Abstract
Manipulating gene expression is critical to exploring gene function and a useful tool for altering commercial traits. Techniques such as hairpin-based RNA interference, virus-induced gene silencing, and artificial microRNAs take advantage of endogenous posttranscriptional gene silencing pathways to block translation of designated transcripts. Here we present a novel gene silencing method utilizing artificial trans-acting small interfering RNAs in Arabidopsis (Arabidopsis thaliana). Replacing the endogenous small interfering RNAs encoded in the TAS1c gene with sequences from the FAD2 gene silenced FAD2 activity to levels comparable to the fad2-1 null allele in nearly all transgenic events. Interestingly, exchanging the endogenous miR173 target sequence in TAS1c with an miR167 target sequence led to variable, inefficient silencing of FAD2, suggesting a specific requirement for the miR173 trigger for production of small interfering RNAs from the TAS1c locus.
Loss-of-function mutations are the most basic tool of genetic analysis: The biological activity of a gene is inferred from the mutant phenotype. In traditional genetic analysis, a plant with a phenotype of interest is identified from a mutagenized population, and the gene responsible for the phenotype is identified by genetic mapping or by means of a T-DNA or transposon tag (Lukowitz et al., 2000). A complementary approach to investigating gene function, often called functional genomics, is to identify genes of interest based on their sequence and/or expression pattern, and then to use a transgenic approach to inactivate the gene to obtain a mutant phenotype. One strategy for functional genomics is to produce massive sequenced-indexed libraries of plants with genes randomly inactivated by T-DNA insertion (Sessions et al., 2002; Alonso et al., 2003). Although this approach has been of great value to the scientific community, T-DNA insertion sites are biased toward 5′ and 3′ untranslated region (UTR) sequences, and T-DNA insertions in a large proportion of genes, especially small genes, have never been recovered (Sessions et al., 2002; Alonso et al., 2003). In addition, indexed T-DNA insertion libraries are not available for many plant species used for basic research or for most agronomically important crop species. Thus, strategies to efficiently silence specific genes are essential.
Many different strategies have been used for gene silencing (for review, see Ossowski et al., 2008). Initial efforts to overexpress genes resulted in cosuppression (Napoli et al., 1990), and overexpression of antisense transcripts was an early method to silence genes, albeit at a low frequency (Baulcombe, 1996). Later studies showed that expression of an inverted repeat of a gene that formed a hairpinRNA was effective at silencing endogenous genes (Waterhouse et al., 1998; Chuang and Meyerowitz, 2000). Subsequently, this method was refined through the use of an intron-spliced hairpin construct that gave more consistent silencing (Smith et al., 2000; Stoutjesdijk et al., 2002). A large-scale evaluation of inverted repeat-based gene silencing found that the success of this method varies greatly from gene to gene (McGinnis et al., 2005). More recently, artificial microRNAs (amiRNAs) have been designed to target endogenous genes, and have also proven to be effective (Alvarez et al., 2006; Schwab et al., 2006).
All of these strategies rely on the genetic machinery of an endogenous phenomenon called posttranscriptional gene silencing (PTGS). PTGS was first recognized as a plant immune response to viral infection (Hamilton and Baulcombe, 1999). In this process, viral RNAs are substrates for an RNA-dependent RNA polymerase that synthesizes a complementary RNA strand. This double-stranded RNA is cut into 21- to 24-bp small interfering RNAs (siRNAs) by an endonuclease called Dicer (Bernstein et al., 2001). One strand of the siRNA is loaded into the RNA-induced silencing complex (RISC). Guided by the associated siRNA, RISC recognizes and destroys complementary target viral RNA via the “slicer” Argonaute protein, the primary enzyme of the RISC complex (Song et al., 2004; Baumberger and Baulcombe, 2005). Another mechanism for gene silencing in plants is mediated by 21- to 24-bp microRNAs (miRNAs). miRNA genes are transcribed to form pre-miRNAs, which fold back on themselves to form stem-loop structures. The functional miRNA is processed from the pre-miRNA by the enzyme Dicer, which cuts the stem of the pre-miRNA to produce a double-stranded miRNA, only one strand of which is active (Park et al., 2002; Reinhart et al., 2002). In plants, miRNAs target endogenous mRNAs, usually in their coding regions, but sometimes in the 5′ or the 3′ UTR (Bartel, 2004; Wu and Poethig, 2006). Typically, an miRNA that is bound to its target mRNA marks it for cleavage by the RISC complex, though there are several cases in which miRNAs have been shown to cause translational inhibition in plants, as they frequently do in animals (Aukerman and Sakai, 2003; Chen, 2004; Arteaga-Vázquez et al., 2006; Gandikota et al., 2007).
Generation of a third class of PTGS-associated small RNAs, called trans-acting siRNAs (ta-siRNAs), involves both siRNAs and miRNAs. ta-siRNAs differ from conventional siRNAs in that they target genes in trans (Peragine et al., 2004; Vazquez et al., 2004). ta-siRNA genes produce nonprotein coding transcripts that are themselves targeted by an miRNA. One of the miRNA cleavage products is converted to double-stranded RNA by RDR6, at which point it can be cleaved into siRNAs by Dicer-like protein DCL4. Upon incorporation into RISC, some of these siRNAs have the ability to target other genes in trans (Allen et al., 2005; Yoshikawa et al., 2005). Thus, all three pathways converge with incorporation of a small RNA into the RISC complex, which then enables cleavage of the target RNA.
In this study, we describe our efforts to extend previous silencing strategies by engineering the TAS1c (trans-acting siRNA1c) locus to silence the FAD2 gene in Arabidopsis (Arabidopsis thaliana). Processing of the TAS1c RNA is initiated by miR173-mediated cleavage, revealing six phased siRNAs downstream of the miR173 target site (Allen et al., 2005). We engineered the TAS1c locus to silence the FAD2 gene by replacing a single native siRNA with an siRNA targeting FAD2 (siFAD2), by replacing five native siRNAs with siFAD2, or by substituting these five native siRNAs with a 210-bp fragment of the FAD2 gene. All three of these strategies resulted in silencing of the FAD2 gene activity similar to levels seen in the fad2-1 null mutant. Furthermore, almost all transgenic events showed a high degree of silencing, and the silencing effect was inherited over four generations. Interestingly, replacement of the miR173 trigger in TAS1c with miR167 did not give effective silencing, suggesting that miR173 may play a specific role in the biology of TAS1c.
RESULTS
Silencing with Artificial trans-Acting siRNAs
To determine if ta-siRNA loci could be engineered to silence genes of interest, we modified the TAS1c sequence to produce siRNAs targeting the FAD2 gene. FAD2 encodes an endoplasmic reticulum-localized Δ12 desaturase required for converting the monounsaturated oleic acid (18:1) to the polyunsaturated linoleic acid (18:2), itself the precursor to linoleic acid (18:3; Okuley et al., 1994). FAD2 is a classic reporter for silencing assays because it is a single-copy, nonessential gene in Arabidopsis with an easily assayed, quantifiable phenotype (Miquel and Browse, 1992). Using FAD2 as a silencing reporter allowed direct comparison between current and previous methods as well (Stoutjesdijk et al., 2002).
We engineered a truncated TAS1c cDNA to silence FAD2 by replacing the sequence encoding the five native TAS1c siRNAs with a sequence encoding five identical siRNAs targeting base pairs 98 to 118 of the FAD2 coding sequence (CDS; 5XsiFAD2; Fig. 1A). As a control, the miR173 binding site of 5XsiFAD2 was mutated to disrupt base pairing at base pairs 4, 10, and 11 of miR173, to produce mut-5XsiFAD2. Loss of the functional miR173 binding site in mut-5XsiFAD2 should prevent production of phased FAD2 siRNAs.
Figure 1.
Silencing of FAD2 with 5XsiFAD2. A, Diagrammatic representation of the 5XsiFAD2 construct. This construct was engineered by replacing the sequence corresponding to five native siRNAs from the TAS1c cDNA at positions 3′D2+ to siR619 with five copies of a 21-bp DNA sequence (shown in gray) complementary to base pairs 98 to 118 of the FAD2 CDS. The name of the native siRNA replaced at each position is shown in parentheses. The miR173 target site is shown in black, and the 3′D1+ siRNA is shown in white. In planta expression of the 5XsiFAD2 construct is driven by the CaMV35S promoter. B, Seed fatty acid analysis of T2 and T5 families harboring the 5XsiFAD2 construct. Fatty acid analysis is expressed as ODP. ODP values (with sd) represent the average of at least five individual seeds. Wild type and mut-5XsiFAD2, a version of the 5XsiFAD2 construct with a mutated miR173 binding site, are shown as controls. C, Leaf fatty acid analysis of wild type, three T2 transgenic lines harboring the 5XsiFAD2 construct (lines 6, 12, and 14), and three T2 transgenic lines (g, h, and i) harboring mut-5XsiFAD2. D, Northern analysis of levels of FAD2 transcripts and siFAD2 siRNAs. RNA samples were extracted from the transgenic plants subjected to fatty acid analysis (shown in C). ACTIN2 is shown as a loading control for FAD2 transcript levels; a picture of the small RNA gel used to separate small RNAs for blotting is shown as a loading control for the siFAD2 hybridization.
FAD2 expression is directly proportional to the amount of oleic to linoleic desaturase activity in a tissue (Okuley et al., 1994). Accordingly, silencing was quantified by expressing the degree of unsaturation of 18:1 to 18:2 and 18:3 fatty acids as oleic desaturation proportion (ODP; see “Materials and Methods”). Wild-type plants had an ODP of approximately 0.7 in seed, meaning that 70% of 18:1 fatty acids in seed are converted to 18:2 and 18:3 fatty acids (Fig. 1B). All 12 transgenic events we obtained for the 5XsiFAD2 construct showed strong silencing of the FAD2 gene. Ten of the 12 families had average ODP values between 0.1 and 0.2, while the remaining two families had ODP values of approximately 0.25 and 0.3 (Fig. 1B). T2 seed from each family showed the same degree of silencing whether they were homozygous or hemizygous for the 5XsiFAD2 transgene. The fad2-1 null allele had an ODP value of about 0.1 (Okuley et al., 1994; Stoutjesdijk et al., 2002), and thus the majority of our transgenic lines showed complete or almost complete silencing of the FAD2 gene, with the remaining two lines showing strong silencing. Plants harboring the mut-5XsiFAD2 construct had ODP values indistinguishable from the wild type (Fig. 1B), indicating that FAD2 silencing initiated by 5XsiFAD2 occurs through an miR173-triggered mechanism. We propagated three lines for several generations more and found that silencing was maintained in the T5 generation (Fig. 1B).
To directly assay FAD2 gene expression in these lines, we determined the levels of FAD2 mRNA and siFAD2 by northern analysis of RNA from leaf tissue (Fig. 1D), and determined leaf fatty acid levels in the plants that we used for northern analysis (Fig. 1C). Using the same transgenic lines subjected to fatty acid analysis, we detected markedly decreased FAD2 mRNA levels in 5XsiFAD2 lines compared to wild-type plants. This decrease is consistent with the large decrease in ODP seen in these lines. Transgenic lines harboring the mut-5XsiFAD2 transgene showed no decrease in FAD2 mRNA levels. In small RNA northerns, siFAD2 siRNAs were observed in 5XsiFAD2 plants, yet they were absent from wild-type and mut-5XsiFAD2 plants (Fig. 1D). In addition, 5′ RACE revealed cleavage at the miR173 site of 5XsiFAD2 (data not shown).
A Single Copy of siFAD2 Is Sufficient for Silencing
Next we sought to determine if a single copy of siFAD2 could effectively silence FAD2. We replaced the sequence encoding the native siRNA at the 3′D2+ position within the full-length TAS1c cDNA with a sequence encoding siFAD2, producing the construct 1XsiFAD2 (Fig. 2A). As a control we engineered mut-1XsiFAD2, identical to 1XsiFAD2 except that the miR173 binding site was mutated to prevent miR173-induced cleavage.
Figure 2.
Silencing of FAD2 with 1XsiFAD2. A, Diagrammatic representation of the 1XsiFAD2 construct. Sequence coding for the 3′D2+ siRNA was replaced with base pairs 98 to 118 of the FAD2 CDS coding for the 21-bp siFAD2 sequence (shown in gray). The miR173 target site is shown in black, and sequences encoding other siRNAs are shown in white. In planta expression of the 1XsiFAD2 construct is driven by the CaMV35S promoter. B, Fatty acid analysis of wild type; T1 and T2 transgenic seed harboring the 1XsiFAD2 construct; and T2 transgenic control seed harboring the mut-1XsiFAD2 construct, a version of the 1XsiFAD2 construct in which the miR173 site has been mutated. Data for T1 seed are the average of 10 individual seeds, and data for T2 seed are the average of four individual seeds each from independent families. Fatty acid profiles are expressed as ODP. The sd is shown at the top of each bar.
When transformed into plants, 1XsiFAD2 decreased fatty acid ODP from about 0.75 in wild-type seed to an average of 0.18 in 10 independent T1 lines (Fig. 2B). All 10 independent transgenic lines showed approximately the same degree of FAD2 sense silencing: The sd in ODP score was only about 0.03. The silencing was heritable, as the majority of the T2 families examined showed ODP scores similar to those of T1 lines. Thus, engineering TAS1c with a single siFAD2 decreased ODP nearly as much as what was achieved with five copies of siFAD2. This level of silencing persisted in almost all lines in the T2 generation (Fig. 2B).
Silencing of the FAD2 Gene with the fragFAD2 Construct
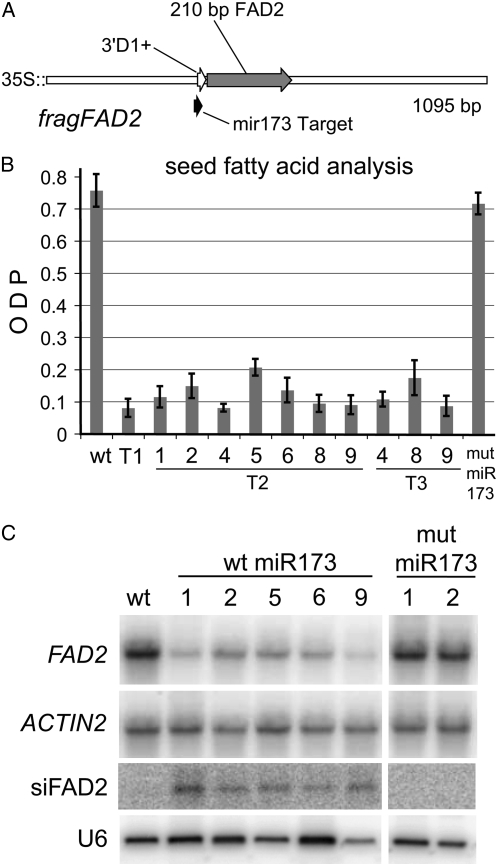
The experiments described above demonstrated that the TAS1c locus can be successfully modified to silence the FAD2 gene by substituting DNA encoding a deliberately designed FAD2 siRNA for a native TAS1c siRNA. However, it may not always be straightforward to design siRNAs targeting a gene of interest, as the characteristics of successful siRNAs are not completely known. For this reason, we determined if inserting a larger fragment of the FAD2 gene into TAS1c would effectively produce siRNAs. A 210-bp sense fragment of the FAD2 gene was inserted into the full-length TAS1c cDNA in place of the segment encoding the phased siRNAs 3′D2+ to 3′D6+ to produce the fragFAD2 construct (Fig. 3A). As a control for miR173-dependent production of phased siRNAs, the miR173 target site was mutated to produce construct mut-fragFAD2.
Figure 3.
Silencing of FAD2 with fragFAD2. A, Diagrammatic representation of the fragFAD2 construct. This construct was engineered by replacing the DNA sequence coding for the native siRNAs from the 3′D2+ to the 3′D6+ positions of the TAS1c cDNA with a 210-bp fragment corresponding to base pairs 1 to 210 of the FAD2 CDS. The miR173 target site is shown in black, and the siRNA at the 3′D1+ position is shown in white. In planta expression of the fragFAD2 construct is driven by the CaMV35S promoter. B, Fatty acid analysis of wild type; T1, T2, and T3 transgenic seed harboring the fragFAD2 construct; and T2 seed harboring the mut-fragFAD2 construct, a version of the fragFAD2 construct with the miR173 binding site mutated. Data for T1 seed are the average of 10 seeds, and data for T2 and T3 seed are the average of four seeds each from independent families. Fatty acid profiles are expressed as ODP. The sd is shown at the top of each bar. C, Northern analysis of levels of FAD2 transcripts and siFAD2 siRNAs of T2 plants. RNA samples were extracted from T2 transgenic plants grown from the same families subjected to fatty acid analysis (shown in B). ACTIN2 is shown as a loading control for FAD2 transcript levels; U6 is shown as a loading control for the siFAD2 hybridization.
As shown in Figure 3B, T1 transgenic plants containing the fragFAD2 construct had an average ODP of less than 0.1, similar to levels seen in the 5XsiFAD2 transgenic lines and in the fad2-1 mutant (Miquel and Browse, 1992). This level of silencing was maintained in six out of seven T2 transgenic lines. We checked ODP levels for three lines in the T3 generation and found that silencing was maintained. Transgenic lines containing the mut-fragFAD2 construct had ODP levels similar to the wild type, indicating that miR173-induced cleavage of fragFAD2 was required for siRNA production.
To verify gene silencing, levels of FAD2 mRNA and FAD2 siRNAs were determined in fragFAD2 and mut-fragFAD2 transgenic plants (Fig. 3C). As expected, FAD2 mRNA levels were markedly decreased in fragFAD2 transgenic plants and were similar to the wild type in mut-fragFAD2 transgenic plants. In addition, FAD2 siRNAs were detected in fragFAD2 plants, but not in wild-type or mut-fragFAD2 transgenic plants. Thus, the substitution of native TAS1c siRNAs with a large fragment of the FAD2 gene leads to efficient and stable silencing of this target gene.
An Alternate miRNA Trigger Leads to Inefficient Silencing
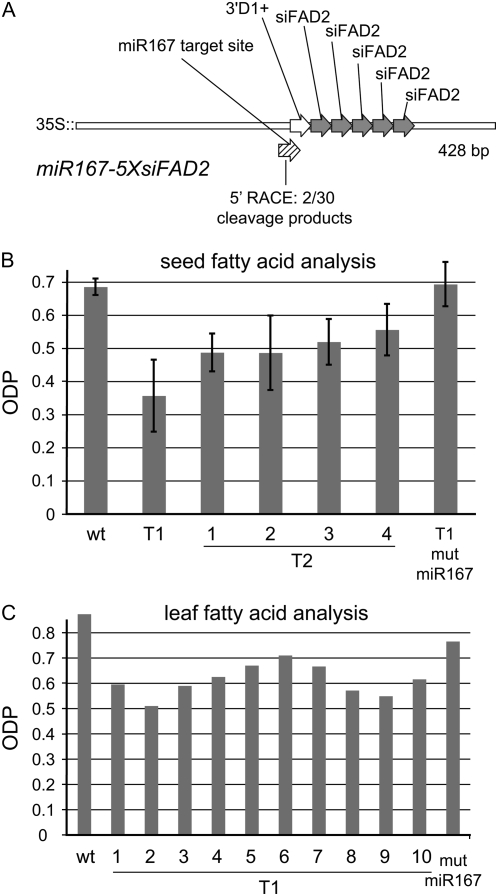
Whether each TAS locus requires its native miRNA trigger for proper function remains an open question. To address this, we substituted the endogenous miR173 target site in the 5XsiFAD2 construct with the target site for miR167, a highly expressed miRNA in leaves (data not shown) to produce miR167-5XsiFAD2 (Fig. 4A). As a control to show that any siRNAs produced from this construct required miR167 for cleavage, we also made mut-miR167-5XsiFAD2, a construct with a mutated miR167 binding site.
Figure 4.
Inefficient silencing of FAD2 by miR167-5XsiFAD2. A, Diagrammatic representation of the miR167-5XsiFAD2 construct. This construct was engineered by replacing the miR173 target site in the 5XsiFAD2 construct with an miR167 target site. The miR167 target is shown with hatched bars, the 3′D1+ siRNA in white, and the siFAD2 siRNAs in gray. The number of cleavage events detected by 5′ RACE at the miR167 target site in miR167-5XsiFAD2 transgenic plants is shown. B, Fatty acid analysis of wild type; T1 and T2 transgenic seed harboring the miR167-5XsiFAD2 construct; and T2 seed harboring the mut-miR167-5XsiFAD2 construct, a version of the miR167-5XsiFAD2 construct in which the miR167 target site has been mutated. Fatty acid profiles are expressed as ODP. Value for wild type is the average of 10 seeds; values for T1 and T2 are the average of four seeds each from independent families. C, Fatty acid analysis of a single leaf of wild-type and T1 plants harboring the miR167-5XsiFAD2 construct and the mut-miR167-5XsiFAD2 construct.
When transformed into plants, miR167-5XsiFAD2 T1 seed had approximately half the ODP of the wild type, although there was considerable variation in the degree of silencing among T1 individuals, as shown by the large sd for the average ODP value of T1 seed (Fig. 4B). ODP levels climbed to approximately 75% of wild type in most T2 families. No difference was observed in ODP levels between mut-miR167-5XsiFAD2 control plants and the wild type. Such moderate and variable silencing was also noted in leaf tissue (Fig. 4C).
Modified 5′ RACE showed a reduced frequency of cleavage at the miR167 site of miR167-5XsiFAD2 mRNA in leaf tissue. Out of 30 sequenced cleavage products, only two were cleaved at the miR167 target site (Fig. 4A). The remaining 28 5′ RACE products showed cleavage throughout the miR167-5XsiFAD2 mRNA. For comparison, we performed 5′ RACE on an endogenous target of miR167, ARF8. We detected robust miR167 cleavage events in ARF8 (data not shown), results similar to those published previously (Yang et al., 2006). The inefficiency of miR167 in triggering ta-siRNA production suggests that miR173 is specially adapted to participating in ta-siRNA biogenesis.
Table I.
Oligonucleotide primers used in this study
*, These oligonucleotides have the attB primer tagged pair to clone the fragment of interest in the pDONR/Zeo vector (Invitrogen) and were used in the three constructs to amplify the 5′ end and the 3′ end.
| Construct Name or Purpose | Primer Name | Sequence |
|---|---|---|
| 5XsiFAD2 | FAD2TAS 5′ sense | GATCACAAGTTTGTACAAAAAAGCAGGCTCATAGAAAGGTACTTTCGTTTACTTCTTTTGAGTATCGAGTAGAGCGTCGTCTATAGTTAGTTTGAGATTGCGTTTGTCAGAAGTTAGGTTCAATGTCCCGGTCCAATTTTCACCAGCCATGTGTCAGTTTCGTTCCTTCCCGTCCTCTTCTTTGATTTCGTTGGGTTACGGATGTTTTCGAGATGAAACAGCATTGTTTTGTTG |
| FAD2TAS 5′ antisense | ATCACAACAAAACAATGCTGTTTCATCTCGAAAACATCCGTAACCCAACGAAATCAAAGAAGAGGACGGGAAGGAACGAAACTGACACATGGCTGGTGAAAATTGGACCGGGACATTGAACCTAACTTCTGACAAACGCAATCTCAAACTAACTATAGACGACGCTCTACTCGATACTCAAAAGAAGTAAACGAAAGTACCTTTCTATGAGCCTGCTTTTTTGTACAAACTTGT | |
| FAD2TAS 3′ sense | TGATTTTTCTCTACAAGCGAATAGACCATTTATTGCTTTCTTCAGATCTCCCATTGCTTTCTTCAGATCTCCCATTGCTTTCTTCAGATCTCCCATTGCTTTCTTCAGATCTCCCATTGCTTTCTTCAGATCTCCCAAAAACAATGAATATTGTTTTGAATGTGTTCAAGTAAATGAGATTTTCAAGTCGTCTAAAGAACAGTTGCTAATACAGTTACTTAACCCAGCTTTCTTGTACAAAGTGGT | |
| FAD2TAS 3′ antisense | AATTACCACTTTGTACAAGAAAGCTGGGTTAAGTAACTGTATTAGCAACTGTTCTTTAGACGACTTGAAAATCTCATTTACTTGAACACATTCAAAACAATATTCATTGTTTTTGGGAGATCTGAAGAAAGCAATGGGAGATCTGAAGAAAGCAATGGGAGATCTGAAGAAAGCAATGGGAGATCTGAAGAAAGCAATGGGAGATCTGAAGAAAGCAATAAATGGTCTATTCGCTTGTAGAGAAAA | |
| FAD2TASmut 3′ sense | TGATTTTTCTAAACAAGAGAATAGACCATTTATTGCTTTCTTCAGATCTCCCATTGCTTTCTTCAGATCTCCCATTGCTTTCTTCAGATCTCCCATTGCTTTCTTCAGATCTCCCATTGCTTTCTTCAGATCTCCCAAAAACAATGAATATTGTTTTGAATGTGTTCAAGTAAATGAGATTTTCAAGTCGTCTAAAGAACAGTTGCTAATACAGTTACTTAACCCAGCTTTCTTGTACAAAGTGGT | |
| FAD2TASmut 3′ antisense | AATTACCACTTTGTACAAGAAAGCTGGGTTAAGTAACTGTATTAGCAACTGTTCTTTAGACGACTTGAAAATCTCATTTACTTGAACACATTCAAAACAATATTCATTGTTTTTGGGAGATCTGAAGAAAGCAATGGGAGATCTGAAGAAAGCAATGGGAGATCTGAAGAAAGCAATGGGAGATCTGAAGAAAGCAATGGGAGATCTGAAGAAAGCAATAAATGGTCTATTCTCTTGTTTAGAAAA | |
| 1XsiFAD2 | TAS1c/FAD2-attB1* | GGGGACAAGTTTGTACAAAAAAGCAGGCTAAACCTAAACCTAAACGGCTAAGCCCG |
| TAS1c/FAD2-2R | GCATATCCTGGAATATGTAGGATCATCTTCTTGATACAGCGATATGTTGAACTTAGAATACGCTATGTTGGACTTAGAATATTGCTTTCTTCAGATCTCCCAAATGGTCTATTCGCTTG | |
| TAS1c/FAD2-3F | CAAGCGAATAGACCATTTGGGAGATCTGAAGAAAGCAATATTCTAAGTCCAACATAGCGTATTCTAAGTTCAACATATCGCTGTATCAAGAAGATGATCCTACATATTCCAGGATATGC | |
| TAS1c/FAD2-attB-4* | GGGGACCACTTTGTACAAGAAAGCTGGGTATTTCACTTTACGATGTGGTGTTC | |
| fragFAD2 | TAS1c/FAD2-frag-2R | GCATTCTTCCACCTGCACCCATTAAATGGTCTATTCGCTTGTAGAG |
| TAS1c/FAD2-frag-3F | CAAGCGAATAGACCATTTAATGGGTGCAGGTGGAAGAATGCCGG | |
| TAS1c/FAD2-frag-4R | CAAAACAATATTCATTGTTTTGGCGACGTAGTAGAAGCATGAGGC | |
| TAS1c/FAD2-frag-5F | CTCATGCTTCTACTACGTCGCCAAAACAATGAATATTGTTTTGAATGTG | |
| miR167-5XsiFAD2 | miR167/TAS1c/5XsiFAD2-attB1 | GGGGACAAGTTTGTACAAAAAAGCAGGCTCATAGAAAGGTACTTTCGTTTAC |
| miR167/TAS1c/5XsiFAD2-2R | GAAAGCAATAAATGGTCTAACAAGCTGCCAGCCTGATCTAAAACAAAACAATGCTGTTTCATCTCGAAAAC | |
| miR167/TAS1c/5XsiFAD2-3F | GTTTTCGAGATGAAACAGCATTGTTTTGTTTTAGATCAGGCTGGCAGCTTGTTAGACCATTTATTGCTTTC | |
| miR167/TAS1c/5XsiFAD2-attB4 | GGGGACCACTTTGTACAAGAAAGCTGGGTTAAGTAACTGTATTAGCAACTGTTC | |
| To mutate miR173 target site | miR173 binding site mut 1for | GTGATTTTTCTAAACAAGAGAATAGACCATTTGG |
| miR173 binding site mut 1rev | CCAAATGGTCTATTCTCTTGTTTAGAAAAATCAC | |
| miR167 binding site mut 1for | TTAGATCAGGCAAACAGCATGTTAGACCATTTA | |
| miR167 binding site mut 1rev | TAAATGGTCTAACATGCTGTTTGCCTGATCTAA | |
| FAD2 northern hybridization | FAD2-3F | TAGGGTGTTCATCGTTATTA |
| FAD2-3R | AAGACCAACTGTGTCATCCA | |
| ACTIN2 northern control | ACTIN2 F | AAGATGACTCAAATCATGTTTGAGAC |
| ACTIN2 R | ACGACCTTAATCTTCATGCTGC | |
| 1XsiFAD2 small RNA northern hybridization | siFAD2 | TGGGAGATCTGAAGAAAGCAA |
| fragFAD2 small RNA hybridization | FAD2 210-bp For | ATGGGTGCAGGTGGAAGAATGCCGG |
| FAD2 210-bp Rev | GGCGACGTAGTAGAAGCATGAGGC | |
| Modified 5′ RACE | TAS1c-2rev | GACGACTTGAAAATCTCATT |
| FAD-TAS 1rev | CAATATTCATTGTTTTTGGGAGATCTGA | |
| ARF8 1Rev | GAGAGAGATGCGAACGAATGGCATAT | |
| ARF8 1Nested | CCCACCACTGCCTTCTCCATGAT |
DISCUSSION
Here we have described a novel method of PTGS utilizing the TAS1c locus in Arabidopsis. Engineering TAS1c to produce single or multiple copies of an artificial trans-acting siRNA (ata-siRNA) targeting the FAD2 gene, or with a 210-bp fragment of the FAD2 gene, resulted in consistent and very efficient silencing of FAD2. Notably, we also showed that miR167 was not an effective trigger for silencing in a TAS1c context, suggesting that miR173 may be specifically required to cleave TAS1c and/or may have a more sophisticated role in initiating phased siRNA from the TAS1c transcript.
TAS1c-mediated silencing consistently reaches complete phenotypic penetration. Nearly all T1 seed for constructs encoding single or multiple ata-siRNAs targeting FAD2 showed FAD2 silencing comparable to that of the fad2-1 null allele. In hairpinRNA-mediated silencing, a continuous spectrum of silencing was obtained in T1 plants with ODP scores ranging from similar to the fad2-1 mutant to as high as 50% of the wild type (Stoutjesdijk et al., 2002). Considering that up to 50% of Arabidopsis genes may be difficult to silence using long hairpin RNA interference constructs (Small, 2007), alternative silencing methods such as engineered ta-siRNA genes represent a valuable resource for functional genomics. Full loss-of-function phenotypes such as those demonstrated here are useful for inferring gene function and for epistasis analysis. Manipulating construct promoters could also produce incomplete silencing or spatially and temporally restricted silencing that would be useful for studying essential genes or moderating gene activity.
In one important aspect, ata-siRNAs are similar to amiRNAs: Both methods produce 21-bp single-stranded RNAs that target a specific sequence. These 21-bp species can be used to target one gene among a cluster of tandemly repeated genes, a specific allele, or a splice variant. This is a distinct advantage over intron-spliced hairpin constructs where much longer sequences are used, and thus more off-target silencing is possible.
It has been suggested that it might be possible to design polycistronic amiRNAs with multiple stem-loops that can encode distinct amiRNAs, in order to target multiple genes from the same construct (Ossowski et al., 2008). Targeting multiple genes with a single construct might be more easily achieved with ata-siRNAs. Instead of designing a precursor with multiple stem-loop structures, designing an ata-siRNA construct to silence multiple genes should be as simple as replacing two or more native sequences encoding siRNAs with artificial siRNAs targeting genes of interest. The main criterion that needs to be considered when designing an ata-siRNA construct is the characteristics of the encoded siRNA. Because ta-siRNAs are presumably derived from double-stranded RNA, not from an intramolecular fold as in miRNA, secondary structure considerations of the ata-siRNA precursor may not be critical.
Substituting miR167 for miR173 in TAS1c greatly reduces the ability of the 5XsiFAD2 construct to silence FAD2 and suggests that an intriguing new layer of ta-siRNA biogenesis remains to be explored. Even in leaves, where miR167 expression is high, the miR167-5XsiFAD2 construct showed inefficient FAD2 repression, precluding the possibility that weak silencing resulted from low miR167 expression. Instead, such poor silencing could indicate that TAS1c transcripts are not accessible to all miRNA. It is also possible that miR173 may have a specific role in promoting ta-siRNA biogenesis by recruiting an RNA-dependent RNA polymerase. In any case, once the hurdles to using miRNAs other than miR173 are overcome, one attractive possibility for silencing genes in a tissue-specific or temporally-specific manner would be to engineer TAS constructs triggered by miRNAs that are expressed in specific tissues or at specific times in development. The expression patterns of trigger miRNAs could be used to confer spatial or temporal specificity on TAS-induced silencing.
In addition to providing an excellent tool for gene silencing, the consistency and efficacy of silencing the FAD2 gene with TAS1c could provide a valuable system for investigating the specificity of sequences and cofactors for the function of ta-siRNAs. We expect that refining the design of ata-siRNAs will go hand in hand with a deepening knowledge of the biology of ta-siRNAs.
MATERIALS AND METHODS
Plant Material and Growth Conditions
All experiments were performed on the Columbia (Col-0) ecotype of Arabidopsis (Arabidopsis thaliana) plants. Plants were grown in long-day conditions (16 h of light at 22°C, 8 h of dark at 20°C).
Transgenic Constructs
The artificial TAS1c constructs targeting the Arabidopsis FAD2 gene were made using a 21-bp or a 210-bp fragment of the 5′ coding region of the gene. The 21-bp siFAD2 siRNA corresponds to base pairs 98 to 118 of the FAD2 CDS. These 21-bp sequences were selected by scanning the CDS for target regions that would be recognized by a ta-siRNA of sequence 5′-(T/A)NNNNNNNNNNNNNNNNN(C/G)NN-3′ with GC content between 30% to 50%. Internal stability was analyzed by running the candidate 21mer through mFold (Zuker, 2003) to look for a secondary structure with a high deltaG (≥1) that had at least three unpaired nucleotides at the 5′ and 3′ ends.
5XsiFAD2 was made as follows: sense and antisense oligos for the 5′ half and for the 3′ half of the 484-nt piece were designed (FAD2TAS 5′ sense, FAD2TAS 5′ antisense, FAD2TAS 3′ sense, FAD2TAS 3′ antisense). The resulting DNA fragment contains the modified TAS1c fragment, flanked by attB sites and single-stranded overhangs (GATC and AATT). Oligos were annealed and ligated into pGEM7 (previously digested with BamHI and EcoRI). The 5XsiFAD2 in pGEM7 was then recombined with pBC vector, using Gateway reactions. This placed the 5XsiFAD2 fragment downstream of the 35S promoter and upstream of the phaseolin terminator. mut-5XsiFAD2 was made as above, except that oligos that contained mutations in the miR173 target site were used for the ligations (FAD2TASmut 3′ sense, FAD2TASmut 3′ antisense).
To build the 1XsiFAD2 construct, we used primers TAS1c/FAD2-attB1 and TAS1c/FAD2-2R to generate the 5′ fragment by PCR, and primers TAS1c/FAD2-3F and TAS1c/FAD2-attB-4 to generate the 3′ fragment by PCR. The TAS1c/FAD2-2R and TAS1c/FAD2-3F fragments overlapped in the region of TAS1c that produces phased siRNAs, and modifications to the siRNAs were introduced using these long primers. We also modified 3′D5+ to target the unrelated AP1 gene. The 21-bp siAP1 sequence corresponds to base pairs 673 to 693 of the AP1 CDS. Silencing of AP1 was ineffective, likely due to the high intramolecular structure of the AP1 siRNA. The 5′ and 3′ fragments were gel purified and used together as a template to amplify the whole fragment by PCR with primers TAS1c/FAD2-attB1 and TAS1c/FAD2-attB-4. In a similar manner, construct miR167-5XsiFAD2 was generated using primers miR167/TAS1c/5XsiFAD2-attB1 and miR167/TAS1c/5XsiFAD2-2R to amplify the 5′ fragment and miR167/TAS1c/5XsiFAD2-3F and miR167/TAS1c/5XsiFAD2-attB4 to amplify the 3′ fragment. Both fragments were purified and used as a template to amplify the final product using primers miR167/TAS1c/5XsiFAD2-attB1 and miR167/TAS1c/5XsiFAD2-attB4.
To build the fragFAD2 construct, we first amplified three separate fragments. Fragment 1 was amplified with primers TAS1c/FAD2-attB1 and TAS1c/FAD2-frag-2R; fragment 2 was amplified with primers TAS1c/FAD2-frag-3F and TAS1c/FAD2-frag-4R; and fragment 3 was amplified with primers TAS1c/FAD2-frag-5F and TAS1c/FAD2-attB-4. Then fragment 1 and fragment 2 were used as templates to amplify fragment 4 using primers TAS1c/FAD2-attB1 and TAS1c/FAD2-frag-4R. The complete construct was then amplified by combining fragment 4 and fragment 3 using primers TAS1c/FAD2-attB1 and TAS1c/FAD2-attB-4. The 210-bp FAD2 fragment corresponds to base pairs 1 to 210 of the FAD2 CDS.
To create the constructs with mutated miRNA binding sites, we used constructs 5XsiFAD2, 1XsiFAD, and fragFAD2 as templates for site-directed mutagenesis (Wang and Malcolm, 1999) using the following oligonucleotides: miR173 binding site mut 1for and miR173 binding site mut 1rev. To mutate the miR167 target site in the construct TAS1c miR167-5XsiFAD2, we used primers miR167 binding site mut1 and miR167 binding site mut1rev, as described previously (Wang and Malcolm, 1999).
The fragments of interest were amplified with attB-tagged primer pairs and cloned into the Gateway pDONR/Zeo (Invitrogen) vector to give rise to the entry clone. Sequences for all primers used in this study are listed in Table I. The pBC Yellow Gateway vector was used as a destination vector to generate the expression clone. pBC Yellow was generated from pBC (Aukerman and Sakai, 2003) by inserting a PvuI-AlwN1 fragment generated by PCR containing the RD29A promoter driving Zs-Yellow YFP (CLONTECH Laboratories) expression into the PvuI/DraIII sites of pBC. This allowed transgenic seed to be selected based on YFP fluorescence. The BAR gene was eliminated from pBC Yellow by restriction digest and religation. Arabidopsis plants were transformed by pipetting Agrobacterium tumefaciens cell suspension directly on inflorescence buds (modified from Clough and Bent, 1998).
Fatty Acid Analysis
Lipid compositional analyses were conducted on mature seeds or rosette leaves, collected from wild-type and transgenic plants. Lipids were extracted from single seeds by homogenization with a pestle in an Eppendorf tube in MeOH:CHCl3 (10 μL; 2:1, v/v). After a 30-min incubation, 50 μL of heptane was added. Samples were mixed thoroughly and spun at 10,000 rpm in a microfuge, and the organic layer was recovered and used for lipid compositional analyses as done by Damude et al. (2006). For leaf tissue, a single fully expanded rosette leaf was ground, and then 1 mL of methanol/sulfuric acid derivitization solution (5% sulfuric acid in anhydrous methanol) was added. The resulting solution was heated at 95°C for 30 min, with vortexing every 10 min. The solution was then cooled to room temp, 1 m NaCl was added, and then 0.5 mL of heptane was added. Solution was then vortexed and centrifuged for 10 min. The resulting supernatant was then used for lipid compositional analysis as done by Damude et al. (2006).
Fatty acid profiles are expressed as ODP. Seed ODP is calculated as the percentage of 18:2 and 18:3 fatty acids out of the total amount of 18:1, 18:2, and 18:3 fatty acids, i.e. seed ODP = (%18:2 + %18:3)/(%18:1 + %18:2 + %18:3). Leaf ODP is calculated as the percentage of 18:2 fatty acids out of the total amount of 18:1 and 18:2 fatty acids, i.e. leaf ODP = (%18:2)/(%18:1 + %18:2). 18:3 fatty acids were not included in leaf ODP because the amount of 18:3 fatty acids in leaves is not dependent on FAD2 (Miquel and Browse, 1992). In leaf chloroplasts, an alternate pathway for 18:1 desaturation exists: The FADC enzyme desaturates 18:1 fatty acids to 18:2, and the FADD enzyme desaturates 18:2 fatty acids to 18:3 (Somerville and Browse, 1991).
Northern Analysis
Total RNA was isolated from aerial tissues of adult wild-type and transgenic plants using TRIZOL reagent (Sigma). RNA was separated on agarose gels, blotted onto a Hybond N+ membrane (Amersham), and probed with 32P-labeled probes randomly primed with the RadPrime DNA labeling system (Invitrogen). Hybridization was carried out at 68°C using PerfectHyb Plus buffer (Sigma). Blots were washed once in 2× SSC and 0.1% SDS for 5 min at room temperature, twice in 0.5× SSC and 0.1% SDS for 20 min at 68°C, and once in 0.1× SSC, 0.1% SDS for 20 min at 68°C. The hybridization signal was detected with a Storm 860 (Molecular Dynamics).
Low-molecular-weight (LMW) RNA was purified from total RNA using the mirVANA miRNA isolation kit (Ambion). LMW RNA was separated on a 15% TBE-Urea Criterion gel (Bio-Rad) and transferred electrophoretically to Hybond N+ membrane (Amersham) using a TransBlot-SD apparatus (Bio-Rad). LMW blots were hybridized at 40°C using ULTRAhyb-oligo buffer (Ambion) with 32P-end-labeled oligonucleotide probes. Probes were labeled with [γ-32P]ATP using OptiKinase (USB). Blots were washed twice in 2× SSC and 0.5% SDS for 30 min at 38°C.
Validation of the miR167 Target Site in the miR167-5XsiFAD2 Construct and in the ARF8 Gene
Modified 5′ RACE with the GeneRacer kit (Invitrogen) was adapted to validate the cleavage site determined by the miR167 target site. Primer TAS1c-2rev and nested primer FAD-TAS 1rev (miR167-5XsiFAD2) or ARF8 1Rev and ARF8 1Nested (ARF8) were used in PCRs, and the cleavage sites were revealed by sequence analyses of the PCR products.
Acknowledgments
Thanks to Leonard Farrell and Howard Damude for gas chromatography analysis of fatty acids, and to Manny Kiflemariam for growing plants for transformation. We greatly appreciate Enno Krebbers, Brian McGonigle, and Carl Falco for many helpful discussions. Additional thanks to Jeanne Wilson for critical comments on this manuscript, and to Mark Mucha for generating the pBC Yellow vector.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Robert W. Williams (robert-w2.williams@cgr.dupont.com).
References
- Allen E, Xie Z, Gustafson AM, Carrington JC (2005) microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121 207–221 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Alvarez JP, Pekker I, Goldshmidt A, Blum E, Amsellem Z, Eshed Y (2006) Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell 18 1134–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arteaga-Vázquez M, Caballero-Pérez J, Vielle-Calzada JP (2006) A family of microRNAs present in plants and animals. Plant Cell 18 3355–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H (2003) Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 11 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe DC (1996) RNA as a target and an initiator of post-transcriptional gene silencing in transgenic plants. Plant Mol Biol 32 79–88 [DOI] [PubMed] [Google Scholar]
- Baumberger N, Baulcombe DC (2005) Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA 102 11928–11933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116 281–297 [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409 363–366 [DOI] [PubMed] [Google Scholar]
- Chen X (2004) A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303 2022–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang CF, Meyerowitz EM (2000) Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc Natl Acad Sci USA 97 4985–4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Damude HG, Zhang H, Farrall L, Ripp KG, Tomb JF, Hollerback D, Yadav NS (2006) Identification of bifunctional delta12/omega3 fatty acid desaturases for improving the ratio of omega3 to omega6 fatty acids in microbes and plants. Proc Natl Acad Sci USA 103 9446–9451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandikota M, Birkenbihl RP, Höhmann S, Cardon GH, Saedler H, Huijser P (2007) The miR156/157 recognition element in the 3′UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J 49 683–693 [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286 950–952 [DOI] [PubMed] [Google Scholar]
- Lukowitz W, Gillmor CS, Scheible W (2000) Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol 123 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis K, Chandler V, Cone K, Kaeppler H, Kaeppler S, Kerschen A, Pikaard C, Richards E, Sidorenko L, Smith T (2005) Transgene-induced RNA interference as a tool for plant functional genomics. Methods Enzymol 392 1–24 [DOI] [PubMed] [Google Scholar]
- Miquel M, Browse J (1992) Arabidopsis mutants deficient in polyunsaturated fatty acid synthesis. Biochemical and genetic characterization of a plant oleoyl-phosphatidylcholine desaturase. J Biol Chem 267 1502–1509 [PubMed] [Google Scholar]
- Napoli C, Lemieux C, Jorgensen R (1990) Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuley J, Lightner J, Feldmann K, Yadav N, Lark E, Browse J (1994) Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell 6 147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S, Schwab R, Weigel D (2008) Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 53 674–690 [DOI] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X (2002) CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12 1484–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS (2004) SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev 18 2368–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP (2002) MicroRNAs in plants. Genes Dev 16 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small I (2007) RNAi for revealing and engineering plant gene functions. Curr Opin Biotechnol 18 148–153 [DOI] [PubMed] [Google Scholar]
- Smith NA, Singh SP, Wang MB, Stoutjesdijk PA, Green AG, Waterhouse PM (2000) Total silencing by intron-spliced hairpin RNAs. Nature 407 319–320 [DOI] [PubMed] [Google Scholar]
- Somerville C, Browse J (1991) Plant lipids: metabolism, mutants, and membranes. Science 252 80–87 [DOI] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L (2004) Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305 1434–1437 [DOI] [PubMed] [Google Scholar]
- Stoutjesdijk PA, Singh SP, Liu Q, Hurlstone CJ, Waterhouse PA, Green AG (2002) hpRNA-mediated targeting of the Arabidopsis FAD2 gene gives highly efficient and stable silencing. Plant Physiol 129 1723–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crété P (2004) Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell 16 69–79 [DOI] [PubMed] [Google Scholar]
- Wang W, Malcolm BA (1999) Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange site directed mutagenesis. Biotechniques 26 680–682 [DOI] [PubMed] [Google Scholar]
- Waterhouse PM, Graham MW, Wang MB (1998) Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc Natl Acad Sci USA 95 13959–13964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Poethig RS (2006) Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 18 3539–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JH, Han SJ, Yoon EK, Lee WS (2006) Evidence of an auxin signal pathway, microRNA167-ARF8-GH3, and its response to exogenous auxin in cultured rice cells. Nucleic Acids Res 34 1892–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M, Peragine A, Park MY, Poethig RS (2005) A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev 19 2164–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]