locates virus assembly adjacent to the arrays, which means the RNA or RNA and coat protein would be transported from their location of synthesis to the edge of the arrays. A second postulates coat protein is synthesized in the vicinity of RNA replication, which occurs at the mitochondria, and that the RNA and coat protein assemble there to form virus particles that are subsequently transported to the paracrystalline arrays (Garzon et al. 1978; Garzon et al. 1990). Recent results suggest that RNA replication and RNA packaging are coupled with both processes occurring in close proximity to each other (Venter and Schneemann 2007). If virus particles assemble close to the mitochondria it is interesting to consider that the large chambers may function to sequester the viral components away from the cytoplasm to increase the local concentrations. RNA produced from spherules outside the chambers and associated assembly processes would not benefit from such confinement.
Distinguishing between these two models requires experimental methods that are able to identify unassembled coat protein and newly assembled virus particles. Previous microscopy studies on Nodaviradae replication have focused on the mitochondria (Garzon et al. 1978; Garzon et al. 1990; Miller et al. 2001; Kopek et al. 2007), the site of RNA replication (Miller et al. 2001), or the arrays of assembled virus particles (Garzon et al. 1978). The large conformational changes of the mitochondria are easily observed and have been reported in numerous studies (Garzon et al. 1978; Garzon et al. 1990; Miller et al. 2001; Kopek et al. 2007). Similarly the arrays of assembled virus particles are easily identifiable and were reported in earlier studies (Garzon et al. 1978). However, it is challenging to distinquish the virus particles in the cytoplasm from ribosomes due to their similar size (~30 nm). Consequently this has made it difficult to determine the location for nodaviradae assembly. The FHV tetracysteine provides an excellent means to circumvent the difficulties in distinguishing between FHV particles and ribosomes in the cytoplasm. Ideally the FHVtetracysteine motif not only allows the location for virus assembly to be determined but also provides a method to determine the location of coat protein synthesis. Using the tetracysteine/FlAsH-ReAsH system, Rodriguez and colleagues were able to identify the site of β-actin protein synthesis and determine the rate at which it trafficked away from these sites (Rodriguez et al. 2006). Similar experiments with FHVtetracysteine 4–9 will allow ReAsH to bind the protein at the earliest moment making it possible to determine the site of protein synthesis. Then the trafficking of the new protein can be determined by sequential labeling with FlAsH-EDT2 and ReAsH-EDT2. These experiments will allow us to assign temporal information to the location of coat protein translation and virus particle assembly.
We have shown here that cryofixation using HPF provides insights into the cellular structure during virus infection, assembly and release. Features we observed in late stages of infection included paracrystalline arrays of viruses localized into domains in the cell, fine substructure in the mitochondrial spherules and even viruses attached to the plasma membrane. However, given the difficulties in identifying single FHV particles in the cytoplasm it is important to combine enhanced and specific staining of coat protein with improvements in ultrastructural preservation. Recent results indicate that photoconverted specimens can be combined with HPF/FS to produce samples with an EM specific labeling and a high quality of preservation (Sosinsky et. al. in preparation). Future experiments will utilize the combination of sequential labeling of coat protein with FlAsH and ReAsH, photoconversion, HPF/FS, and EMT will allow us to determine if coat protein expression occurs inside these mitochondrial chambers or if the protein is expressed elsewhere.
ACKNOWLEDGEMENTS
We are very thankful for the help and assistance provided by Dr. Malcolm Wood (EM facility, The Scripps Research Institute) with preparing samples for EM and Sassan Ghassemzadeh (National Center for Microscopy and Imaging Research, Center for Research on Biological Structure) with image processing of the tomographic data sets. This work was supported by a National Institute of Health training grant GM074536 (to J.K.L.), National Institute of Health grants GM53491 (to A.S.), GM072881 (to G.E.S.), RR004050 (to M.H.E.), GM034220 (to J.E.J.), and a National Science Foundation grant MCB0543934 (to G.E.S.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams SR, Campbell RE, Gross LA, Martin BR, Walkup GK, Yao Y, Llopis J, Tsien RY. New biarsenical ligands and tetracysteine motifs for protein labeling in vitro and in vivo: synthesis and biological applications. Journal of the American Chemical Society. 2002;124:6063–6076. doi: 10.1021/ja017687n. [DOI] [PubMed] [Google Scholar]
- Fisher AJ, Johnson JE. Ordered duplex RNA controls capsid architecture in an icosahedral animal virus. Nature. 1993;361:176–179. doi: 10.1038/361176a0. [DOI] [PubMed] [Google Scholar]
- Friesen PD, Rueckert RR. Synthesis of Black Beetle Virus Proteins in Cultured Drosophila Cells: Differential Expression of RNAs 1 and 2. Journal of virology. 1981;37:876–886. doi: 10.1128/jvi.37.3.876-886.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaietta G, Deerinck TJ, Adams SR, Bouwer J, Tour O, Laird DW, Sosinsky GE, Tsien RY, Ellisman MH. Multicolor and electron microscopic imaging of connexin trafficking. Science. 2002;296:503–507. doi: 10.1126/science.1068793. [DOI] [PubMed] [Google Scholar]
- Gallagher TM, Rueckert RR. Assembly-dependent maturation cleavage in provirions of a small icosahedral insect ribovirus. Journal of virology. 1988;62:3399–3406. doi: 10.1128/jvi.62.9.3399-3406.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon S, Charpentier G, Kurstak E. Morphogenesis of the nodamura virus in the larbae of the lepidopteran Galleria mellonella (L.) Archives of virology. 1978;56:61–76. doi: 10.1007/BF01317283. [DOI] [PubMed] [Google Scholar]
- Garzon S, Strykowski H, Charpentier G. Implication of mitochondria in the replication of Nodamura virus in larvae of the Lepidoptera, Galleria mellonella (L.) and in suckling mice. Archives of virology. 1990;113:165–176. doi: 10.1007/BF01316670. [DOI] [PubMed] [Google Scholar]
- Hoffmann C, Gaietta G, Bunemann M, Adams SR, Oberdorff-Maass S, Behr B, Vilardaga JP, Tsien RY, Ellisman MH, Lohse MJ. A FlAsH-based FRET approach to determine G protein-coupled receptor activation in living cells. Nat Methods. 2005;2:171–176. doi: 10.1038/nmeth742. [DOI] [PubMed] [Google Scholar]
- Kopek BG, Perkins G, Miller DJ, Ellisman MH, Ahlquist P. Three-Dimensional Analysis of a Viral RNA Replication Complex Reveals a Virus-Induced Mini-Organelle. PLoS Biol. 2007;5:e220. doi: 10.1371/journal.pbio.0050220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. Journal of structural biology. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Krishna NK, Schneemann A. Formation of an RNA heterodimer upon heating of nodavirus particles. Journal of virology. 1999;73:1699–1703. doi: 10.1128/jvi.73.2.1699-1703.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence A, Bouwer JC, Perkins G, Ellisman MH. Transform-based backprojection for volume reconstruction of large format electron microscope tilt series. Journal of structural biology. 2006;154:144–167. doi: 10.1016/j.jsb.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Miller DJ, Schwartz MD, Ahlquist P. Flock house virus RNA replicates on outer mitochondrial membranes in Drosophila cells. Journal of virology. 2001;75:11664–11676. doi: 10.1128/JVI.75.23.11664-11676.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchal RG, Ruthel G, Kenny TA, Kallstrom GH, Lane D, Badie SS, Li L, Bavari S, Aman MJ. In vivo oligomerization and raft localization of Ebola virus protein VP40 during vesicular budding. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:15936–15941. doi: 10.1073/pnas.2533915100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez AJ, Shenoy SM, Singer RH, Condeelis J. Visualization of mRNA translation in living cells. J Cell Biol. 2006;175:67–76. doi: 10.1083/jcb.200512137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SL, Rogers GC, Sharp DJ, Vale RD. Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J Cell Biol. 2002;158:873–884. doi: 10.1083/jcb.200202032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner L, Nydegger S, Coren LV, Nagashima K, Thali M, Ott DE. Dynamic fluorescent imaging of human immunodeficiency virus type 1 gag in live cells by biarsenical labeling. Journal of virology. 2005;79:4055–4065. doi: 10.1128/JVI.79.7.4055-4065.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneemann A, Marshall D. Specific encapsidation of nodavirus RNAs is mediated through the C terminus of capsid precursor protein alpha. Journal of virology. 1998;72:8738–8746. doi: 10.1128/jvi.72.11.8738-8746.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneemann A, Zhong W, Gallagher TM, Rueckert RR. Maturation cleavage required for infectivity of a nodavirus. Journal of virology. 1992;66:6728–6734. doi: 10.1128/jvi.66.11.6728-6734.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter PA, Krishna NK, Schneemann A. Capsid protein synthesis from replicating RNA directs specific packaging of the genome of a multipartite, positive-strand RNA virus. Journal of virology. 2005;79:6239–6248. doi: 10.1128/JVI.79.10.6239-6248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter PA, Schneemann A. Assembly of two independent populations of flock house virus particles with distinct RNA packaging characteristics in the same cell. Journal of virology. 2007;81:613–619. doi: 10.1128/JVI.01668-06. [DOI] [PMC free article] [PubMed] [Google Scholar]