Abstract
The analysis of gene expression in Arabidopsis (Arabidopsis thaliana) using cDNA microarrays and reverse transcription-polymerase chain reaction showed that AtOSA1 (A. thaliana oxidative stress-related Abc1-like protein) transcript levels are influenced by Cd2+ treatment. The comparison of protein sequences revealed that AtOSA1 belongs to the family of Abc1 proteins. Up to now, Abc1-like proteins have been identified in prokaryotes and in the mitochondria of eukaryotes. AtOSA1 is the first member of this family to be localized in the chloroplasts. However, despite sharing homology to the mitochondrial ABC1 of Saccharomyces cerevisiae, AtOSA1 was not able to complement yeast strains deleted in the endogenous ABC1 gene, thereby suggesting different function between AtOSA1 and the yeast ABC1. The atosa1-1 and atosa1-2 T-DNA insertion mutants were more affected than wild-type plants by Cd2+ and revealed an increased sensitivity toward oxidative stress (hydrogen peroxide) and high light. The mutants exhibited higher superoxide dismutase activities and differences in the expression of genes involved in the antioxidant pathway. In addition to the conserved Abc1 region in the AtOSA1 protein sequence, putative kinase domains were found. Protein kinase assays in gelo using myelin basic protein as a kinase substrate revealed that chloroplast envelope membrane fractions from the AtOSA1 mutant lacked a 70-kD phosphorylated protein compared to the wild type. Our data suggest that the chloroplast AtOSA1 protein is a new factor playing a role in the balance of oxidative stress.
Heavy metals like Cu2+, Zn2+, and Mn2+ in trace amounts play an essential role in many physiological processes but can be toxic if accumulated at high concentrations. In contrast, other heavy metals such as Cd2+ and Pb2+ have no biological functions and can be extremely toxic. Cadmium is a nonessential heavy metal widespread in the environment, being an important pollutant and known to be toxic for plants not only at the root level where Cd2+ is taken up but also in the aerial part. It can be transported from root to shoot via the xylem (Salt et al., 1995; Verret et al., 2004). Cadmium has been reported to interfere with micronutrient homeostasis (Clemens, 2001; Cobbett and Goldsbrough, 2002). It might replace Zn2+ in the active site of some enzymes, resulting in the inactivation of the enzymatic activity. Cadmium also strongly reacts with protein thiols, potentially inactivating the corresponding enzymes. To overcome this problem, cells produce excess quantities of chelating compounds containing thiols, such as small proteins called metallothioneins (Cobbett and Goldsbrough, 2002) or peptides like glutathione and phytochelatins (Clemens et al., 2002), which limit the damage induced by Cd2+. In addition, several types of transport systems have been shown to contribute to heavy metal resistance, including P-type ATPases and ABC transporters. They transport either free or ligand-bound heavy metals across biological membranes, extruding them into the apoplast or into the vacuole (Kim et al., 2007).
In response to heavy metals, diverse signal transduction pathways are activated, including mitogen-activated protein kinases, transcription factors, and stress-induced proteins (Jonak et al., 2004). Our knowledge concerning components of these pathways is growing but still incomplete.
The Abc1 protein family originates from the Saccharomyces cerevisiae ABC1 gene, which has been isolated as a suppressor of a cytochrome b mRNA translation defect (Bousquet et al., 1991). The mitochondrial ABC1 in yeast was suggested to have a chaperone-like activity essential for a proper conformation of cytochrome b complex III (Brasseur et al., 1997). However, more recent data suggest that the ABC1 protein might be implicated in the regulation of coenzyme Q biosynthesis (Hsieh et al., 2004). The Abc1 family has also been described as a new family of putative kinases (Leonard et al., 1998), and it has been suggested that the putative kinase function of Abc1-like proteins is related to the regulation of the synthesis of ubiquinone (Poon et al., 2000). Homologs of yeast ABC1 have been isolated in higher eukaryotes. In Arabidopsis (Arabidopsis thaliana), the only studied ABC1-like protein has been predicted to be localized in the mitochondria. It partially restored the respiratory complex deficiency when expressed in S. cerevisiae (Cardazzo et al., 1998). In humans, a homolog of the Abc1 proteins (CABC1) has been identified and it is possibly involved in apoptosis (Iiizumi et al., 2002). The human CABC1 protein has 47% and 46% similarity to ABC1 of Arabidopsis and Schizosaccharomyces pombe, respectively.
The data presented in this study suggest that the chloroplast AtOSA1 (A. thaliana oxidative stress-related Abc1-like protein), an Arabidopsis protein belonging to the Abc1 protein family, is implicated in the plant response to oxidative stress that can be generated by Cd2+, hydrogen peroxide (H2O2), and light. Our results show that AtOSA1 is functioning differently from Abc1; hence, the proteins of the Abc1 family can fulfill diverse functions.
RESULTS
AtOSA1 Transcript Levels Change in Response to Cadmium Exposure
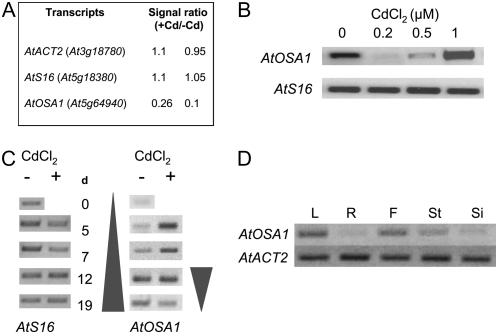
The elucidation of the physiological functions of gene products that transcript levels are up- or down-regulated by Cd2+ in the model plant Arabidopsis is of major interest to understand response of plants to Cd2+. Several transcriptomic analyses have been performed using a subarray spotted with a large number of different cDNA sequences. cDNA microarrays from two independent experiments revealed that transcript levels of AtOSA1 (At5g64940) were down-regulated after the treatment with 0.2 μm CdCl2 for 21 d (Fig. 1A). The microarray data were confirmed by reverse transcription (RT)-PCR using the same mRNA template used for the microarray analyses and RNA isolated from plants exposed to 0.5 and 1 μm CdCl2. After the 1 μm CdCl2 treatment, the transcript level of AtOSA1 was up-regulated (Fig. 1B). Additionally, a time-course experiment was carried out with 1-week-old plants exposed to 0.5 μm CdCl2 (Fig. 1C). The data showed that AtOSA1 was up-regulated in the leaves after 5 d of Cd2+ exposure, then stably expressed until day 12 and, finally, down-regulated. In the absence of Cd2+, an increase in the expression of AtOSA1 was found to be correlated with plant aging. The analysis of AtOSA1 transcript levels in the major plant organs of 6-week-old flowering plants revealed that this gene is expressed particularly in leaves, but also in flowers and slightly in stems (Fig. 1D). Under normal growth conditions, we found only a very low level of AtOSA1 transcripts in roots. Expression of AtOSA1 is in all likelihood related to the green tissues, because in this experiment the flowers were not dissected and still contained green sepals. However, we cannot exclude that AtOSA1 is also expressed in petals, stamen, and pistils. The data collected for the At5g64940 entry in the digital northern program Genevestigator (www.genevestigator.ethz.ch; Zimmermann et al., 2004) confirm predominant expression of AtOSA1 in leaves and flowers and that the transcript level of AtOSA1, which is age dependent (Fig. 1C), is also down-regulated in the night (circadian rhythm dependencies) and senescent leaves. We confirmed these two last results experimentally (data not shown).
Figure 1.
AtOSA1 gene expression in Arabidopsis. A, Analysis of the transcript levels of AtOSA1 in leaves after exposure to 0.2 μm CdCl2 for 3 weeks under hydroponic growth conditions using cDNA spotted arrays. The data presented show the +Cd to −Cd ratio obtained from spotted array replicates. B, Confirmation of the chip data and cadmium dose-dependent experiment using semiquantitative RT-PCR (35 cycles). C, Time-dependent (days) regulation of AtOSA1 in leaves of Arabidopsis in the presence (+) or absence (−) of 0.5 μm CdCl2. D, RT-PCR analysis of AtOSA1 in plant organs: leaf (L), root (R), flower (F), stem (St), and silique (Si).
AtOSA1 Has Homology to the Abc1-Like Protein Family
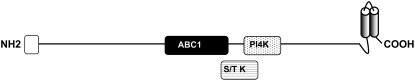
The protein sequence of AtOSA1 possesses a conserved region of around 120 to 130 amino acids (according to the Conserved Domain Database for protein classification; Marchler-Bauer et al., 2005) that is characteristic for the so-called Abc1 protein family (Fig. 2; Supplemental Fig. S1). Using the Conserved Domain Database search engine at the National Center for Biotechnology Information (Marchler-Bauer et al., 2003), putative kinase domains were detected within the AtOSA1 protein sequence (Fig. 2). Similar domains were found in phosphoinositide 4-kinase and Mn2+-dependent Ser/Thr protein kinase.
Figure 2.
Schematic illustration of the AtOSA1 protein topology. Identified domains are depicted as follows: white box, chloroplast targeting peptide; black box, ABC1; horizontal stripe box, Mn2+-dependent Ser/Thr protein kinase (S/T K); dotted box, phosphoinositide 4-kinase (PI4K); and shaded barrel, a region with predicted transmembrane spans.
The hydropathy plot made with TMpred (Hofmann and Stoffel, 1993) revealed the presence of two transmembrane spans within the C-terminal part of AtOSA1 (Supplemental Fig. S1). Similar results were obtained using the DAS transmembrane prediction server (http://www.sbc.su.se/∼miklos/DAS/).
The members of Abc1 protein family have been identified in both pro- and eukaryota, for example, AarF from Escherichia coli (Macinga et al., 1998) and ABC1 from yeast (Bousquet et al., 1991). It is worth emphasizing that the Abc1 protein family is not related to ABC transporters despite the fact that AtOSA1 has been previously described as an ABC transporter belonging to the ATH (ABC2) subfamily (Sanchez Fernandez et al., 2001). AtOSA1 does not possess any typical features, including, for instance, the most characteristic sequence of ABC transporters known as signature motif [LIVMFY]S[SG]GX3[RKA][LIVMYA]X[LIVFM][AG] (Bairoch, 1992).
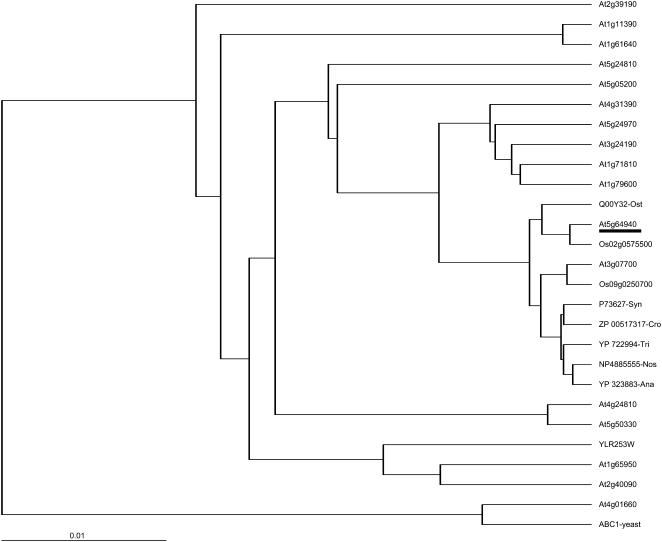
In Arabidopsis, the sole ABC1-like protein (At4g01660) studied so far has been predicted to be localized in mitochondria and can partially restore the respiratory complex deficiency when expressed in S. cerevisiae (Cardazzo et al., 1998). This protein has 32% amino acid identity with AtOSA1. The Arabidopsis genome contains 17 putative Abc1-like genes. Based on the aligned translated products, a phylogenetic tree has been drawn (Fig. 3). The closest Arabidopsis homolog is At3g07700, which shares 45% amino acid identity with AtOSA1. To date, nothing is known about the localization and potential function of both gene products, although the expression of an apparent homolog of At3g07700 in Brassica juncea (DT317667) has also been found to be regulated by cadmium (Fusco et al., 2005). Two translated gene products from rice (Oryza sativa), Os02g0575500 and Os09g0250700, share high homologies with AtOSA1. In prokaryotes, the closest homologs of AtOSA1 are the members of the Abc1 family found in different cyanobacteria like Nostoc (NP_4885555) and Synechocystis sp. (P73627), sharing, respectively, 45% and 44% identity at the amino acid level. Prokaryotic Abc1 proteins also have been detected in E. coli and Clostridium. Interestingly, these organisms lack complex III (Trumpower, 1990; Unden and Bongaerts, 1997), suggesting that the possible function for Abc1-like proteins may not be exclusively linked to the transfer of electrons in membranes.
Figure 3.
Phylogenetic tree of Arabidopsis Abc1 proteins (accession nos. according to TAIR http://www.arabidopsis.org/): rice Os02g0575500 and Os09g0250700, S. cerevisiae ABC1 (CAA41759) and YLR253W, Ostreococcus tauri Q00Y32, Crocosphaera watsonii ZP_00517317, Trichodesmium erythraeum YP_722994, Anabaena variabilis YP_323883, Nostoc NP4885555, and Synechocystis P73627. Protein sequences were aligned using the program DIALIGN (Morgenstern, 2004) and the phylogenetic tree was drawn with the TreeView32 software (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html). Scale bar indicates distance values of 0.01 substitutions per site.
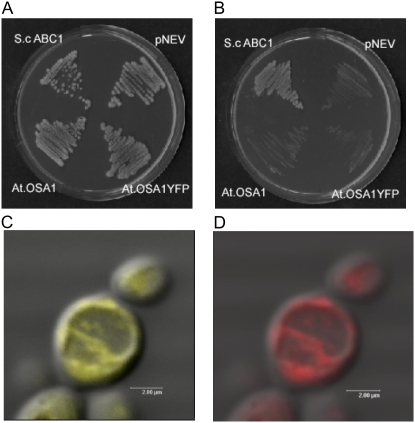
Identification of the Abc1 domain within the AtOSA1 sequence prompted us to determine the functional homology of AtOSA1 with Abc1 proteins. For this purpose, we used the yeast S. cerevisiae deletion mutant W303-1A abc1∷HIS3 deficient in the endogenous ABC1 activity (Hsieh et al., 2004). Deletion of the ABC1 gene in yeast disturbs the function of the respiratory chain and prevents growth of this mutant strain on media containing nonfermentable carbon sources such as glycerol (Bousquet et al., 1991). The expression of the entire AtOSA1 gene, including its targeting presequence in the W303-1A abc1∷HIS3 strain, did not restore growth of this mutant on glycerol-containing media. Neither AtOSA1-EYFP nor AtOSA1 restored growth. As a control, the growth of the same strain was restored after complementation with yeast ABC1 gene (Fig. 4, A and B), suggesting functional divergence between AtOSA1 and the yeast ABC1. We included the targeting presequence, because chloroplast proteins tend to be targeted to the mitochondria when expressed in fungal cells (Pfaller et al., 1989; Brink et al., 1994). We monitored the expression of AtOSA1-EYFP by confocal microscopy. The signal emitted by the strains expressing TPAtOSA1-YFP (Fig. 4C) was similar to that of the Rhodamine HexylB used for staining mitochondria (Fig. 4D), confirming localization of AtOSA1 in yeast mitochondria or mitochondrion in the presence of the chloroplast targeting presequence.
Figure 4.
Complementation test of the S. cerevisiae mutant W303-1A abc1∷HIS3. Yeast strains S. cerevisiae ABC1 (pRS316 harboring S. cerevisiae ABC1), pNEV (vector only), AtOSA1 (pNEV harboring AtOSA1), and AtOSA1YFP (pNEV harboring AtOSA1 with YFP) were streaked on plates containing minimal medium lacking uracil for selection (A) and onto minimal medium containing glycerol as a sole carbon source (B). Plates were incubated for 4 d at 28°C. C, Superposition of a confocal and a bright field image of W303-1A abc1∷HIS3 expressing AtOSA1YFP. D, Superposition of a confocal image and a bright field image of the same cell stained with Rhodamine HexylB.
AtOSA1 Is Localized in the Chloroplast
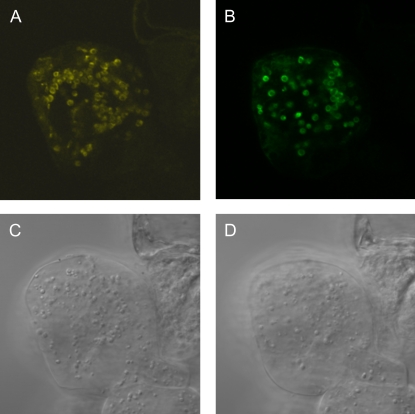
Sequence analysis of the AtOSA1 protein with Target P (http://www.cbs.dtu.dk/services/TargetP/; Emanuelsson et al., 2000), used for proteomic analyses and theoretical predictions of protein localization (Koo and Ohlrogge, 2002; Peltier et al., 2002), revealed the presence of a 28-amino acid N-terminal chloroplast targeting presequence (Supplemental Fig. S1). Both rice sequences Os02g0575500 and Os09g0250700 also have such putative chloroplast transit peptide regions of 56 and 39 amino acids, respectively. To verify its subcellular localization, AtOSA1 was fused (C terminal) with EYFP and transiently expressed under the control of the cauliflower mosaic virus 35S promoter in Arabidopsis suspension cell culture (Fig. 5A). The signal was visualized by confocal microscopy. The observed localization was identical with that obtained for the Tic110-GFP (Fig. 5B), an integral inner envelope membrane protein of the chloroplast import machinery (Inaba et al., 2003). Our results confirm in silico and proteomic data, suggesting a localization of the AtOSA1 protein in the chloroplast envelope of Arabidopsis (Froehlich et al., 2003).
Figure 5.
Confocal laser scanning microscopic analysis of an Arabidopsis suspension cell culture transiently expressing EYFP-tagged AtOSA1 (A) and Tic110-GFP (B) and the corresponding bright field images (C and D).
Cadmium Effect on AtOSA1 Mutants
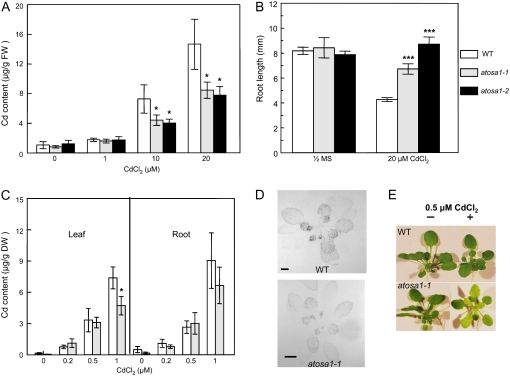
The identification of mutants for AtOSA1 was possible from T-DNA insertion lines of the SALK Institute (SALK 045739) and GABI Kat (GABI, 132G06). To find the homozygote lines for both mutants, we screened the F3-F4 generation by PCR using RP, LP, and LB T-DNA primers designed by SIGnAL T-DNA Express (http://signal.salk.edu). The mutants were named atosa1-1 (SALK 045739) and atosa1-2 (GABI 132G06), respectively (Supplemental Fig. S2). In both mutants, T-DNA insertions are located toward the 3′ end, thereby excluding the presence of a membrane anchor in case truncated transcripts are translated (Supplemental Fig. S3). Seedlings of both mutants accumulated less cadmium than the wild type at 10 and 20 μm CdCl2 (Fig. 6A). Therefore, we investigated cadmium tolerance in AtOSA1 T-DNA insertion mutants in 1-week-old seedlings grown on bactoagar plates containing 20 μm CdCl2. Interestingly, roots of atosa1-1 and atosa1-2 mutant seedlings were longer than that of wild-type seedlings (Fig. 6B), thereby suggesting that root growth is less affected by cadmium toxicity in AtOSA1 than in the wild type. Under hydroponic culture conditions, leaves from wild-type Arabidopsis plants took up significantly more cadmium than atosa1-1, confirming the data obtained in seedlings (Fig. 6C). A similar picture could be observed in the autoradiograms from 4-week-old plants exposed to 0.04 MBq 109CdCl2 for 4 h (Fig. 6D), in which higher radioactivity was detected in wild-type plants. Surprisingly, despite the fact that the mutant plants took up less cadmium, they exhibited a marked chlorotic phenotype when exposed to 0.5 μm CdCl2 for 7 d (Fig. 6E).
Figure 6.
Cadmium tolerance and accumulation in atosa1. A, Determination of cadmium accumulation by AAS in 10 seedlings exposed to 0 (one-half-strength MS), 1, 10, or 20 μm CdCl2 on agar plates (n = 5). B, The effect of cadmium on root growth of atosa1-1, atosa1-2, and Col-0 (WT) in the absence (one-half-strength MS) or presence of 20 μm CdCl2. Root length of 8-d-old seedlings (10 < n < 20, representative results from four independent experiments). C, Determination of cadmium content in leaves and roots in wild type (Col-0) and atosa1-1 grown under hydroponic conditions (n = 8; mean ± se; t test: *, P = 0.1; **, P = 0.05; ***, P = 0.01). D, Autoradiography of plant roots labeled with 0.04 MBq 109CdCl2 in one-eighth-strength MAMI for 4 h. E, Phenotype of atosa1 grown in the absence or presence of 0.5 μm CdCl2.
Superoxide Dismutase Activity and Gene Expression in the AtOSA1 T-DNA Mutants
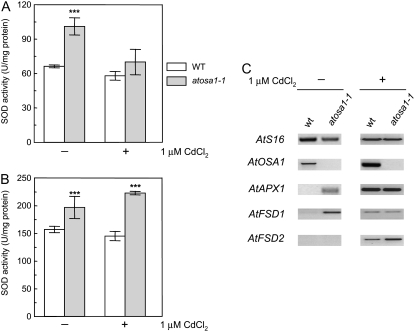
Leaf chlorosis observed in the AtOSA1 T-DNA insertion mutants but not in wild-type plants after cadmium treatment prompted us to determine whether atosa1-1 is more sensitive to oxidative stress than wild type and whether some of the genes involved in reactive oxygen species (ROS) scavenging are regulated differently in mutants. A suitable approach to determine sensitivity to ROS is measurement of the activity of superoxide dismutase (SOD), an essential enzyme to attenuate plant oxidative stress. In the first approach, we determined the overall SOD activity in the leaves of wild type and atosa1-1 exposed or not to 1 μm CdCl2. The AtOSA1 mutant plants showed increased SOD activity compared to wild-type plants both in absence and presence of Cd2+. The effect was particularly marked in the absence of Cd2+ treatment (Fig. 7A). To determine whether chloroplasts also exhibit an increased SOD activity, we isolated chloroplasts from plants grown in the presence or absence of Cd2+. The data showed that chloroplasts isolated from the AtOSA1 deletion mutant displayed a slight but consistently higher SOD activity compared to the wild-type chloroplasts. This effect was independent of whether the plants were exposed to Cd2+ or not (Fig. 7B).
Figure 7.
SOD activity and AtOSA1 expression. A, Comparison of the total SOD activities between Arabidopsis wild-type Col-0 (WT) and atosa1-1 under normal growth conditions (−) and after treatment with 1 μm CdCl2 (+; n = 4). B, Measurement of SOD activity in intact chloroplasts obtained from wild-type Col-0 (WT) and the AtOSA1 T-DNA-inserted mutant (atosa1-1) treated (+) or not (−) with 1 μm CdCl2 (n = 4; mean ± se; t test: *, P = 0.1; **, P = 0.05; ***, P = 0.01). C, Analyses of the expression of AtOSA1, AtAPX1, AtFSD1, and AtFSD2 in wild-type Col-0 (WT) and atosa1-1 by RT-PCR in the absence (−) or presence (+) of 1 μm CdCl2 under light superior to 100 μmol m−2 s−1. AtS16 was used as control (30 cycles).
Transcript levels of genes (AtAPX1, At1g07890; AtFSD1, At4g25100; AtFSD2, At5g51100) responding to oxidative stress (Kliebenstein et al., 1998; Ball et al., 2004) were investigated. AtAPX1, AtFSD1, AtFSD2, as well as AtOSA1 were found to be up-regulated in wild type after 1 μm cadmium treatment. In atosa1-1, only AtFSD2 was comparatively induced by cadmium. Interestingly, AtFSD1 was more expressed in atosa1.1 under control conditions when compared to wild type and induction of AtFSD2 was stronger in the mutant (Fig. 7C).
H2O2, known as an ROS inducer, reduced the growth of the seedling roots more in the mutants than in the wild type (Fig. 8A). The effect of H2O2 was also more pronounced in atosa1-1 leaves compared to the wild-type leaves. Indeed, after spraying leaves of wild-type and mutant plants with 300 μm H2O2 in 0.2% (v/v) Tween 20, we observed a rapid appearance of necrotic spots in the mutant, already 1 d after spraying (Fig. 8, B and C). In contrast, only a very few or no spots were found in the wild-type plants 4 d after spraying with H2O2 (Fig. 8B). No necrotic spots were detected when both the wild type and AtOSA1 T-DNA-inserted mutant were sprayed with 0.2% (v/v) Tween 20 only (data not shown).
Figure 8.
Effect of oxidative stress. A, After treatment with 1 mm H2O2 on agar plates, root length of 8-d-old seedlings was measured (10 < n < 20, representative results from four independent experiments; mean ± se; t test: *, P = 0.1; **, P = 0.05; ***, P = 0.01). B, Five-week-old Col-0 (WT) and atosa1-1 plants were sprayed with 300 μm H2O2 and 0.2% (v/v) Tween 20 at day 0. Plants were photographed at days 0, 1, and 4 following the treatment. C, Magnification of atosa1 leaves 4 d after treatment with H2O2.
The Effect of Light on AtOSA1 T-DNA-Inserted Mutants
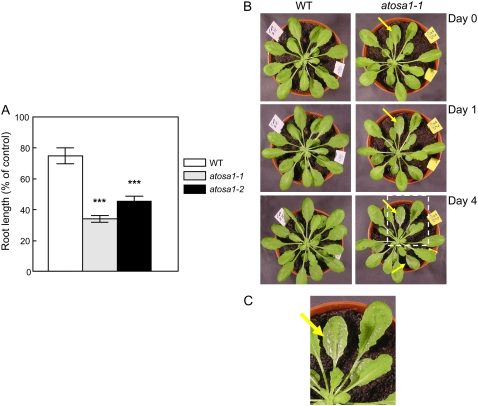
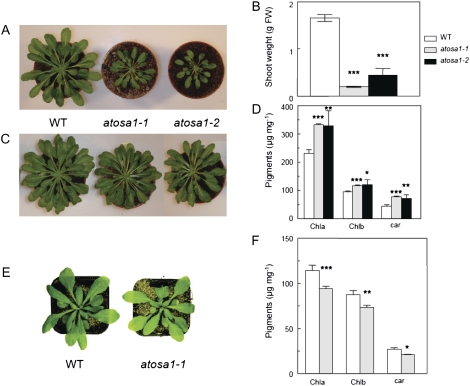
Light has a complex effect on AtOSA1 mutants depending on light intensities. At a low light regime (50 μmol m−2 s−1) for 8 h during 4 weeks, the shoot growth of atosa1-1 and atosa1-2 was significantly altered compared to the wild type (Fig. 9, A and B). After an additional 4 weeks of growth in the same experimental conditions, leaf sizes were still different, and based on fresh weight, chlorophyll a (Chla), chlorophyll b (Chlb), and carotenoid contents were higher in the mutants compared to the wild type (Fig. 9, C and D). Under a light regime of 120 to 150 μmol m−2 s−1 for 8 and 16 h, no visible phenotypes were found in the AtOSA1 mutants. Surprisingly, under 16 h of high light (350 μmol m−2 s−1), atosa1-1 exhibited a pale-green phenotype (Fig. 9E). In this case, the analyses of pigments showed slightly less chlorophyll and carotenoids in atosa1-1 compared to the wild type (Fig. 9F).
Figure 9.
Effects of light on pigments and shoot growth of atosa1-1 and atosa1-2. Plants were grown at 8 h light (50 μmol m−2 s−1) for 4 weeks (A) or 8 weeks (C). B, Shoot weight of 4-week-old plants (n = 10). D, Contents of Chla and Chlb and carotenoids of 8-week-old plants (n = 10). E, Plants grown at 16 h light (350 μmol m−2 s−1) for 5 weeks. F, Contents of Chla and Chlb and carotenoids in plants depicted in E (n = 10; mean ± se; t test: *, P = 0.1; **, P = 0.05; ***, P = 0.01).
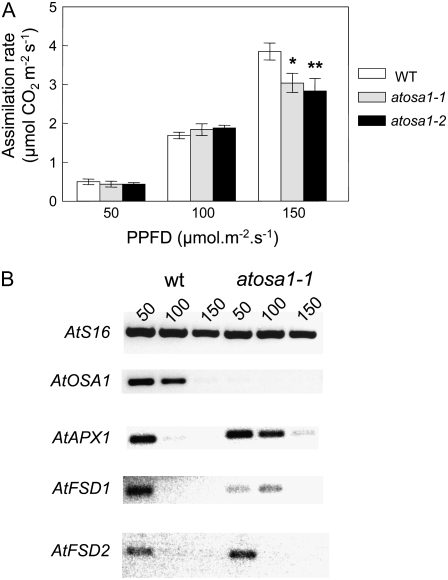
Analysis of photosynthetic activities in terms of net CO2 assimilation rate also revealed differences between Atosa1 mutants and the wild type depending on the light intensities. Under higher light intensities, mutants were more affected than the wild type (Fig. 10A). Increasing light intensities from 50 to 150 μmol m−2 s−1 led to a reduction of AtOSA1, AtAPX1, AtFSD1, and AtFSD2 transcript levels in wild-type plants. A similar reduction of AtAPX1, AtFSD1, and AtFSD2 could be observed in the atosa1-1 mutant, but this effect was visible only under higher light intensities (Fig. 10B).
Figure 10.
Effect of light intensity on gas exchange and expression of oxidative stress-related genes. Analyses of CO2 assimilation rate (A) of Col-0 (WT), atosa1-1, and atosa1-2. Measurements were performed in plants grown at 8 h light at a photosynthetic photon flux density of 50, 100, or 150 μmol m−2 s−1 (n = 10; mean ± se; t test: *, P = 0.1; **, P = 0.05; ***, P = 0.01). B, RT-PCR expression analysis of AtS16 (housekeeping gene), AtOSA1, AtAPX1, AtFSD1, and AtFSD2 in plants used for the determination of gas exchange measurements (A; 28 cycles).
No significant differences were found by the electron microscopic analysis in chloroplast structures (stroma lamellae, grana stacks, and envelope membranes) between the atosa1-1 and wild type. In addition, the inductively coupled plasma mass spectrometry data showed that the content in essential metals and heavy metals was not changed by the AtOSA1 T-DNA insertion (data not shown).
Because possible connections between Abc1 proteins, electron transport, and ubiquinone (plastoquinone and phylloquinone) synthesis have been postulated (Poon et al., 2000), we performed analysis of electron transport in AtOSA1 mutants. The kinetic measurements of Chla fluorescence probing the redox state of the primary quinone acceptor of PSII and 820 nm transmission probing the redox state of mainly plastocyanin and P700 (reaction center chlorophylls of PSI) revealed no differences between atosa1-1 and the wild type (data not shown). This indicates that the electron transport functioned well in atosa1-1 and that the number of oxidized electron acceptors per chain at the beginning of the measurement was similar to the wild type at “standard” light regime (120 μmol m−2 s−1).
Detection of Protein Kinase Activities in Gelo
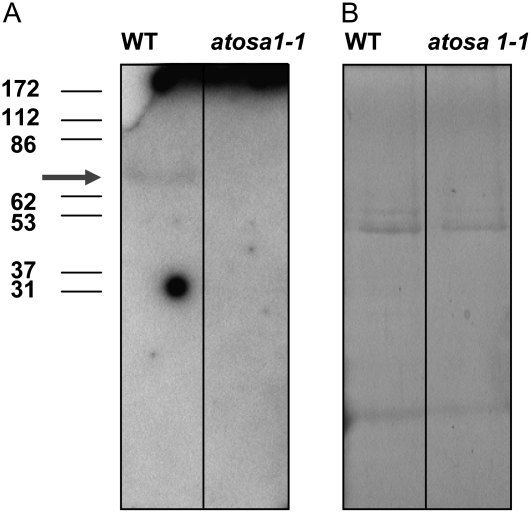
In addition to the Abc1 protein family, AtOSA1 contains motifs found in eukaryotic-type protein kinases. Therefore, we decided to examine protein kinase activities in the AtOSA1 mutant by in gelo phosphorylation assays using myelin basic protein as a substrate. Because we localized AtOSA1 in chloroplasts and the proteomic analysis identified AtOSA1 in the envelope fraction (Froehlich et al., 2003), we isolated and used this fraction for the assay. In-gel protein kinase assay allowed us to detect one chloroplast envelope protein kinase of about 70 kD in the Columbia (Col-0) ecotype of Arabidopsis (Fig. 11A). Interestingly, this labeled band was not present in the envelope membranes isolated from the AtOSA1 T-DNA-inserted plants. This might indicate that the AtOSA1 mutant lacks this protein kinase. The labeled bands with a similar Mr were not detected in thylakoid membranes, and a more complex phosphorylation pattern, which, however, did not show the absence of a labeled band, was obtained with Histone III-S as a substrate (data not shown). The envelope protein profile after Coomassie Blue staining of the SDS gel did not show marked differences between the mutant and the wild type (Fig. 11B).
Figure 11.
Protein kinase activity. A, Detection of protein kinase activity in chloroplast envelope membranes isolated from leaves of wild type and atosa1-1. The arrow indicates the position of the phosphorylated myelin basic protein at around 70 kD in the wild type. B, Coomassie Blue staining of the gel shown in A. For details, see “Materials and Methods.”
DISCUSSION
We performed microarray chip analyses to identify genes up- and down-regulated in response to cadmium stress. Among the genes exhibiting an altered transcript level in response to Cd2+, we identified AtOSA1 (At5g64940) as a member of the Abc1 family. In Arabidopsis, 17 genes contain a typical Abc1 motif and hence constitute a small gene family. The sole Abc1 representative described so far in plants (At4g01660) is a homolog to the yeast ABC1 (Cardazzo et al., 1998). Both are localized in mitochondria (Bousquet et al., 1991; Cardazzo et al., 1998), in contrast to AtOSA1, which is targeted to the chloroplast and does not subcluster with them. AtOSA1 transcript level followed a complex kinetics in response to Cd2+ during dose-dependent and time-course experiments. In the absence of cadmium treatment, its expression in leaves increased during the life of Arabidopsis, and it has been reported that plant aging increases oxidative stress in chloroplasts (Munne-Bosch and Alegre, 2002).
Two independent T-DNA insertion mutants, lacking functional AtOSA1, exhibited a complex behavior toward cadmium. Indeed, the seedling roots of AtOSA1 deletion mutants were less affected by Cd2+ than those of the wild-type plants, possibly due to a reduced Cd2+ uptake.
The increased cadmium tolerance of wild type compared to atosa1 mutants is very likely not supported by the direct binding of cadmium to AtOSA1. Indeed, AtOSA1 lacks of sequence motifs containing cysteins, involved in the binding of heavy metal ions (Zn2+, Cd2+, Pb2+, Co2+, Cu2+, Ag+, or Cu+), like CXXC and CPC. Such motifs have been found, for example, in members of the subclass of heavy metal-transporting P-type ATPases (P1B-type ATPases; Eren and Argüello, 2004). In addition, AtOSA1 is likely not a heavy metal (cadmium) transporter, because vesicles isolated from YMK2 yeast (Klein et al., 2002) transformed with AtOSA1 did not show any cadmium transport (data not shown).
The pale phenotype of leaves was more pronounced in the case of mutant plants exposed even to a low dose of Cd2+ despite the fact that lack of AtOSA1 results in lower Cd2+ uptake rates in shoots. Such a chlorotic phenotype of leaves was not correlated with an elevated accumulation of cadmium and was also observed under high light conditions. This pale phenotype might be a consequence of Cd2+ toxic effect due to a modification of the cellular cadmium distribution (Ranieri et al., 2001) and an increased Cd2+ sensitivity related to an increased production of ROS in the AtOSA1 mutants, similarly to those described in Euglena gracilis (Watanabe and Suzuki, 2002) or yeast (Brennan and Schiestl, 1996).
Although the mechanism of oxidative stress induction by Cd2+ is still obscure, Cd2+ can inhibit electron transfer and induces ROS formation (Wang et al., 2004). It has been also suggested that Cd2+ can interfere in living cells with cellular redox reactions and displaces or releases other active metal ions (e.g. Zn2+) from various biological complexes, thereby causing a reduction of the capacity of the antioxidant system (Jonak et al., 2004).
Besides cadmium, the AtOSA1 T-DNA-inserted mutants actually showed a phenotype illustrated by a reduced tolerance to H2O2 and light. At 150 μmol m−2 s−1, we observed the same transpiration rate for wild type, atosa1-1, and atosa1-2. Nevertheless, stomatal conductance and CO2 assimilation were higher in wild type than in mutants (data not shown). This observation suggests that, at this light intensity (150 μmol m−2s−1), transpiration occurs not only at the stomatal level but also directly through the epidermis. This hypothesis is supported by the experiments showing increased sensitivity of atosa1 toward H2O2 (Fig. 8B). Indeed, it is still possible that the AtOSA1 mutation also affects the epidermal cell wall and the cuticule. At low light intensity and period, atosa1 exhibited retardation in growth correlated with an increase in pigment production (Chla, Chlb, and carotenoids). Under higher light intensity and period, a pale-green phenotype correlated with a decrease in pigment contents when compared with the wild type. In addition, changes of light intensities influenced photosynthetic activities. These data suggest participation of the chloroplast AtOSA1 in light-generated stress (ROS) and pigment response.
Obtained results suggest that AtOSA1 mutants have a hypersensitivity to broad abiotic stresses, including photooxidative stress. RT-PCR analyses in atosa1 plants showed different behavior for transcripts of genes responding to oxidative stress. For instance, it was shown that AtFSD1 transcript in Arabidopsis is high at 60 μmol m−2 s−1 and then down-regulated under increasing light fluences (Kliebenstein et al., 1998). A similar tendency could be observed for atosa1 but under higher light intensity. The lack of AtOSA1 caused a global shift under increasing light conditions. This might indicate necessity to compensate increased oxidative stress level in the mutant by the expression of components of the antioxidant network like AtAPX1 and AtFSD1 and permanent SOD activities (Ball et al., 2004). Interestingly, the increased SOD activity detected in the isolated chloroplasts was not enhanced by Cd2+ treatments, thereby confirming the data reported by Fornazier et al. (2002) showing that the Cd2+ treatment did not enhance SOD activities, possibly by displacing Fe2+, Zn2+, or Cu2+ required for the SOD activity. Most presumably, these results indicate that AtOSA1 deletion mutants permanently suffer from oxidative stress and compensate it to a certain level under controlled growth conditions; however, these plants are apparently not able to do it when environmental parameters like ROS inducers, light regime, or nutrient supply vary.
AtOSA1 is probably not directly induced by external oxidative stress but acts in a more complex manner, for example, as a part of a signal transduction pathway related to oxidative stress. Indeed, the Abc1 family has been described as a family of putative kinases (Leonard et al., 1998), and it is possible that AtOSA1 exhibits protein kinase activity, because the predicted molecular mass of mature AtOSA1 (83 kD) is close to the phosphorylated polypeptide detected in the autoradiography (approximately 70 kD) after in-gel assay. The phosphorylated polypeptide is not present in the envelope membranes derived from AtOSA1 mutant. Nevertheless, we cannot exclude that the protein kinase detected within the gel matrix is a member of a signal transduction cascade, which is not active in the AtOSA1 mutant. Further studies are required to elucidate the role of this protein kinase within the chloroplast.
Based on the phylogenetic tree, cell localization, and involvement in oxidative stress response, AtOSA1 is rather not a functional homolog of the yeast ABC1 and At4g01660 (Cardazzo et al., 1998). As a chloroplast protein, AtOSA1 is more closely related to prokaryotic Abc1 proteins from cyanobacteria like Synechocystis or Nostoc than to those of mitochondria. These ABC1 proteins have not been characterized so far. Therefore, our data are in agreement with the studies on evolutionary relations between different Abc1 proteins, which led to the conclusion that Abc1 proteins from cyanobacteria and chloroplasts, on the one hand, and from mitochondria on the other have independent origins (Leonard et al., 1998). To date, it has been suggested that Abc1 proteins control the biogenesis of respiratory complexes in mitochondria. The yeast ABC1 knockout mutants are unable to grow on glycerol, making the exact molecular functions of these proteins still a matter of debate (Do et al., 2001).
In Arabidopsis, AtOSA1 (At5g64940) clusters together with Abc1-like gene At3g07700. Interestingly, a homolog of this gene in B. juncea is also cadmium regulated and possibly localized in the chloroplast (Fusco et al., 2005). Concerning the other Abc1 sequence-related genes in Arabidopsis, four of them (At5g5200, At4g31390, At1g79600, and At1g71810) have been recently found to be localized in plastoglobules in a proteomic study and are possibly involved in the regulation of quinine monooxygenases (Ytterberg et al., 2006). As illustrated by the pleiotropic effect and permanent oxidative stress caused by deletion of AtOSA1, despite the fact that our knowledge about Abc1-related proteins is still scarce, our results indicate this gene family triggers essential regulatory functions.
MATERIALS AND METHODS
cDNA Microarrays
The mRNAs were isolated as described at http://www.unil/ibpv. Fluorescent labeling of cDNAs, hybridization on homemade DNA microarray slides spotted with ESTs and 3′ end coding sequences (corresponding to putative ABC transporter proteins [124 of 127] and other protein families), and fluorescence analyses (Scanarray 4000) were performed as described by Bovet et al. (2005).
Semiquantitative PCR
For semiquantitative RT-PCR, the housekeeping genes AtACT2 (actin; At3g18780) and AtS16 (At5g18380) were amplified using the primers actin2-S (5′-TGGAATCCACGAGACAACCTA-3′) and actin2-AS (5′-TTCTGTGAACGATTCCTGGAC-3′) and S16-S (GGCGACTCAACCAGCTACTGA) and S16-AS (CGGTAACTCTTCTGGTAACGA), respectively. For the ascorbate peroxidase 1 (AtAPX1) gene (At1g07890), Fe-SOD 1 (AtFSD1) gene (At4g25100), and Fe-SOD 2 (AtFSD2) gene (At5g51100), we designed the following primers: APX1-S (5′-GCATGGACATCAAACCCTCTA-3′) and APX1-AS (5′-TTAAGCATCAGCAAACCCAAG-3′); FSD1-S (5′-GGAGGAAAACCATCAGGAGAG-3′) and FSD1-AS (5′-TCCCAGACATCAATGGTAAGC-3′); and FSD2-S (5′-CCACTCCCTCGTCTCTCTTG-3′) and FSD2-AS (5′-CCACCTCCAGGTTGGATAGA-3′). The primers for AtOSA1 were AtOSA1-S (5′-GACAGGCAATCACAAGCATTC-3′) and AtOSA1-AS (5′-CGATTAGAACTTGGAGGCTGA-3′), respectively. For the selection of the atosa1-1 T-DNA insertion homozygote lines (SALK 045739), the primers were: RP (5′-AACGCGTTGAAATGCCCTCTC-3′), LP (5′-CTTGCTTCTTATCCATCGAGC-3′), and LB T-DNA (5′-GCGTGGACCGCTTGCTGCAACT-3′). For the selection of the atosa1-2 T-DNA insertion homozygote lines (GABI 132G06), the primers were: RP (5′-TTTGTTGGAGGCATTTTATGG-3′), LP (5′-GAATGCTTGTGATTGCCTGTC-3′), and LB T-DNA (5′-ATTTGGACGTGAATGTAGACA-3′). The primers for the verification of truncated transcript were: 1-S (5′-AATCGCCGGGATCTTCTTAC-3′) and 1-AS (5′-TTGTCACTTCCTCCGTTTCC-3′), 2-S (5′-TTTGTTGGAGGCATTTTATGG-3′) and 2-AS (5′-AACGCGTTGAAATGCCCTCTC-3′), and 3-S (5′-GACAGGCAATCACAAGCATTC-3′) and 3-AS (5′-CGATTAGAACTTGGAGGCTGA-3′). The PCR reactions were performed in a final volume of 25 μL containing the following mixture: PCR buffer, 0.2 mm dNTPs, 0.5 μm of both 5′ and 3′ primers, 1 unit Taq DNA polymerase (Promega), and adjusted amounts of cDNA. DNA was isolated using NUCLOSPIN plant (Macherey-Nagel). Total RNA was purified from the plants using the RNeasy Plant Mini kit (Qiagen) and stored at −80°C following quantification by spectrophotometry. After DNAse treatment (DNase, RQ1, RNase free, Promega), cDNAs were prepared using Moloney murine leukemia virus reverse transcriptase, RNaseH minus, point mutant (Promega) as indicated by the manufacturer and stored at −20°C. cDNAs were diluted approximately 10 times for the PCR reaction. After denaturation at 95°C for 3 min, 35 PCR cycles (94°C for 45 s, 58°C for 45 s, and 72°C for 1 min) were run.
Complementation of Yeast
For complementation of W303-1A abc1∷HIS3 (Hsieh et al., 2004) deficient in the endogenous Abc1 gene, we used AtOSA1 sequence with the chloroplast targeting presequence. Two constructs were tested with and without EYFP. The construct with EYFP was obtained by recloning of AtOSA1-EYFP from pRT vector into pNEV (Sauer and Stolz, 1994) via NOTI site. The construct without YFP and with targeting presequence was obtained by PCR (5NOTAtOSA1-S, 5′-TGCTACCGGTGCGGCCGCATGGCGACTTCTTCTTCTTCATCG-3′; and 3′-NOTAtOSA1-AS, 5′-ATAAGAATGCGGCCGCTTAAGCTGTTCCAGTGATTAGTTTTTCC-3′) using pRT-AtOSA1-EYFP as a template. PCR product was sequenced to avoid errors. Yeast transformation was performed using standard protocols. Transformants were growing on the synthetic dextrose medium (2% [w/v] Glc, 0.7% [w/v] yeast nitrogen base, and required amino acids) with Glc or glycerol as a source of carbon. Cells were analyzed by confocal laser scanning microscopy (TCS SP2 Leica).
Localization of AtOSA1
The AtOSA1 cDNA was PCR amplified (AtOSA1-S, 5′-TGCTACCGGTGCGGCCGCATGGCGACTTCTTCTTCTTCATCG-3′; and AtOSA1-AS, 5′-TCGTCCATGGAAGCTGTTCCAGTGATTAGTTTTTCC-3′) to introduce appropriate restriction sites and cloned into AgeI/NcoI from vector pEYFP (BD Biosciences) to fuse it with EYFP. We used cDNA prepared as described above as a template for the PCR. The resulting AtOSA1-EYFP was cut off by NotI and cloned into vector pRT (Überlacker and Werr, 1996), resulting in pRT-AtOSA1-EYFP. The entire gene fusion product was sequenced to verify the absence of PCR errors. The Tic110-GFP construct was kindly provided by F. Kessler, University of Neuchatel.
Arabidopsis (Arabidopsis thaliana) suspension cell cultures were grown as described in Millar et al. (2001). Three days after culture dilution, the cells were transferred onto solid medium, and 48 h later the plants were transfected with appropriate constructs using a particle inflow gun (PDS1000He; Bio-Rad) with 0.6-μm particles and 1,300 psi pressure. The transfected Arabidopsis cells were analyzed by confocal laser scanning microscopy (TCS SP2 Leica) 24 and 48 h after bombardment.
Chloroplast and Envelope Membrane Preparation
First, the mesophyll protoplasts were prepared from leaves according to the protocol described in Cosio et al. (2004) and subsequently, the intact chloroplasts were obtained according to the method of Fitzpatrick and Keegstra (2001). The collected protoplast pellet was resuspended briefly in 300 mm sorbitol, 20 mm Tricine-KOH, pH 8.4, 10 mm EDTA, 10 mm NaHCO3, 0.1% (w/v) bovine serum albumin, and forced twice through 20- and 11-μm nylon mesh. Released chloroplasts were immediately purified on an 85%/45% (v/v) Percoll gradient and collected by centrifugation at 250g. The chloroplast envelope membranes were isolated from purified chloroplasts as described by Froehlich et al. (2003).
Plant Growth
Arabidopsis (Col-0) called above wild-type and AtOSA1 T-DNA-inserted mutant (SALK 045739, GABI 132G06) plants were grown on soil in a growth chamber (8-h-light period, 22°C; 16-h-dark period, 21°C; 70% relative humidity) and at a light intensity of 140 to 160 μmol m−2 s−1.
For sterile growth after sterilization, the seeds (approximately 20) were placed on 0.8% (w/v) agar plates containing one-half-strength Murashige and Skoog (MS; Duchefa) or MAMI and 1% (w/v) Suc. MAMI medium is: KH2PO4 (200 mg/L); MgSO4.7H2O (187.5 mg/L); Ca(NO3).4H2O (79.25 mg/L); KNO3 (22 mg/L); Fe-EDDHA sequestren (17.5 mg/L); MnCl2.4H2O (48.75 μg/L); H3BO3 (76.25 μg/L); ZnSO4.7H2O (12.25 μg/L); CuSO4.5H2O (6.875 μg/L); NaNoO4.2H2O (12.5 μg/L); and Ni(NO3)2.6H2O (3.75 μg/L). The plates were stored at 4°C for 24 h for synchronization of seed germination and then placed vertically in the phytotron (25°C, 16 h light, and 70% humidity) at light intensity of 80 to 120 μmol m−2 s−1. For treatments, seeds were germinated and grown vertically on one-half-strength MS bactoagar plates in the presence or absence of 1, 10, or 20 μm CdCl2 or 1 mm H2O2 at 16 h light for 7 d. Hydroponic culture: Seeds were first germinated and grown vertically on one-half-strength MAMI bactoagar plates at 8 h light for 2 weeks. Seedlings were then transferred in one-half-strength MAMI liquid medium under the same growth conditions for 2 weeks. Plants were finally cultivated for an additional 3 weeks in the presence or absence of 0.2, 0.5, or 1 μm CdCl2 in one-half-strength MAMI. Cadmium was desorbed after 10 min of root incubation in 1 mm CaCl2 cold solution. The cadmium content was determined by atomic absorption spectroscopy (AAS) in shoots and roots.
Plant Labeling
The plants were root labeled with 0.04 MBq 109CdCl2 in one-eighth-strength MAMI for 4 h. After washing with cold distilled water, plants were grown in one-half-strength MS for an additional 3 d, dried, and subjected to autoradiography.
Determination of SOD Activity
For the SOD activity measurements without any treatment, we used 4-week-old plants grown on soil. For measurement following a Cd2+ application, plants were germinated on 0.8% (w/v) agar plates containing one-half-strength MS (Duchefa) and 1% (w/v) Suc. The plates were stored at 4°C for 16 h for synchronization of seed germination, then placed vertically in the phytotron (22°C, 8 h light, and 70% humidity). Two-week-old seedlings were transferred to liquid medium and cultivated under hydroponic conditions for 3 weeks on MAMI medium. CdCl2 was added to the medium to a final concentration of 1 μm and the samples were taken 24 h later. The activity of SOD was measured as described by Hacisalihoglu et al. (2003). Leaves were homogenized briefly with 50 mm HEPES buffer, pH 7.6, containing 0.1 mm Na2EDTA, 1 mm phenylmethylsulfonyl fluoride, 1% (w/v) PEG4000, and 1% (w/v) polyvinylpolypyrrolidone (Sigma) and centrifuged at 14,000 rpm for 10 min at 4°C. The supernatant was desalted on a Biospin column P6 (Bio-Rad) according to the supplier's protocol and used for protein and SOD assays. The assay determines the inhibition of the photochemical reduction of nitroblue tetrazolium (NBT) as described by Giannopolitis and Ries (1977). The 1-mL reaction mixture for the SOD assay contained 50 mm HEPES, pH 7.6, 0.1 mm EDTA, 50 mm Na2CO3, pH 10.4, 13 mm Met, 75 μm NBT, 0.5 mL of enzyme extract, and 2 μm riboflavin. The reaction mixtures were illuminated for 15 min at 250 μmol m−2 s−1 light intensity or kept in the dark (negative control). One unit of SOD activity was defined as the amount of enzyme required to cause 50% inhibition of the reduction of NBT measured at 560 nm. Protein content was determined according to Bradford (1976) using bovine serum albumin as a standard.
Gas Exchange
Photosynthetic gas exchange measurements were performed on attached leaves before plants flowered using an open infrared gas analyzer system (CIRAS-1; PP-Systems). Measurements were made on plants grown at 8 h light at a photosynthetic photon flux density of 50, 100, or 150 μmol m−2 s−1, and CO2 concentration of 350 μmol. Leaf temperature was adjusted to the desired level using the internal heating/cooling system of the analyzer.
Detection of Protein Kinase Activity in Gelo
The method for detecting protein kinases in gelo was adapted from Mori and Muto (1997). Chloroplast envelope membranes were isolated from wild type and the AtOSA1 mutant and separated by SDS-PAGE. In this experiment, 350 μg of myelin basic protein (M1891; Sigma) was used as protein kinase substrate and incorporated in the running gel solution before polymerization. After electrophoresis, polypeptides were renaturated for 12 h in 50 mm MOPS-KOH, pH 7.6, changing the buffer four times during this period. The gel was then labeled with 1.6 MBq [γ-32P]ATP (AA0068; Amersham-Bioscience) in 5 mL of 50 mm MOPS-KOH, pH 7.6, 10 mm MgCl2, 0.5 mm CaCl2 for 3 h following a 45-min preincubation in the same buffer without the labeled ATP. The gel was then rapidly washed with deionized water and incubated in 100 mL of 50 mm MOPS-KOH, pH 7.6, containing 10 g of a strong basic anion exchanger (Amberlit IRA-410; Sigma) for 3 h. The removal of unbound 32P was terminated by incubation of the gel in 50 mm MOPS-KOH, pH 7.6, supplemented with 1% (w/v) sodium pyrophosphate for 3 h. The polypeptides were then fixed in the gel in 10% (v/v) 2-propanol, 5% (v/v) acetic acid, and 1% (w/v) sodium pyrophosphate. The gel was finally dried and subjected to autoradiography.
Pigment Analyses
The plants were grown at 8 h light (50 μmol m−2 s−1) for 8 weeks. From these plants, leaf samples (50 mg) were collected and analyzed for the content of Chla and Chlb, as well as carotenoids (n = 10). Plants were grown at 16 h light (350 μmol m−2 s−1) for 5 weeks. From these plants, leaf samples (50 mg) were collected and analyzed for the content of Chla and Chlb, as well as carotenoids. Pigments were measured using the method described by Pruzinska et al. (2005).
Statistics
Each value represents the mean of n replicates. Error bars represent se. Significant differences from wild type as determined by Student's t test are indicated as follows: *, P < 0.1; **, P < 0.05; and ***, P < 0.001, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of predicted Abc1 proteins related to AtOSA1.
Supplemental Figure S2. AtOSA1 T-DNA insertion mutants.
Supplemental Figure S3. Verification of a truncated transcript in atosa1 mutants.
Supplementary Material
Acknowledgments
We thank Prof. F. Kessler for providing us with Tic110-GFP construct, and E. Hsieh for the kind gift of W303-1A abc1∷HIS3 and W303-1A strains as well as p3HN4 plasmid. We acknowledge Amélie Fragnière, Regis Mark, and Esther Vogt for technical assistance; Dr. Daniel Studer, University of Bern, for electron microscopy; Prof. Detlef Günther, Swiss Federal Institute of Technology, for inductively coupled plasma mass spectrometry measurements; Dr. Stefan Hortensteiner, University of Bern, for phylogenetic tree; and Prof. Urs Feller, University of Bern, and Dr. Sonia Plaza, University of Fribourg, for AAS measurements.
This work was supported by the Bundesamt für Bildung und Wissenschaft (grant nos. 01.0599 and EU HPRNT–CT–2002–00269 to E.M. and to L.B. under COST Action E28 [Genosylva: European Forest Genomic Network] and COST 859 [Phytotechnologies to promote sustainable land use management and improve food chain safety]). M.J. was a Marie Curie fellow (HPRN–CT–2002–00269).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Lucien Bovet (lucien.bovet@pmintl.com).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Bairoch A (1992) PROSITE: a dictionary of sites and patterns in proteins. Nucleic Acids Res 20 2013–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball L, Accotto GP, Bechtold U, Creissen G, Funck D, Jimenez A, Kular B, Leyland N, Mejia-Carranza J, Reynolds H, et al (2004) Evidence for a direct link between glutathione biosynthesis and stress defense gene expression in Arabidopsis. Plant Cell 16 2448–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet I, Dujardin G, Slonimski PP (1991) ABC1, a novel yeast nuclear gene has a dual function in mitochondria: it suppresses a cytochrome b mRNA translation defect and is essential for the electron transfer in the bc 1 complex. EMBO J 10 2023–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovet L, Feller U, Martinoia E (2005) Possible involvement of plant ABC transporters in cadmium detoxification: a c-DNA sub-microarray approach. Environ Int 31 263–267 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Brasseur G, Tron G, Dujardin G, Slonimski PP, Brivet-Chevillotte P (1997) The nuclear ABC1 gene is essential for the correct conformation and functioning of the cytochrome bc1 complex and the neighbouring complexes II and IV in the mitochondrial respiratory chain. Eur J Biochem 15 103–111 [DOI] [PubMed] [Google Scholar]
- Brennan RJ, Schiestl RH (1996) Cadmium is an inducer of oxidative stress in yeast. Mutat Res 356 171–178 [DOI] [PubMed] [Google Scholar]
- Brink S, Flugge UI, Chaumont F, Boutry M, Emmermann M, Schmitz U, Becker K, Pfanner N (1994) Preproteins of chloroplast envelope inner membrane contain targeting information for receptor-dependent import into fungal mitochondria. J Biol Chem 269 16478–16485 [PubMed] [Google Scholar]
- Cardazzo B, Hamel P, Sakamoto W, Wintz H, Dujardin G (1998) Isolation of an Arabidopsis thaliana cDNA by complementation of a yeast abc1 deletion mutant deficient in complex III respiratory activity. Gene 221 117–125 [DOI] [PubMed] [Google Scholar]
- Clemens S (2001) Molecular mechanisms of plant metal tolerance and homeostasis. Planta 212 475–486 [DOI] [PubMed] [Google Scholar]
- Clemens S, Palmgren MG, Kramer U (2002) A long way ahead: understanding and engineering plant metal accumulation. Trends Plant Sci 7 309–315 [DOI] [PubMed] [Google Scholar]
- Cobbett C, Goldsbrough P (2002) Phytochelatins and metallothioneins: roles in heavy metal detoxification and homeostasis. Annu Rev Plant Biol 53 159–182 [DOI] [PubMed] [Google Scholar]
- Cosio C, Martinoia E, Keller C (2004) Hyperaccumulation of cadmium and zinc in Thlaspi caerulescens and Arabidopsis halleri at the leaf cellular level. Plant Physiol 134 716–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do TQ, Hsu AY, Jonassen T, Lee PT, Clarke CF (2001) A defect in coenzyme Q biosynthesis is responsible for the respiratory deficiency in Saccharomyces cerevisiae abc1 mutants. J Biol Chem 276 18161–18168 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300 1005–1016 [DOI] [PubMed] [Google Scholar]
- Eren E, Argüello JM (2004) Arabidopsis HMA2, a divalent heavy metal-transporting P(IB)-type ATPase, is involved in cytoplasmic Zn2+ homeostasis. Plant Physiol 136 3712–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick LM, Keegstra K (2001) A method for isolating a high yield of Arabidopsis chloroplasts capable of efficient import of precursor proteins. Plant J 27 59–65 [DOI] [PubMed] [Google Scholar]
- Fornazier RF, Ferreira RR, Vitoria AP, Molina SMG, Lea PJ, Azevedo RA (2002) Effects of cadmium on antioxidant enzyme activities in sugar cane. Biol Plant 45 91–97 [Google Scholar]
- Froehlich JE, Wilkerson CG, Ray WK, McAndrew RS, Osteryoung KW, Gage DA, Phinney BS (2003) Proteomic study of the Arabidopsis thaliana chloroplastic envelope membrane utilizing alternatives to traditional two-dimensional electrophoresis. J Proteome Res 2 413–425 [DOI] [PubMed] [Google Scholar]
- Fusco N, Micheletto L, Dal Corso G, Borgato L, Furini A (2005) Identification of cadmium-regulated genes by cDNA-AFLP in the heavy metal accumulator Brassica juncea L. J Exp Bot 56 3017–3027 [DOI] [PubMed] [Google Scholar]
- Giannopolitis CN, Ries SK (1977) Superoxide dismutases-occurrence in higher plants. Plant Physiol 59 309–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacisalihoglu G, Hart JJ, Wang YH, Cakmak I, Kochian LV (2003) Zinc efficiency is correlated with enhanced expression and activity of zinc-requiring enzymes in wheat. Plant Physiol 131 595–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W (1993) TMbase: a database of membrane spanning proteins segments. Biol Chem Hoppe Seyler 374: 166
- Hsieh EJ, Dinoso JB, Clarke CF (2004) A tRNA(TRP) gene mediates the suppression of cbs2-223 previously attributed to ABC1/COQ8. Biochem Biophys Res Commun 317 648–653 [DOI] [PubMed] [Google Scholar]
- Iiizumi M, Arakawa H, Mori T, Ando A, Nakamura Y (2002) Isolation of a novel gene, CABC1, encoding a mitochondrial protein that is highly homologous to yeast activity of bc1 complex. Cancer Res 62 1246–1250 [PubMed] [Google Scholar]
- Inaba T, Li M, Alvarez-Huerta M, Kessler F, Schnell DJ (2003) atTic110 functions as a scaffold for coordinating the stromal events of protein import into chloroplasts. J Biol Chem 278 38617–38627 [DOI] [PubMed] [Google Scholar]
- Jonak C, Nakagami H, Hirt H (2004) Heavy metal stress. Activation of distinct mitogen-activated protein kinase pathways by copper and cadmium. Plant Physiol 136 3276–3283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DY, Bovet L, Maeshima M, Martinoia E, Lee Y (2007) The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant J 50 207–218 [DOI] [PubMed] [Google Scholar]
- Klein M, Mamnun YM, Eggmann T, Schuller C, Wolfger H, Martinoia E, Kuchler K (2002) The ATP-binding cassette (ABC) transporter Bpt1p mediates vacuolar sequestration of glutathione conjugates in yeast. FEBS Lett 520 63–67 [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, Monde RA, Last RL (1998) Superoxide dismutase in Arabidopsis: an eclectic enzyme family with disparate regulation and protein localization. Plant Physiol 118 637–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo AJK, Ohlrogge JB (2002) The predicted candidates of Arabidopsis plastid inner envelope membrane proteins and their expression profiles. Plant Physiol 130 823–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard CJ, Aravind L, Koonin EV (1998) Novel families of putative protein kinases in bacteria and archaea: evolution of the “eukaryotic” protein kinase superfamily. Genome Res 8 1038–1047 [DOI] [PubMed] [Google Scholar]
- Macinga DR, Cook GM, Poole RK, Rather PN (1998) Identification and characterization of aarF, a locus required for production of ubiquinone in Providencia stuartii and Escherichia coli and for expression of 2′-N-acetyltransferase in P. stuartii. J Bacteriol 180 128–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWeese-Scott C, Geer LY, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, et al (2005) CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res 33 D192–D196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, DeWeese-Scott C, Fedorova ND, Geer LY, He S, Hurwitz DI, Jackson JD, Jacobs AR, Lanczycki CJ, et al (2003) CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res 31 383–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Sweetlove LJ, Giege P, Leaver CJ (2001) Analysis of the Arabidopsis mitochondrial proteome. Plant Physiol 127 1711–1727 [PMC free article] [PubMed] [Google Scholar]
- Morgenstern B (2004) DIALIGN: multiple DNA and protein sequence alignment at BiBiServ. Nucleic Acids Res 32 W33–W36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori IC, Muto S (1997) Abscisic acid activates a 48-kDa protein kinase in guard cell protoplasts. Plant Physiol 113 833–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munne-Bosch S, Alegre L (2002) Plant aging increases oxidative stress in chloroplasts. Planta 214 608–615 [DOI] [PubMed] [Google Scholar]
- Peltier JB, Emanuelsson O, Kalume DE, Ytterberg J, Friso G, Rudella A, Liberles DA, Soderberg L, Roepstorff P, von Heijne G, et al (2002) Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell 14 211–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller R, Pfanner N, Neupert W (1989) Mitochondrial protein import. Bypass of proteinaceous surface receptors can occur with low specificity and efficiency. J Biol Chem 264 34–39 [PubMed] [Google Scholar]
- Poon WW, Davis DE, Ha HT, Jonassen T, Rather PN, Clarke CF (2000) Identification of Escherichia coli ubiB, a gene required for the first monooxygenase step in ubiquinone biosynthesis. J Bacteriol 182 5139–5146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruzinska A, Tanner G, Aubry S, Anders I, Moser S, Müller T, Ongania KH, Kräutler B, Youn JY, Liljegren SJ, et al (2005) Chlorophyll breakdown in senescent Arabidopsis leaves. Characterization of chlorophyll catabolites and of chlorophyll catabolic enzymes involved in the degreening reaction1. Plant Physiol 139 52–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranieri A, Castagna A, Baldan B, Soldatini GF (2001) Iron deficiency differently affects peroxidase isoforms in sunflower. J Exp Bot 52 25–35 [PubMed] [Google Scholar]
- Salt DE, Prince RC, Pickering IJ, Raskin I (1995) Mechanisms of cadmium mobility and accumulation in Indian mustard. Plant Physiol 109 1427–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Fernandez R, Davies TG, Coleman JO, Rea PA (2001) The Arabidopsis thaliana ABC protein superfamily, a complete inventory. J Biol Chem 276 30231–30244 [DOI] [PubMed] [Google Scholar]
- Sauer N, Stolz J (1994) SUC1 and SUC2: two sucrose transporters from Arabidopsis thaliana; expression and characterization in baker's yeast and identification of the histidine-tagged protein. Plant J 6 67–77 [DOI] [PubMed] [Google Scholar]
- Trumpower BL (1990) Cytochrome bc1 complexes of microorganisms. Microbiol Rev 54 101–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Überlacker B, Werr W (1996) Vectors with rare-cutter restriction enzyme sites for expression of open reading frames in transgenic plants. Mol Breed 2 293–295 [Google Scholar]
- Unden G, Bongaerts J (1997) Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim Biophys Acta 1320 217–234 [DOI] [PubMed] [Google Scholar]
- Verret F, Gravot A, Auroy P, Leonhardt N, David P, Nussaume L, Vavasseur A, Richaud P (2004) Overexpression of AtHMA4 enhances root-to-shoot translocation of zinc and cadmium and plant metal tolerance. FEBS Lett 576 306–312 [DOI] [PubMed] [Google Scholar]
- Wang Y, Fang J, Leonard SS, Rao KMK (2004) Cadmium inhibits the electron transfer chain and induces reactive oxygen species. Free Radic Biol Med 36 1434–1443 [DOI] [PubMed] [Google Scholar]
- Watanabe M, Suzuki T (2002) Involvement of reactive oxygen stress in cadmium-induced cellular damage in Euglena gracilis. Comp Biochem Physiol 131 491–500 [DOI] [PubMed] [Google Scholar]
- Ytterberg AJ, Peltier JB, van Wijk KJ (2006) Protein profiling of plastoglobules in chloroplasts and chromoplasts. A surprising site for differential accumulation of metabolic enzymes. Plant Physiol 140 984–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.