Abstract
Histone deacetylases (HDACs) regulate many important physiological processes and the discovery of small molecules that modulate HDAC activity has both academic and clinical relevance. HDAC inhibitors, most notably SAHA, have been pursued as cancer chemotherapeutics but may be useful in treating psychiatric disorders, malaria, and other diseases. Herein, we describe an inexpensive and robust assay, based on fluorescence polarization, for HDAC ligand discovery. The assay is well suited for high-throughput screening and enzyme kinetic studies.
The reversible acetylation of lysine residues within histone tails is regulated by histone acetyltransferase (HAT) and histone deacetylase (HDAC) activity. HATs catalyze the acetylation of histone tails, which generally results in transcriptional activation of nearby genes. In contrast, HDACs catalyze the deacetylation of acetylated histones, which causes transcriptional repression.1–3 The dynamic equilibrium that exists between the activity of HAT and HDAC must be maintained for proper transcriptional activity and cellular function. HDACs are classified into four major classes based upon size, cellular localization, catalytic domain, and mechanism of action. Class I, II and IV are zinc dependent hydrolases, whereas class III enzymes, also called sirtuins, form an unrelated NAD-dependent subfamily. Class I HDACs are generally located in the nucleus and are relatively small in size; Class II HDACs are present both in the nucleus and cytoplasm and are generally larger.4–6
HDAC activity has been shown to repress the transcription of tumor suppressor genes associated with the progression of various leukemias.7 As a result, HDAC inhibitors are being developed to reverse tumor progression by inducing cell differentiation and apoptosis.8–10 Suberoylanilide hydroxamic acid (SAHA; Zolinza) is the best characterized of these inhibitors and was recently approved by the Food and Drug Administration for the treatment of cutaneous T-cell lymphoma.11, 12 Although SAHA is an effective HDAC inhibitor, it shows little selectivity for the Class I, II, and IV enzymes. More selective HDAC inhibitors would be used to deconvolute the biology of individual HDACs and potentially develop better therapeutic agents.
Finding such HDAC inhibitors requires assays that are both cost-effective and amenable to high-throughput screening (HTS). Thus far, only a few in vitro assays have been described to evaluate HDAC inhibitors. Early approaches simply incubated enzyme with in situ acetate-radiolabeled histones and measured the kinetics of acetate release.13, 14 This method is very accurate but not suitable for HTS because of the use of radiolabeled reagents. Another assay utilizes N-(4-methyl-7-coumarinyl)-N-α-(tert-butyloxy-carbonyl)-N-ε-acetyllysinamide as an artificial substrate where the formation of the deacetylated product is monitored, after an extraction step, by fluorescence detection or HPLC.15 Although this assay avoids the use of radiolabeled reagents, the artificial substrate may not reveal catalysis by many HDACs and the requisite extraction step is not conducive to HTS. The most widely used in vitro HDAC assay for drug discovery employs an ε-acetylated lysine based substrate that is C-terminally coupled with a 4-methyl-coumarin-7-amide residue.16, 17 Once deacetylated, trypsin recognizes the product as a substrate and releases 7-amino-4-methylcoumarin (AMC). Free AMC, in contrast to acylated AMC, is highly fluorescent and easily monitored with a fluorescence microtiter plate reader.18 Although the assay is readily adapted to HTS it does have some drawbacks. Most importantly, the artificial substrate is not recognized by all HDACs, and many HDACs require prolonged incubation times due to low catalytic turnover.19 Also, trypsin inhibitors are often identified as false positives in an HTS campaign with this assay.27
This report describes a single-step, fluorescence polarization (FP) based displacement assay that is suitable for HTS of compounds targeting HDACs. While the HDAC assays described above measure the inhibition of HDAC activity, this assay detects molecules that compete for the substrate binding site. The assay measures ligand binding under equilibrium conditions and does not suffer from product inhibition. The amount of HDAC required per well is comparable to the amount of HDAC used with the fluorogenic substrate. The Ki values for various literature described HDAC inhibitors correlate well with the IC50 values derived from enzymatic turnover screens.
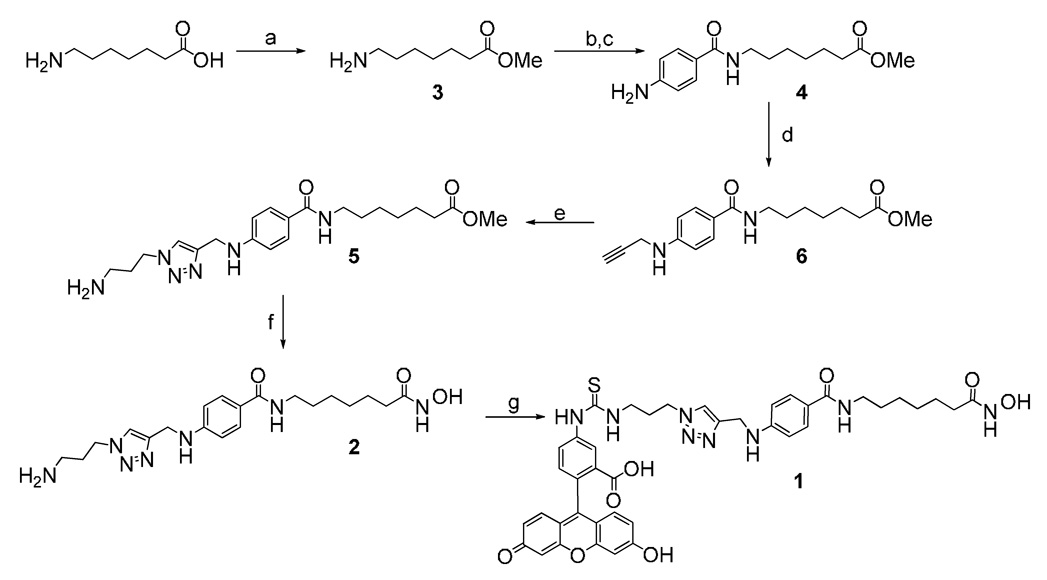
We have designed the FP-HDAC probe 1 based on the unselective HDAC inhibitor M344,20 which has been coupled through a short linker element to FITC. The synthesis was accomplished in 8 steps starting from commercially available 7-amino heptanoic acid in 40% overall yield (Scheme 1).
Scheme 1.
Synthesis of fluorescence polarization assay probe 1 Reagents and conditions: (a) methanol, SOCl2, rt, 5h; (b) 4-Nitrobenzoic acid, isobutyl chloroformate, N-methylmorpholine, THF, 0 C to room temperature; (c) MeOH, Pd/C, H2; (d) propargyl bromide, K2CO3, DMA, 16h, 50C; (d) Cu(MeCN)4PF6, ligand, 3-azidopropyl amine, MeCN; (e) KCN, H2NOH, MeOH, THF; (f) FITC, DMF
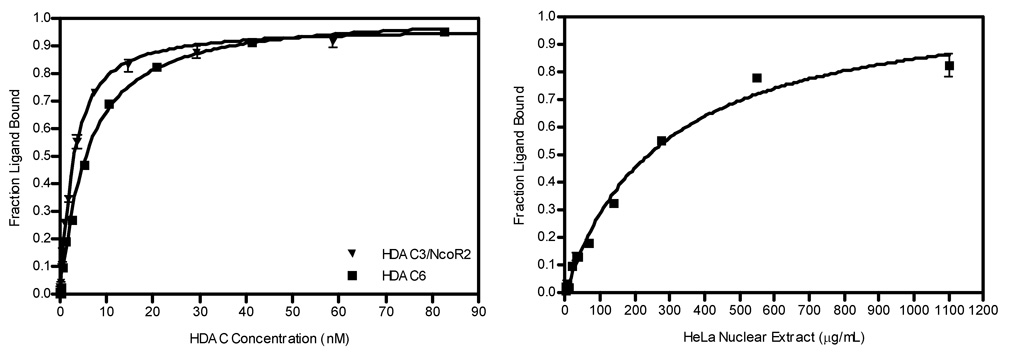
Direct binding of 1 to purified Class I HDAC3/NcoR2, Class II HDAC6 or crude HeLa nuclear extract was measured by a change in FP. The fraction of bound 1 relative to protein concentration is plotted in Figure 1. The binding isotherm data were analyzed by nonlinear regression to calculate the dissociation equilibrium constant (Kd).21, 22 The binding affinity of 1 to HDACs within the HeLa cell nuclear extract is a composite value based upon both Class I and Class II HDACs. Since the relative HDAC concentration and composition within the HeLa nuclear extract is unknown, the equilibrium Kd could not be calculated; however, half maximal binding was seen at 247.6±1.2 µg/mL. Compound 1 bound tightly to both HDAC3/NcoR2 and HDAC6 with a Kd = 2.0±0.2 nM and 4.6±0.2 nM, respectively.
Figure 1.
Binding curve of tracer to HeLa nuclear extract and recombinant HDAC3, HDAC6 and HeLa nuclear extract.
A displacement assay was then used to assess the affinity of various known HDAC inhibitors to HDACs within HeLa nuclear extract as well as purified HDAC3/NcoR2 and HDAC6. Protein was diluted in assay buffer and 1 was added to 2 nM. Increasing concentrations of 2, apicidin, TSA, FK228, or unlabeled SAHA were added to the HDAC/1 complex and the fluorescence anisotropy was measured at equilibrium as described in the supporting information section. Half maximal inhibitory concentrations (IC50) were determined by non-linear regression analysis and the dissociation constant for inhibitor binding (Ki) was calculated (Table 1).23 As expected, the Class III specific HDAC inhibitor splitomicin did not displace 1. The binding affinity for SAHA, Apicidin, TSA, and FK228 correlate well to IC50 values determined by the fluorogenic substrate assay18.
Table 1.
Ligand affinities of known HDAC inhibitors
| Compounds | HeLa Nuclear Extract | HDAC3/NcoR | HDAC6 | ||
|---|---|---|---|---|---|
| IC50 (nM) | IC50 (nM) | Ki (nM) | IC50 (nM) | Ki (nM) | |
| 2 | 58.0±1.4 | 137.0±1.0 | 23.1±0.2 | 50.8±1.2 | 6.6±0.2 |
| SAHA | 23.9±1.3 | 18.8±1.1 | 2.0±0.2 | 21.6±1.0 | 1.0±0.1 |
| Apicidin | 8.0±1.6 | 15.8±1.1 | 1.5±0.1 | 665.1±1.3 | 123.4±0.3 |
| TSA | 1.5±2.3 | 9.0±1.5 | 0.26±0.1 | 16.8±1.6 | 0.13±0.1 |
| FK228 | 107.2±1.2 | 146.9±1.0 | 24.9±0.2 | 1595±1.3 | 300.1±0.3 |
| Splitomicin | >75,000 | >75,000 | >75,000 | >75,000 | >75,000 |
The adaptability of the FP based 1 displacement assay for HTS against human HDAC3/NcoR2 and HDAC6 was assessed using a statistical measure described by Zhang et al.24 A Z-factor between 0.5 and 1 is generally favorable for a HTS assay. The minimum amount of HDAC protein required to yield a statistically significant dynamic range was determined by keeping the concentration of 1 constant at 2 nM while varying the receptor concentration in a manner similar to that described in the previous section. The positive control wells where 1 was completely displaced from the receptor contained 10 µM SAHA, while the negative control samples were mixed with an equal volume of DMSO. In order to maintain a Z-factor > 0.5, 1.9 nM HDAC3/NcoR2 and 5.7 nM HDAC6 were necessary to generate HTS data of high quality.
We have synthesized a fluorescently-labeled, unselective HDAC inhibitor that measures binding to HDACs by fluorescence polarization. This assay can be readily used for HTS for the identification of active site directed inhibitors of HDACs. Although the assay does not allow for the direct readout of enzymatic activity, it correlates well with data derived from assays that are based on the enzymatic turnover of a fluorogenic substrate. The amount of protein required for the FP assay is comparable to the traditional enzymatic assay. Issues associated with the prolonged time necessary for sufficient substrate turnover and the subsequent coupled enzymatic reaction to develop the substrate are circumvented with the FP assay. Additionally, all measurements are conducted at an equilibrium state and the potential for substrate inhibition is avoided.
Supplementary Material
Acknowledgement
The authors thank James E. Bradner for reference data. This work was supported by the NIH (P01 CA078048) and CA24487 (JC)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Pogo BG, Pogo AO, Allfrey VG, Mirsky AE. Proc. Natl. Acad. Sci. U.S.A. 1968;59:1337. doi: 10.1073/pnas.59.4.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reeves R, Candido EP. Biochem. Biophys. Res. Commun. 1979;89:571. doi: 10.1016/0006-291x(79)90668-5. [DOI] [PubMed] [Google Scholar]
- 3.Taunton J, Hassig CA, Schreiber SL. Science. 1996;272:408. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 4.Gregoretti IV, Lee YM, Goodson HV. J. Mol. Biol. 2004;338:17. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Nature. 2000;403:795. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 6.Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. Proc. Natl. Acad. Sci. U.S.A. 2000;97:5807. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhalla KN. J. Clin. Oncol. 2005;23:3971. doi: 10.1200/JCO.2005.16.600. [DOI] [PubMed] [Google Scholar]
- 8.Minucci S, Pelicci PG. Nat. Rev. Cancer. 2006;6:38. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 9.Richon VM, Emiliani S, Verdin E, Webb Y, Breslow R, Rifkind RA, Marks PA. Proc. Natl. Acad. Sci. U.S.A. 1998;95:3003. doi: 10.1073/pnas.95.6.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosato RR, Almenara JA, Grant S. Cancer Res. 2003;63:3637. [PubMed] [Google Scholar]
- 11.Marks PA, Breslow R. Nat. Biotechnol. 2007;25:84. doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- 12.Ruefli AA, Bernhard D, Tainton KM, Kofler R, Smyth MJ, Johnstone RW. Int. J. Cancer. 2002;99:292. doi: 10.1002/ijc.10327. [DOI] [PubMed] [Google Scholar]
- 13.Inoue A, Fujimoto D. Biochem. Biophys. Res. Commun. 1969;36:146. doi: 10.1016/0006-291x(69)90661-5. [DOI] [PubMed] [Google Scholar]
- 14.Nohara H, Takahashi T, Ogata K. Biochim. Biophys. Acta. 1966;127:282. doi: 10.1016/0304-4165(66)90511-3. [DOI] [PubMed] [Google Scholar]
- 15.Hoffmann K, Brosch G, Loidl P, Jung M. Nucleic Acids Res. 1999;27:2057. doi: 10.1093/nar/27.9.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wegener D, Hildmann C, Riester D, Schwienhorst A. Anal. Biochem. 2003;321:202. doi: 10.1016/s0003-2697(03)00426-3. [DOI] [PubMed] [Google Scholar]
- 17.Wegener D, Wirsching F, Riester D, Schwienhorst A. Chem. Biol. 2003;10:61. doi: 10.1016/s1074-5521(02)00305-8. [DOI] [PubMed] [Google Scholar]
- 18.Riester D, Hildmann C, Grunewald S, Beckers T, Schwienhorst A. Biochem. Biophys. Res. Commun. 2007;357:439. doi: 10.1016/j.bbrc.2007.03.158. [DOI] [PubMed] [Google Scholar]
- 19.Lahm A, Paolini C, Pallaoro M, Nardi MC, Jones P, Neddermann P, Sambucini S, Bottomley MJ, Lo Surdo P, Carfi A, Koch U, De Francesco R, Steinkuhler C, Gallinari P. Proc. Natl. Acad. Sci. U.S.A. 2007;104:17335. doi: 10.1073/pnas.0706487104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jung M, Brosch G, Kolle D, Scherf H, Gerhauser C, Loidl P. J. Med. Chem. 42:4669. doi: 10.1021/jm991091h. [DOI] [PubMed] [Google Scholar]
- 21.Dandliker WB, Hsu ML, Levin J, Rao BR. Methods Enzymol. 1981;74 Pt C:3. doi: 10.1016/0076-6879(81)74003-5. [DOI] [PubMed] [Google Scholar]
- 22.Kenakin TP. Pharmacologic analysis of drug-receptor interaction. 3rd ed. Philadelphia: Lippincott-Raven Publishers; 1997. p. p xiii. [Google Scholar]
- 23.Munson PJ, Rodbard D. J. Recept. Res. 1988;8:533. doi: 10.3109/10799898809049010. [DOI] [PubMed] [Google Scholar]
- 24.Zhang JH, Chung TD, Oldenburg KR. J. Biomol. Screen. 1999;4:67. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 25.Checovich WJ, Bolger RE, Burke T. Nature. 1995;375:254. doi: 10.1038/375254a0. [DOI] [PubMed] [Google Scholar]
- 26.Jameson DM, Sawyer WH. Methods Enzymol. 1995;246:283. doi: 10.1016/0076-6879(95)46014-4. [DOI] [PubMed] [Google Scholar]
- 27.Data derived from screening campaigns of HDAC-unbiased small molecule libraries performed at the Broad Institute (unpublished data).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.