Abstract
Transcription corepressors play important roles in animal and plant development. In Arabidopsis (Arabidopsis thaliana), LEUNIG (LUG) and LEUNIG_HOMOLOG (LUH) encode two highly homologous proteins that are similar to the animal and fungal Gro/Tup1-type corepressors. LUG was previously shown to form a putative corepressor complex with another protein, SEUSS (SEU), and to repress the transcription of AGAMOUS in floral organ identity specification. However, the function of LUH is completely unknown. Here, we show that single luh loss-of-function mutants develop normal flowers, but lug; luh double mutants are embryo lethal, uncovering a previously unknown function of LUG and LUH in embryonic development. In addition, luh/+ enhances the floral phenotype of lug, revealing a minor role of LUH in flower development. Functional diversification between LUH and LUG is evidenced by the inability of 35S∷LUH overexpression to rescue lug mutants and by the opposite expression trends of LUG and LUH in response to biotic and abiotic stresses. The luh-1 mutation does not enhance the defect of seu in flower development, but LUH could directly interact with SEU in yeast. We propose a model that explains the complex relationships among LUH, LUG, and SEU. As most eukaryotes have undergone at least one round of whole-genome duplication during evolution, gene duplication and functional diversification are important issues to consider in uncovering gene function. Our study provides important insights into the complexity in the relationship between two highly homologous paralogous genes.
Transcription repression plays a key regulatory role in cell fate specification, hormone signaling, and plant stress responses. LEUNIG (LUG) was first identified in Arabidopsis (Arabidopsis thaliana) based on its role in regulating the stage- and domain-specific expression of the C class floral homeotic gene AGAMOUS (AG) in flower development (Liu and Meyerowitz, 1995). In lug mutants, ectopic AG expression in the outer two whorls of a flower leads to homeotic transformation of sepals into carpels and petals into stamens as well as a reduction of floral organs. LUG protein is similar in domain structure and biochemical function to the Groucho (Gro), Transducin-Like Enhancer of Split, and Tup1 family corepressors in Drosophila, mammals, and yeast, respectively (Liu and Karmarkar, 2008). These corepressors do not possess a DNA-binding domain and are recruited to their regulatory targets by interacting with DNA-bound transcription factors.
The N terminus of LUG possesses a conserved domain, the LUFS domain, named after the four founding members LUG, LUH (for LEUNIG_HOMOLOG), yeast Flo8, and human SSDP (for single-stranded DNA-binding protein). The LUFS domain of LUG is essential for the direct interaction with its cofactor SEUSS (SEU; Sridhar et al., 2004). SEU encodes a Gln (Q)-rich protein with a centrally positioned dimerization domain also present in the LIM domain-binding family of transcriptional coregulators in mammals and Drosophila (Franks et al., 2002). Recruitment of the LUG/SEU corepressor complex by the MADS box proteins APETALA1 (AP1) and SEPALLATA3 (SEP3) was shown to target the LUG/SEU corepressors to the AG cis-regulatory element, leading to repressed chromatin at the AG locus (Sridhar et al., 2006). The repressor activity of LUG was shown to depend on histone deacetylase (HDAC) activity, and LUG was shown to directly interact with HDAC19 (Sridhar et al., 2004; Gonzalez et al., 2007), suggesting that the plant Gro/Tup1 family corepressors mediate transcription repression by histone modification and chromatin reorganization. Recently, LUG was shown to repress gene expression via a HDAC-independent but mediator-dependent mechanism (Gonzalez et al., 2007).
Like corepressors in animals and fungi, LUG/SEU possesses multiple functions. lug mutants showed defects in gynoecium development, female and male fertility, leaf and floral organ shape, and vasculature (Liu and Meyerowitz, 1995; Chen et al., 2000; Liu et al., 2000; Cnops et al., 2004; Franks et al., 2006). Antirrhinum mutants of STYLOSA, a LUG ortholog, not only showed abnormal flower development but also exhibited hypersensitivity toward auxin and polar auxin inhibitors (Navarro et al., 2004). A transcriptome study identified LUG-regulated genes in abiotic and biotic stress response, meristem function, and transport (Gonzalez et al., 2007). Therefore, LUG likely encodes a global regulator for multiple developmental processes and signal pathways.
In Arabidopsis, LUG belongs to a small family of about 13 genes (http://smart.embl-heidelberg.de/), including TOPLESS (TPL), TOPLESS-RELATED, and WUSCHEL-INTERACTING PROTEINs (WSIPs; Kieffer et al., 2006; Long et al., 2006; Liu and Karmarkar, 2008). These genes are involved in regulating embryonic shoot-root axis determination and appear to repress auxin-mediated signaling events during embryogenesis (Szemenyei et al., 2008). The TPL/WSIP genes are also involved in mediating the effect of WUSCHEL in target gene repression to maintain the stem cell pool at the shoot apical meristem (Kieffer et al., 2006). Therefore, the plant Gro/Tup1 family corepressors are emerging as a fundamentally important class of regulators in plant development.
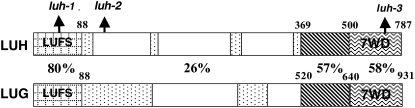
Among the 13 Arabidopsis Gro/Tup1 corepressor-like proteins, LUH (At2g32700) is most similar to LUG (Conner and Liu, 2000). Both proteins possess an N-terminal LUFS domain that is 80% identical (Fig. 1). In addition, both proteins possess seven WD repeats at the C terminus that show 58% identity to each other. A third domain that immediately precedes the WD repeats (residues 369–500) also shows a high level of sequence similarity (57%). Both LUG and LUH have centrally located Q-rich regions, but the Q-rich regions in LUH are less continuous and less extensive than those in LUG.
Figure 1.
Schematic diagram showing the protein domains of LUH and LUG. Numbers represent amino acid positions, and percentage values indicate the percentage identity between LUH and LUG. In addition to the N-terminal LUFS domain and the C-terminal seven WD repeats, a region immediately preceding the WD repeats (box with diagonal lines) is also highly conserved. The dotted boxes represent Q-rich regions. The location of each luh allele is indicated by an arrow.
Despite the significant sequence similarity, almost nothing is known about LUH function or expression. This article presents data indicating that LUH possesses both unique and overlapping functions with LUG and that LUH activity is required for proper embryo and flower development. We propose a model that explains the complex relationship between LUH and LUG.
RESULTS
luh-1 Mutants Exhibit Vegetative Defects
Through the Arabidopsis TILLING Project (McCallum et al., 2000), we obtained several luh mutations (see “Materials and Methods”). luh-1 (luh_172H3) is caused by a G-to-A change resulting in the conversion of Trp (W-55) to a STOP codon (Fig. 1; Supplemental Fig. S1), truncating the protein at the 55th residue. luh-2 (luh_147A6) changes C to T, resulting in an amino acid substitution from Ser (S-123) to Phe (F). luh-3 (SALK_107245C) is caused by a T-DNA insertion in the third to the last exon (Supplemental Fig. S1), disrupting the last three WD repeats. We have focused on luh-1 as it likely represents a null or a strong loss-of-function mutation.
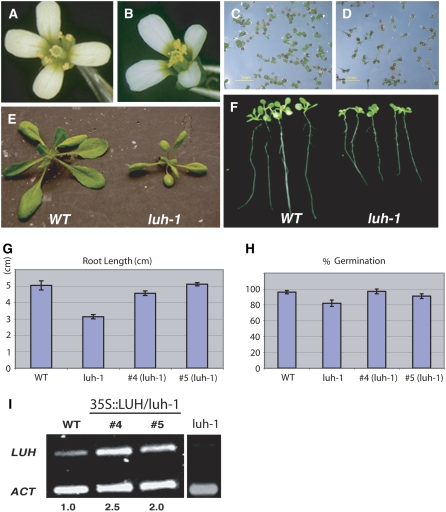
luh-1 single mutants did not exhibit any abnormality in flowers (Fig. 2, A and B). Nevertheless, luh-1 mutant seedlings showed slower and poorer germination on Murashige and Skoog (MS) medium (Fig. 2, C and D), with a germination rate of about 80% of wild type (Fig. 2H). In addition, luh-1 mutants grew slower compared with the wild type (Fig. 2E) but eventually caught up. Finally, the roots of luh-1 seedlings were significantly shorter than wild-type roots (Fig. 2, F and G). To test whether these phenotypes are caused by the luh-1 mutation, 35S∷LUH cDNA was transformed into luh-1 mutants. Eight transgenic plants were obtained, and two of these transgenic lines (lines 4 and 5) showed a higher level of LUH mRNA (Fig. 2I) and were further analyzed. The developmental defects of luh-1 described above were rescued by the 35S∷LUH transgene. Figure 2, G and H, illustrate the rescue of root length and germination rate, respectively.
Figure 2.
luh-1 develops normal flowers but exhibits defects in vegetative growth. A, A wild-type (Columbia erecta-105) flower. B, A luh-1 flower in the Columbia erecta-105 background. C, Germination of wild-type seeds on MS medium at 5 d. D, Germination of luh-1 seeds on MS medium at 5 d. E, Three-week-old wild-type and luh-1 plants. F, Root elongation of wild-type and luh-1 seedlings on MS medium. Germinated seedlings were transferred to MS plates and grown on vertical plates. Photographs were taken after 7 d. G, 35S∷LUH complemented the luh-1 defect in root elongation as shown in transgenic lines 4 and 5. Root length, measured after 7 d, was expressed as mean ± se. H, Germination phenotype of luh-1 is complemented by 35S∷LUH. Germination is expressed as mean ± se. I, Semiquantitative RT-PCR showing the expression of LUH mRNA in the wild type (Columbia erecta-105), luh-1, and two different 35S∷LUH transgenic lines (lines 4 and 5) in the luh-1 background. The ratio between LUH band intensity and that of the ACT2 control band, which is set to 1 in wild type, is shown below each lane.
luh-1/+ Enhances Defects of lug in Flowers
It is possible that the function of LUH in flowers is not necessary when LUG is intact but becomes necessary when LUG is absent or reduced. If this is the case, luh-1 may enhance the phenotype of lug. To test this, both the weak lug-16 and the strong lug-3 were crossed into luh-1 to construct lug-16; luh-1 and lug-3; luh-1 double mutants. F2 progeny segregated lug single mutants as well as mutants with a more severe phenotype than lug single mutants. Allele-discriminating single nucleotide polymorphism (SNP) assays (see “Materials and Methods”) were used to genotype F2 plants with a more severe phenotype than lug single mutants, and they were found to be homozygous for lug and heterozygous for luh-1. Specifically, while lug-16 single mutants developed elongated siliques (Fig. 3A), the lug-16/lug-16; luh-1/+ plants did not show any silique elongation and were completely sterile (Fig. 3B). However, the floral phenotype of lug-16/lug-16; luh-1/+ was similar to that of lug-16 single mutants (Fig. 3, C and D).
Figure 3.
luh-1 enhances lug-16 and lug-3 during flower development. A, An inflorescence shoot of lug-16. Note the elongating siliques. B, An inflorescence shoot of lug-16/lug-16; luh-1/+. Note the absence of silique development. C, A lug-16 flower with narrow sepals and petals. D, A lug-16/lug-16; luh-1/+ flower. E, A lug-3 flower. F, A lug-3/lug-3; luh-1/+ flower. G, A lug-16; luh-1 double mutant. Note the extreme small stature; the mature plant is smaller than a rosette leaf. H, A close-up of an inflorescence shoot of the lug-16; luh-1 double mutant. I, A seu-1 flower. Note the reduced stamen number. J, A seu-1; luh-1 double mutant flower.
lug-3/lug-3; luh-1/+ flowers exhibited a more severe phenotype than lug-3 single mutant flowers (Fig. 3, E and F). lug-3/lug-3; luh-1/+ mutant flowers consisted of only a few carpelloid sepals and sepal-like organs topped with horns, suggesting a more severe homeotic transformation, possibly caused by more extensive ectopic expression of AG. There was no petal, and stamens were either completely absent or partially fused to first whorl organs (Fig. 3F). The lug-3/lug-3; luh-1/+ flowers are completely sterile, and they resemble lug; seu double mutant flowers (Franks et al., 2002).
Among the F2 progeny of the lug-3 and luh-1 cross, lug-3; luh-1 double mutants were never found, although they should occur at a frequency of one in 16. Only two lug-16; luh-1 double homozygous mutants were found after screening several hundred F2 progeny of the lug-16 and luh-1 cross. The lug-16; luh-1 double mutants were extremely small in stature; the entire mature plant is smaller than a single rosette leaf (Fig. 3G). The inflorescence meristem only bears three to five flowers consisting of only carpels (Fig. 3H).
Since SEU acts as an adaptor for LUG and the seu mutation enhances lug mutants (Franks et al., 2002; Sridhar et al., 2004), we tested the genetic interaction between luh and seu. seu-1; luh-1 double mutants were identified by genotyping F2 as well as F3 plants of the seu-1 and luh-1 cross. The seu-1; luh-1 double mutants are morphologically indistinguishable from seu-1 single mutants (Fig. 3, I and J).
Most luh; lug Double Mutants Are Embryo Lethal
The absence of lug-3; luh-1 double mutants and a significant reduction of lug-16; luh-1 double mutants among the F2 progeny suggested that most lug; luh-1 double mutants die prematurely. The complete sterility of lug-16/lug-16; luh-1/+ plants (Fig. 3, A and B) makes it impossible to identify lug-16; luh-1 double mutants in the next generation. Instead, we identified several luh-1/luh-1; lug-16/+ plants through genotyping F2 progeny of the lug-16 and luh-1 cross. Surprisingly, these luh-1/luh-1; lug-16/+ plants developed wild-type-like flowers, albeit at a slightly smaller size (Fig. 4, A and D). Thus, it appears that LUG is more critical for proper flower development than LUH, as luh-1/luh-1; lug-16/+ plants with only one copy of wild-type LUG are capable of normal floral development but lug-16/lug-16; luh-1/+ plants with only one copy of wild-type LUH fail to develop normal flowers (Fig. 3).
Figure 4.
luh-1; lug-16 double mutants are embryo lethal. A, luh-1 flower. B, An open silique of luh-1 showing green seeds inside. C, An open silique of lug-16 showing green seeds inside. D, A luh-1/luh-1; lug-16/+ flower. Note the smaller flower size. E, An open luh-1/luh-1; lug-16/+ silique showing white seeds among green seeds. F, Nomarski image of a green seed in a silique derived from a luh-1/luh-1; lug-16/+ plant. The arrow indicates the embryo proper at the torpedo stage. G, Nomarski image of a white seed from the same silique as F. The arrow indicates an embryo at the late globular stage.
When luh-1/luh-1; lug-16/+ plants were self-fertilized and their siliques were examined, white and abnormal seeds occurred at a frequency of about 36% in a silique (Fig. 4E; Supplemental Table S1). This is in contrast to luh-1 and lug-16 single mutants, whose siliques contain only about 5% of white seeds (Fig. 4, B and C; Supplemental Table S1). To verify the genotype of white and green seeds segregated by luh-1/luh-1; lug-16/+ plants, eight white seeds and 10 greens seeds (collected from several different siliques) were individually genotyped. All eight white seeds were found to be luh-1/luh-1; lug-16/+. Among the 10 green seeds, six were luh-1/luh-1; +/+ and four were luh-1/luh-1; lug-16/+. This suggests that luh-1/luh-1; lug-16/+ seeds could develop into either normal green seeds or abnormal white seeds. An absence of luh-1/luh-1; lug-16/lug-16 genotype among the eight white and 10 green seeds indicated that the luh-1/luh-1; lug-16/lug-16 embryos died early during embryogenesis, before visible seeds were formed. Therefore, significant functional redundancy must exist between LUH and LUG during early embryo development.
To better pinpoint the stage at which embryo development is affected in the white seeds, we examined the white and green seeds dissected from the same siliques of luh-1/luh-1; lug-16/+ plants. While the green embryos were already at the torpedo stage (Fig. 4F), the white embryos from the same silique were arrested at the late globular stage (Fig. 4G). In some of the white seeds, the globular embryos appeared disintegrated (data not shown).
LUH and LUG Exhibit Divergent Functions and Expression Patterns
Functional diversification between LUH and LUG could result from their differences in expression or in protein-coding sequences or both. To test this, we transformed 35S∷LUH into lug-16 mutants. lug-16 is the most fertile allele and can be easily transformed. If overexpressing LUH could rescue lug-16, the LUH coding region may be equivalent to that of LUG. None of the 12 T1 transformants was able to rescue lug-16. On the contrary, five of these 12 lines showed an enhanced phenotype, with more carpelloid sepals and a greatly reduced organ number (Fig. 5, C and D). We hypothesized that these five lines exhibited cosuppression and silencing of the endogenous LUH. Semiquantitative reverse transcription (RT)-PCR was performed for three of the five lines, revealing that that the LUH mRNA level in these lines was approximately half of that in wild-type plants (Fig. 5E) and supporting the cosuppression hypothesis.
Figure 5.
35S∷LUH failed to rescue lug-16 mutants. A, A lug-16 mutant flower. B, A lug-16 inflorescence. C, A 35S∷LUH; lug-16 flower, where the 35S∷LUH appears to enhance the defect of lug-16, probably by silencing endogenous LUH. D, An inflorescence of a 35S∷LUH; lug-16 transgenic plant similar to that in C. E, RT-PCR result showing reduced LUH mRNA in the 35S∷LUH; lug-16 transgenic lines 1, 2, and 3. The numbers below represent the relative mRNA level normalized to ACT2 and compared with the wild type, which is taken as 1. F and G, 35S∷LUH line 5 in the wild type, causing no obvious phenotype. H and I, 35S∷LUH line 5 in lug-16, showing phenotypes identical to those of the lug-16 mutants shown in A and B. [See online article for color version of this figure.]
The remaining seven 35S∷LUH; lug-16 lines did not show a cosuppressed phenotype, but the 35S∷LUH transgene did not rescue lug-16 (data not shown). Either they did not express high enough levels of LUH or LUH protein is not equivalent to LUG in function. To distinguish these alternative explanations, 35S∷LUH; luh-1 transgenic line 5, previously shown to rescue luh-1 phenotype (Fig. 2), was crossed into lug-16. The F2 plants harboring 35S∷LUH line 5 and carrying wild-type LUH and LUG are wild type in phenotype (Fig. 5, F and G). However, the F2 35S∷LUH line 5 plants carrying the wild-type LUH allele but the lug-16/lug-16 mutant allele exhibited phenotypes identical to the lug-16 single mutants (Fig. 5, H and I), suggesting that increasing (and ectopic expressing) LUH transcripts could not substitute for LUG.
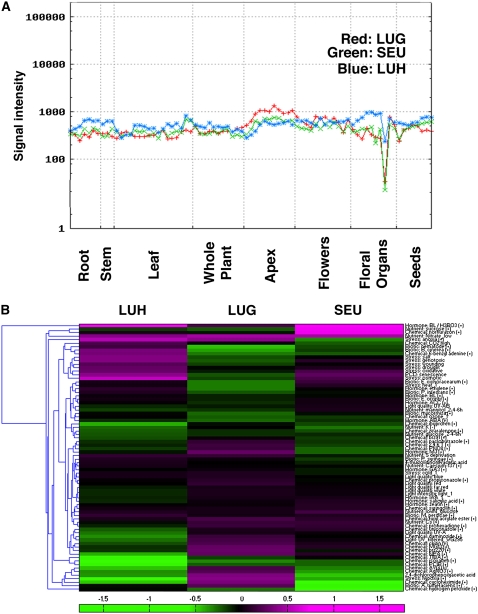
To compare the expression patterns of LUG and LUH during development, we utilized the AtGenExpress atlas that compares the expression profiles of 22,746 probe sets on the Affymetrix ATH1 array using triplicate expression estimates from 79 diverse development samples ranging from embryogenesis to senescence and from roots to flowers (Schmid et al., 2005). LUG and LUH were shown to be expressed in all 79 samples at comparable levels (Fig. 6A). Interestingly, SEU, the partner of LUG, showed an almost identical expression profile to LUG, supporting that proteins present in the same complex are likely expressed in similar profiles (Schmid et al., 2005). A comparison between LUH and SEU revealed highly similar but not identical profiles (Fig. 6A).
Figure 6.
LUH expression in comparison with LUG and SEU. A, Expression profile of LUH, LUG, and SEU in different developmental tissues and stages. The data were generated by AtGenExpress (Development) and presented using the AtGenExpression Visualization Tool (Schmid et al., 2005). LUG (red) and SEU (green) showed almost complete coexpression in all tissues. LUH (blue) closely resembled but was not identical to the LUG/SEU profile. B, Hierarchal cluster analysis of environmental regulation of LUH, LUG, and SEU expression using AtGenExpression (Abiotic, Light, Hormone, Pathogen) estimates by gcRMA (Kilian et al., 2007; www.weigelworld.org/resources/microarray/AtGenExpress/). The clustergram was generated with the Matlab RC13 (Mathworks) Bioinformatics Toolbox. An increase in expression is indicated by magenta, and a decrease in expression is indicated by green (see bar at bottom).
In addition, the expression profiles of LUG, SEU, and LUH were compared using the AtGenExpress data with Arabidopsis samples challenged with biotic and abiotic stresses, hormones, lights, and nutrients (www.weigelworld.org/resources/microarray/AtGenExpress/; Kilian et al., 2007). As shown in Figure 6B, an increased expression upon a particular treatment is indicated by magenta and a decreased expression upon a treatment is indicated by green. The clustergram showed that LUH and LUG exhibited almost opposite expression trends upon treatment with similar conditions. For example, LUH transcription is induced by exposures to biotic stress (nematode and Botrytis cinerea) and abiotic stress (salt, genotoxic, wounding, drought, oxidative); LUG transcription, on the contrary, is reduced or unchanged under these same conditions. Additionally, certain chemicals (cycloheximide, 2,4-dichlorophenoxyacetic acid, AgNO3, aminoethoxyvinylglycine), biotic stress (Agrobacterium tumefaciens), and abiotic stress (hypoxia) caused increased LUG expression but reduced LUH expression. SEU appears to exhibit an expression pattern more similar to that of LUH. This analysis suggests that LUG and LUH are utilized to play opposite regulatory roles in different stress signal pathways.
LUH Interacts Directly with SEU But Not with LUG
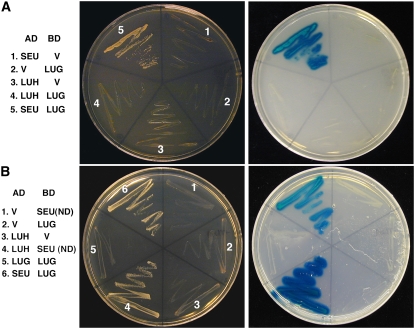
If LUH and LUG have both overlapping and unique functions, do their proteins interact directly with each other to form heterodimers? Yeast two-hybrid assays failed to detect an interaction between LUH-AD (full-length LUH fused to the GAL4 activation domain) and LUG-BD (full-length LUG fused to the GAL4 DNA-binding domain; Fig. 7A), nor could LUG homodimerize (Fig. 7B). Thus LUG and LUH likely act independently or in parallel to regulate common as well as unique target genes.
Figure 7.
LUH interacts with SEU but not with LUG in yeast. A, Yeast two-hybrid assay showing a lack of interaction between LUH and LUG. Positive interaction is indicated by the activation of HIS3 and ADE2 reporter genes allowing colony growth on −Trp, −Leu, −His, and −Ade plates containing 3 mM 3-amino-1,2,4-triazole (left). The activity of a third reporter gene, LacZ, encoding β-galactosidase was tested by the X-Gal overlay assay (right); blue color indicates a positive interaction. B, Yeast two-hybrid assay showing a positive interaction between LUH and SEU(ND). V, Vector. [See online article for color version of this figure.]
Previously, LUG was reported to interact directly with SEU via the N-terminal LUFS domain (Sridhar et al., 2004; Fig. 7, A and B). Because of the 80% sequence identity between LUG and LUH at the LUFS domain, we tested whether LUH could also interact with SEU. A strong interaction was detected between full-length LUH-AD and SEU(ND)-BD (Fig. 7B). SEU(ND) is a truncated SEU with its C-terminal domain (capable of self-activation) removed. Therefore, like LUG, LUH is likely able to form a corepressor complex with SEU.
DISCUSSION
LUG and LUH Exhibit Partially Redundant But Not Identical Functions
In this study, we investigated the function of LUH, the closest homolog of LUG in Arabidopsis. We discovered that these two genes play redundant roles during embryo development, revealing a previously unknown role of LUG during embryonic development. Second, we identified a relatively minor role of LUH compared with LUG in flower development, as the luh-1 single mutation does not affect flower development but luh-1/+ can enhance the floral phenotype of lug. Third, since overexpressing LUH could not rescue the weak lug-16 mutation, the divergence in their coding sequences rather than their expression level or pattern likely contributes to their functional differences.
LUH and SEU Likely Act in the Same Pathway to Regulate Flower Development
We showed that both LUG and LUH interact physically with SEU in yeast, suggesting the possibility of forming both LUG/SEU and LUH/SEU corepressor complexes. Interestingly, lug and luh mutations exhibited drastically different genetic interactions with seu. Specifically, lug; seu double mutants exhibited a synergistic genetic interaction (Conner and Liu, 2000; Franks et al., 2002). In contrast, seu-1 mutant flowers are indistinguishable from seu-1; luh-1 double mutant flowers, suggesting that seu-1 is completely epistatic to luh-1 and implying that the function of LUH in flower development is entirely dependent on SEU. The yeast two-hybrid interactions detected between LUH and SEU suggest that LUH likely functions exclusively in a LUH/SEU complex.
LUG and LUH May Have Divergent Functions in Environmental Stress Responses
Recent genome-wide transcriptome studies comparing wild-type and lug-3 mutant tissues revealed dramatic changes in the expression of genes involved in abiotic and biotic stress responses (Gonzalez et al., 2007). It is thus of particular interest to note the almost opposite expression trends between LUG and LUH under different biotic and abiotic challenges. This difference in gene expression patterns between LUH and LUG upon exposure to different environmental conditions (Fig. 6B) is in sharp contrast to the highly similar gene expression patterns between LUH and LUG in different tissues and developmental stages (Fig. 6A). This suggests that substantial differences may have occurred in the cis-regulatory elements of LUH and LUG involving responses to environmental signals.
Gene duplications are important evolutionary strategies in facilitating species adaptation, buffering deleterious mutations, subdividing their function, or evolving new functions (Lynch and Force, 2000; Lynch et al., 2001). Based on analyses of 2,022 recent duplicated gene pairs in Arabidopsis, duplicate genes with functions in developmental processes were found to be largely coregulated, while duplicate genes acting in abiotic or biotic stress responses were found to exhibit divergent expression profiles (Ha et al., 2007). This is consistent with our finding that LUG and LUH showed similar expression profiles during development but exhibited almost opposite expression trends when challenged with various environmental stresses. Our observation suggests that LUG and LUH may have substantially divergent functions when they act in stress response pathways.
A Proposed Model
Previous molecular and genetic characterizations of lug and seu mutants have been focused on flower development, which revealed important mechanistic insights into how LUG and SEU negatively regulate AG expression (Liu and Meyerowitz, 1995; Conner and Liu, 2000; Franks et al., 2002; Sridhar et al., 2004, 2006). SEU was shown to function as an adaptor protein bridging the interaction between the LUG corepressor and two MADS box transcription factors, AP1 and SEP3. AP1 and SEP3 recruit the SEU/LUG complex to AG by binding directly to the enhancer elements located in the second intron of AG. These previous studies laid important groundwork for our study and served as the basis for the model proposed below. Because of limited data on the function of LUG, LUH, and SEU in nonfloral tissues, the model (Fig. 8) is focused on the regulation of AG in flower development.
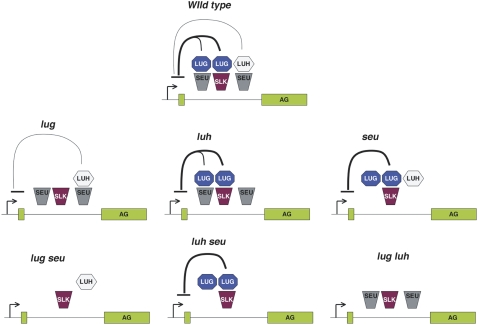
Figure 8.
A model of the repression of AG by LUG, LUH, and SEU during flower development. The LUG/SEU, LUG/SLK, and LUH/SEU putative repressor complexes all act through the second intron of AG. The arrows indicate the transcription initiation from the AG promoter. Bars indicate repressor activity. Thick lines connecting to the bars indicate stronger repressor activity than thin lines. The specific repressor complexes are indicated in single and double mutant combinations. [See online article for color version of this figure.]
Previous synergistic genetic interactions between lug and seu in flowers (Franks et al., 2002) suggested that SEU serves as an adaptor for LUG as well as for other corepressors and that LUG may utilize SEU as well as other adaptor proteins. Thus, removing both SEU and LUG in seu; lug double mutants has a more severe effect. In contrast, the similar phenotype between seu single and seu; luh-1 double mutants suggests that the function of LUH in flower development is dependent entirely on forming a complex with SEU. Therefore, we propose that LUG could form a corepressor complex with SEU as well as SEUSS-like (SLK) proteins (Franks et al., 2002) but that LUH could only pair with SEU. LUG/SEU and LUG/SLK complexes are either more prevalent or exhibit a higher repressor activity or both than LUH/SEU. As a result, LUG plays a more prominent role than LUH in the negative regulation of AG. This is illustrated in Figure 8, where in luh mutants, the LUG/SEU and LUG/SLK complexes are sufficient to cover the loss of LUH/SEU. In seu single or seu; luh double mutants, the LUG/SLK complex can still provide most if not all of the function. In lug mutants, LUH-SEU can also perform most of the jobs. In seu; lug, or luh; lug double mutants, however, none of the LUH/SEU, LUG/SEU, or LUG/SLK complex is formed, leading to a much enhanced defect in the repression of AG and explaining the similar mutant floral phenotype between seu-1; lug-3 and luh-1/+; lug-3/lug-3 (Fig. 3; Franks et al., 2002).
CONCLUSION
Plant Gro/Tup1-type corepressors constitute an important class of regulatory molecules with roles in embryo shoot-root axis determination, stem cell pool maintenance, and floral homeotic gene regulation. Among the 13 Gro/Tup1-type corepressors in Arabidopsis, LUG and LUH are most similar to each other. We show that LUH and LUG exhibit both redundant and divergent functions in embryonic development, floral homeotic gene regulation, and plant biotic and abiotic stress responses. Gene duplication and functional diversification are important for species adaptation. Our study provides important insights into the complexity in the relationship between two highly homologous paralogous genes.
MATERIALS AND METHODS
Plant Growth and Mutant Identification
Arabidopsis (Arabidopsis thaliana) plants were grown on Sun Shine professional soil in controlled growth chambers at 20°C and 55% humidity under long-day (16 h of light) conditions. Seeds used in germination and root elongation assays were sterilized with 70% ethanol and 0.6% hypochlorite (bleach), plated on MS basal medium plates, incubated in the dark for 3 d at 4°C, and then grown for 5 d at 20°C under long days before transferring to another MS plate for root analyses.
lug-3, lug-16, and seu-1 were generated in the Landsberg erecta background and were described previously (Conner and Liu, 2000; Franks et al., 2002). luh-1 (luh_172H3; Arabidopsis Biological Resource Center stock no. CS91893) and luh-2 (luh_147A6; Arabidopsis Biological Resource Center stock no. CS91036) were generated by the Arabidopsis TILLING Project by ethyl methanesulfonate mutagenesis in the Columbia erecta-105 background (McCallum et al., 2000). luh-3 (SALK_107245) was generated by T-DNA insertion (Alonso et al., 2003).
Double Mutant Construction and Genotyping
To generate double mutants, luh-1 pollen was used to pollinate lug-16, lug-3, or seu-1. F2 plants were analyzed by PCR-based genotyping methods. The luh-1 dCAPS marker uses primers 5′-GCACCTGGAGGGTTTCCATTTGAGTG-3′ and 5′-CGCTTTACCTTGTTGTGCCTAAAATT-3′ in 35 cycles of PCR at 94°C for 30 s, 50°C for 30 s, and 72°C for 30 s. PCR products (6 μL) were digested with BstXI at 55°C and analyzed on 2.5% agarose gels. luh-1 PCR products were resistant to BstXI. seu-1 dCAPS primers (Franks et al., 2002) amplified genomic DNA at 94°C for 30 s, 50°C for 30 s, and 72°C for 30 s for 35 cycles. The PCR products were digested with RsaI. seu-1 PCR products are resistant to the RsaI digestion.
Since the dCAPS assay was not always reliable, an alternative fluorescence-based SNP assay (Amplifluor SNP Genotyping) was adopted for luh-1 and lug-16. An individual leaf or single embryo was pressed onto FTA MicroCard (Whatman). A 0.2-mm-diameter disc was punctured out of the FTA MicroCard and served as a template for PCR following the manufacturer's protocol. Primers for lug-16 (5′-GTTAAGTAGGAAGTTAAGCCC-3′ and 5′-GAGAACACCATTCAACTGTAC-3′) and luh-1 (5′-GTTTGGGCTTTTATTCAGGTT-3′ and 5′-GCACTAGCATTAGACTGCCC-3′) were first used in a conventional PCR. Then, 25 ng of diluted PCR products served as templates for the Amplifluor SNP Genotyping system (assay development kit from Chemicon International, a subsidiary of Serologicals) using Platinum Taq DNA polymerase (Invitrogen). The Amplifluor AssayArchitect program (Chemicon International) was used for primer design; the allele-specific primer has a tail sequence complementary to either fluorescent FAM- or JOE-labeled primer. For the lug-16 locus (tail sequence underlined), the wild-type-specific primer is 5′-GAAGGTGACCAAGTTCATGCTTCACCAGGTGCGTCAATAGCT-3′ and the lug-16-specific primer is 5′-GAAGGTCGGAGTCAACGGATTTCCACCAGGTGCGTCAATAGT-3′. Both allele-specific primers pair with the same reverse primer, 5′-CTGCAGTTGCTCTGTTTCCTAA-3′. All three primers were used in the same PCR genotyping procedure. For the luh-1 locus (tails underlined), the wild-type-specific primer is 5′-GAAGGTCGGAGTCAACGGATTTGTCCCAAAACACAGACCAC-3′ and the luh-1-specific primer is 5′-GAAGGTGACCAAGTTCATGCTAATGTCCCAAAACACAGACCAT-3′. The reverse primer is 5′-GCACCTGGAGGGTTTCTTTTT-3′. PCR was run on a conventional PCR machine programmed as follows: (1) 96°C for 4 min; (2) 96°C for 12 s; (3) 57°C for 5 s; (4) 72°C for 10 s; (5) repeat steps 2 to 4 for 15 cycles; (6) 96°C for 12 s; (7) 55°C for 20 s; (8) 72°C for 40 s; (9) repeat steps 6 to 8 for 19 cycles; (10) 72°C for 3 s; and (11) hold at 20°C. Allelic discrimination was determined by reading FAM and JOE fluorophore signals using the Bio-Rad iQ5 PCR machine.
Microscopy and Photography
Floral, silique, and seedling photographs were captured with a Nikon SMZ1000 microscope equipped with a Nikon digital camera. The green and white seeds were dissected from siliques and fixed in Hoyer's solution for 15 min (Liu and Meinke, 1998) and then examined and photographed with a Nikon ECL1PSE E600W microscope with Nomarski optics and equipped with a DXM1200 digital still camera. Images were processed with Adobe Photoshop version 7.0.
Molecular Analyses of LUH
LUH (At2g32700) has 17 exons. 5′ RACE was performed to verify the LUH transcript using the Generacer kit (version F; Invitrogen) and total RNA from Arabidopsis flowers. The 5′ nested primer 5′-GGACACTGACATGGACTGAAGGAGTA-3′ was used, and the RACE products were cloned in pCRII TOPO (Invitrogen) and sequenced. LUH full-length cDNA (RAFL09-12-E08 [R12254]) was obtained from RIKEN Genomic Sciences Center and sequenced for confirmation.
To generate 35S∷LUH, the full-length LUH cDNA from RIKEN was amplified by PCR with primers 35SLUH-F (5′-ATTACCCGGGGATGGCTCAGAGTAATTGGGAAG-3′) and 35SLUH-R (5′-TCCCCCGGGCTACTTCCAAATCTTTACGGA-3′) containing engineered XmaI sites with the high-fidelity Taq polymerase (Roche). The PCR product was cloned in the pBI121 vector at the XmaI site and verified by sequencing. Plasmids were transformed into Agrobacterium tumefaciens GV3101 through electroporation. luh-1 and lug-16 plants were transformed by the floral dip method (Clough, 2005). Kanamycin-resistant T1 seedlings were identified on MS plates containing 50 μm kanamycin and transferred to soil.
For RT-PCR, RT was performed with oligo(dT) and SuperScript II reverse transcriptase enzyme (Invitrogen). All RT-PCR procedures were carried out for 25 cycles and were repeated at least twice. Primers used were LUH (5′-TGGCTCAGAGTAATTGGGAAG-3′ and 5′-CCAGGCTTTGATTGCAGA-3′) and ACT2 (5′-GTTGGGATGAACCAGAAGGA-3′ and 5′-CTTACAATTTCCCGCTCTTC-3′). The primers were designed to span introns to avoid amplification from contaminated genomic DNA. ACT2 was used as a loading control. The RT-PCR procedures were quantified using ImageQuant 1.1 (National Institutes of Health) software, based on the intensity of the ethidium bromide staining.
Yeast Two-Hybrid Assay
Full-length LUH cDNA was amplified by PCR using high-fidelity Taq polymerases (Roche) and the RIKEN (RAFL09) cDNA as a template with engineered primers. PCR products were cloned into the pCRII TOPO vector (Invitrogen). The clone was sequenced to verify amplification accuracy. The primers LUH-BD-f and LUH-AD-f, which are the same (5′-ATTACCCGGGGATGGCTCAGAGTAATTGGGAAG-3′), LUH-BD-r (5′-ACGCGTCGACATCTACTTCCAAATCTTTACGGA-3′), and LUH-AD-r (5′-ATTCTCGAGCTACTTCCAAATCTTTACGGA-3′) contain SalI and XmaI sites for the BD fusion and XhoI and XmaI sites for the AD fusion. The LUH fragments were excised from corresponding pCRII TOPO vectors and inserted into pGBKT7 and pGADT7 (Clontech), respectively, at corresponding enzyme sites. The yeast host (PJ69-4A), genotype MATa trp1-901 leu2-3,112 ura3-52 his3-200 gal4delta gal80delta GAL2-ADE2 LYS2∷GAL1-HIS3 met2∷GAL7-lacZ (James et al., 1996), was used for transformation as described previously (Sridhar et al., 2004).
For the X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) overlay assay, 0.125 g of agarose was dissolved in 25 mL of sterilized Z-buffer (60 mm Na2HPO4·2H2O, 40 mm NaH2PO4·H2O, 10 mm KCl, and 1 mm MgSO4·7H2O, pH 7.0) by heating in a microwave oven. After cooling to 50°C, 0.5 mL of 10% SDS and X-Gal dissolved in N,N-dimethylformamide (final concentration, 2 mg/mL) were added. The molten agarose solution was poured over one to two plates containing yeast colonies. After 30 min of incubation at 37°C, the plates were photographed.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Sequence of LUH cDNA and deduced amino acid sequence.
Supplemental Table S1. Percentage of abnormal seeds (white seeds) in the wild type and mutants.
Supplementary Material
Acknowledgments
We thank Parsa Hosseini for help with analyzing LUG and LUH expression data, the Arabidopsis Biological Resource Center and the Arabidopsis TILLING Project for the LUH cDNA clones and luh mutant seeds, and Robert Franks, Paja Sijacic, and Courtney Hollender for critical comments of the manuscript. We are grateful to Dr. Heven Sze for her support in difficult times and Dr. William Higgins for his advice and mentoring (to M.B.).
This work was supported by the National Science Foundation (grant no. IOB0616096 to Z.L.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Zhongchi Liu (zliu@umd.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Chen C, Wang S, Huang H (2000) LEUNIG has multiple functions in gynoecium development in Arabidopsis. Genesis 26 42–54 [DOI] [PubMed] [Google Scholar]
- Clough SJ (2005) Floral dip: Agrobacterium-mediated germ line transformation. Methods Mol Biol 286 91–102 [DOI] [PubMed] [Google Scholar]
- Cnops G, Jover-Gil S, Peters JL, Neyt P, De Block S, Robles P, Ponce MR, Gerats T, Micol JL, Van Lijsebettens M (2004) The rotunda2 mutants identify a role for the LEUNIG gene in vegetative leaf morphogenesis. J Exp Bot 55 1529–1539 [DOI] [PubMed] [Google Scholar]
- Conner J, Liu Z (2000) LEUNIG, a putative transcriptional corepressor that regulates AGAMOUS expression during flower development. Proc Natl Acad Sci USA 97 12902–12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks RG, Liu Z, Fischer RL (2006) SEUSS and LEUNIG regulate cell proliferation, vascular development and organ polarity in Arabidopsis petals. Planta 224 801–811 [DOI] [PubMed] [Google Scholar]
- Franks RG, Wang C, Levin JZ, Liu Z (2002) SEUSS, a member of a novel family of plant regulatory proteins, represses floral homeotic gene expression with LEUNIG. Development 129 253–263 [DOI] [PubMed] [Google Scholar]
- Gonzalez D, Bowen AJ, Carroll TS, Conlan RS (2007) The transcription corepressor LEUNIG interacts with the histone deacetylase HDA19 and mediator components MED14 (SWP) and CDK8 (HEN3) to repress transcription. Mol Cell Biol 27 5306–5315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Li WH, Chen ZJ (2007) External factors accelerate expression divergence between duplicate genes. Trends Genet 23 162–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer M, Stern Y, Cook H, Clerici E, Maulbetsch C, Laux T, Davies B (2006) Analysis of the transcription factor WUSCHEL and its functional homologue in Antirrhinum reveals a potential mechanism for their roles in meristem maintenance. Plant Cell 18 560–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D'Angelo C, Bornberg-Bauer E, Kudla J, Harter K (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50 347–363 [DOI] [PubMed] [Google Scholar]
- Liu CM, Meinke DW (1998) The titan mutants of Arabidopsis are disrupted in mitosis and cell cycle control during seed development. Plant J 16 21–31 [DOI] [PubMed] [Google Scholar]
- Liu L, Karmarkar V (2008) Gro-Tup1 family co-repressors in plant development. Trends Plant Sci 13 137–144 [DOI] [PubMed] [Google Scholar]
- Liu Z, Franks RG, Klink VP (2000) Regulation of gynoecium marginal tissue formation by LEUNIG and AINTEGUMENTA. Plant Cell 12 1879–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Meyerowitz EM (1995) LEUNIG regulates AGAMOUS expression in Arabidopsis flowers. Development 121 975–991 [DOI] [PubMed] [Google Scholar]
- Long JA, Ohno C, Smith ZR, Meyerowitz EM (2006) TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312 1520–1523 [DOI] [PubMed] [Google Scholar]
- Lynch M, Force A (2000) The probability of duplicate gene preservation by subfunctionalization. Genetics 154 459–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, O'Hely M, Walsh B, Force A (2001) The probability of preservation of a newly arisen gene duplicate. Genetics 159 1789–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum CM, Comai L, Greene EA, Henikoff S (2000) Targeted screening for induced mutations. Nat Biotechnol 18 455–457 [DOI] [PubMed] [Google Scholar]
- Navarro C, Efremova N, Golz JF, Rubiera R, Kuckenberg M, Castillo R, Tietz O, Saedler H, Schwarz-Sommer Z (2004) Molecular and genetic interactions between STYLOSA and GRAMINIFOLIA in the control of Antirrhinum vegetative and reproductive development. Development 131 3649–3659 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37 501–506 [DOI] [PubMed] [Google Scholar]
- Sridhar VV, Surendrarao A, Gonzalez D, Conlan RS, Liu Z (2004) Transcriptional repression of target genes by LEUNIG and SEUSS, two interacting regulatory proteins for Arabidopsis flower development. Proc Natl Acad Sci USA 101 11494–11499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar VV, Surendrarao A, Liu Z (2006) APETALA1 and SEPALLATA3 interact with SEUSS to mediate transcription repression during flower development. Development 133 3159–3166 [DOI] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA (2008) TOPLESS mediates IAA12/BODENLOS transcriptional repression during embryogenesis in Arabidopsis. Science 319 1384–1386 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.