Abstract
Recently, we showed that microRNA399s (miR399s) control inorganic phosphate (Pi) homeostasis by regulating the expression of PHO2 encoding a ubiquitin-conjugating E2 enzyme 24. Arabidopsis (Arabidopsis thaliana) plants overexpressing miR399 or the pho2 mutant overaccumulate Pi in shoots. The association of Pi translocation and coexpression of miR399s and PHO2 in vascular tissues suggests their involvement in long-distance signaling. In this study, we used reciprocal grafting between wild-type and miR399-overexpressing transgenic plants to dissect the systemic roles of miR399 and PHO2. Arabidopsis rootstocks overexpressing miR399 showed high accumulation of Pi in the wild-type scions because of reduced PHO2 expression in the rootstocks. Although miR399 precursors or expression was not detected, we found a small but substantial amount of mature miR399 in the wild-type rootstocks grafted with transgenic scions, which indicates the movement of miR399 from shoots to roots. Suppression of PHO2 with miR399b or c was less efficient than that with miR399f. Of note, findings in grafted Arabidopsis were also discovered in grafted tobacco (Nicotiana benthamiana) plants. The analysis of the pho1 mutant provides additional support for systemic suppression of PHO2 by the movement of miR399 from Pi-depleted shoots to Pi-sufficient roots. We propose that the long-distance movement of miR399s from shoots to roots is crucial to enhance Pi uptake and translocation during the onset of Pi deficiency. Moreover, PHO2 small interfering RNAs mediated by the cleavage of miR399s may function to refine the suppression of PHO2. The regulation of miR399 and PHO2 via long-distance communication in response to Pi deficiency is discussed.
Mineral nutrients are acquired by roots from the rhizosphere and are subsequently distributed to shoots. The demand of shoots and supply from roots must be balanced and coordinated to maintain normal growth and development of plants. Therefore, communication between roots and shoots is indispensable; long-distance signals are required to report the nutrient status in these tissues. Local and long-distance signaling pathways are triggered when the nutrient concentration fluctuates (Forde, 2002; Atkins and Smith, 2007; Schachtman and Shin, 2007). The local signaling pathway is restricted to roots directly exposed to nutrient solutions, such as in the localized proliferation of branch roots and root hairs in response to the availability of nitrate, inorganic phosphate (Pi), and iron (Robinson, 1994; Bates and Lynch, 1996; Schikora and Schmidt, 2001). However, many adaptive responses are generated through the transmission of long-distance signals. The early warning of alteration in the external nutrient status sensed by shoots is achieved by long-distance signals from roots to shoots. Additional long-distance signals from shoots to roots are necessary to ensure the integration of changes in root physiology and development with the nutritional demand of shoots. Many nutrient uptake systems and enzymes involved in nutrient assimilation and mobilization in roots are regulated by demand from the shoot via shoot-derived signals (Marschner, 1995).
Hormones and nutrients themselves or their derivates could be potential candidates of long-distance signals (Forde, 2002; Atkins and Smith, 2007; Schachtman and Shin, 2007). In the case of phosphorus, Pi itself does not appear to be a long-distance signal from shoots; instead, allocation or recycling of Pi between shoots and roots is considered to provide the systemic signals (Drew and Saker, 1984; Jeschke et al., 1997; Burleigh and Harrison, 1999). Plants have developed a series of adaptive responses to Pi deficiency to overcome the low availability of Pi (Raghothama, 1999; Poirier and Bucher, 2002). These responses include conservation of internal Pi resources and enhancement of external Pi acquisition by roots, both depending on coordinated regulation by local and systemic signals.
Identification and characterization of several mutants have advanced our understanding of the local and systemic regulation of Pi homeostasis. The pdr2 mutant is unable to maintain the root meristem activity specifically under Pi-deficient conditions because of the disruption of local Pi sensing (Ticconi et al., 2004). The pho1 mutant is impaired in the loading of Pi into xylem, which results in Pi-deficient symptoms in shoots (Poirier et al., 1991; Hamburger et al., 2002). In contrast to pho1, the pho2 mutant overaccumulates Pi in shoots as a result of enhanced Pi uptake and translocation (Delhaize and Randall, 1995; Dong et al., 1998). Recently, PHO2 was identified as a target gene of microRNA399 (miR399), encoding a ubiquitin-conjugating E2 enzyme 24 (Aung et al., 2006; Bari et al., 2006).
miRNAs are a group of small RNAs processed from the stem-loop region of single-stranded endogenous transcripts and are involved in posttranscriptional gene regulation (Bonnet et al., 2006; Jones-Rhoades et al., 2006; Mallory and Vaucheret, 2006; Sunkar et al., 2007). Such small RNA-mediated regulation is crucial for mRNAs or proteins to accumulate preferentially in specific tissues, at specific growth stages, or in response to environmental stimuli. The expression of miR399 was up-regulated by Pi deprivation in accordance with reduced PHO2 mRNA level (Fujii et al., 2005; Chiou et al., 2006; Chiou, 2007). Up-regulation of miR399 is a specific response to deficiency of Pi but not other nutrients such as nitrogen, potassium, sulfate, or carbohydrates (Fujii et al., 2005; Bari et al., 2006). Transgenic plants overexpressing miR399, in which PHO2 is suppressed, resemble pho2 mutant and PHO2 T-DNA knockout lines. These plants all display Pi toxicity in the shoots as a consequence of enhanced Pi uptake, Pi translocation from roots to shoots, and retention of Pi in old leaves (Aung et al., 2006; Chiou et al., 2006). These observations reveal a crucial role of interaction between miR399 and PHO2 in the maintenance of Pi homeostasis. This regulation appears to be conserved among different plant species because of the conservation of miR399 and PHO2 genes (Bari et al., 2006; Chiou et al., 2006). Recently, AT4 and IPS1, members of a noncoding RNA class highly induced by Pi starvation (Burleigh and Harrison, 1999; Martin et al., 2000), were found to inhibit the cleavage of miR399 on the PHO2 transcript through the mimicry of the target sequence (Franco-Zorrilla et al., 2007). Of interest, up-regulation of miR399, At4, and IPS1 by Pi deprivation is mediated by phosphate starvation response 1, a MYB transcription factor (Rubio et al., 2001; Bari et al., 2006), which represents a regulatory circuit in controlling Pi homeostasis during the fluctuation of available Pi.
The vascular expression pattern of PHO2 and miR399 and their systemic role in Pi uptake and translocation (Aung et al., 2006; Chiou et al., 2006) prompted us to examine their systemic functions. Moreover, the recent identification of a diverse and dynamic population of small RNAs in phloem sap of many plant species suggested the potential function of these small RNAs as information molecules (Yoo et al., 2004; Lough and Lucas, 2006; Buhtz et al., 2008). In this study, we performed reciprocal grafting between wild-type and miR399-overexpressing Arabidopsis (Arabidopsis thaliana) and tobacco (Nicotiana benthamiana) plants to address the systemic regulation of PHO2 by miR399. We analyzed quantitatively the level of miR399 precursors and mature miR399 and PHO2 mRNA, as well as Pi accumulation, in scions and rootstocks of Arabidopsis and tobacco and revealed systemic regulation of PHO2 by the movement of miR399.
Recently, Pant et al. (2008) reported miR399 as a long-distance signal for the regulation of Pi homeostasis on the basis of the authors' detection of miR399 in phloem sap of rapeseed and pumpkin (Cucurbita maxima) and the movement of miR399 in grafted Arabidopsis plants, research that is similar to the work we describe here. Our results not only support the Pant et al. (2008) findings but also provide additional and profound information about this systemic signaling. We show the common phenomena for the long-distance movement of miR399s in Arabidopsis and tobacco plants and the differential efficiency of miR399 species in targeting the PHO2 transcript and identify miR399-mediated PHO2 small interfering RNAs (siRNAs). Furthermore, we elucidate the biological significance for shoot-to-root movement of miR399s in the initial response to Pi deprivation and in the pho1 mutant.
RESULTS
Systemic Movement of miR399 in Arabidopsis
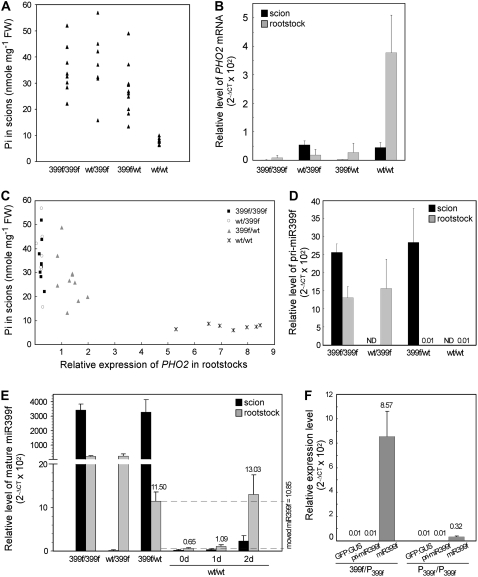
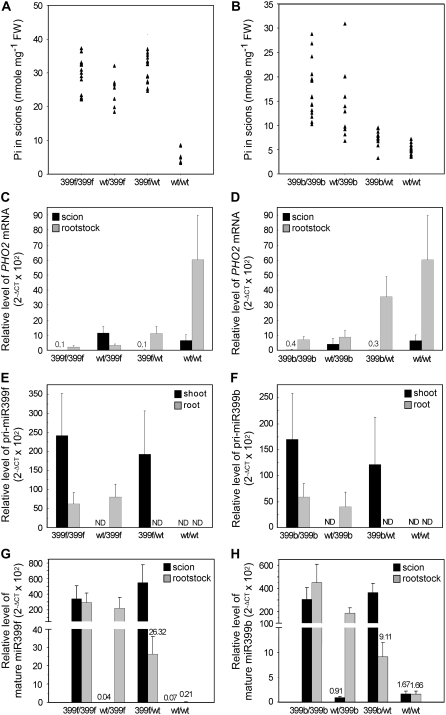
We performed reciprocal micrografting using wild-type and miR399f-overexpressing transgenic Arabidopsis plants (Chiou et al., 2006). Four combinations of grafted plants were generated, including 2 self-grafted controls, wild-type to wild-type (designated as wt/wt for scion/rootstock) and transgenic to transgenic plants (399f/399f), and two reciprocal grafts, wild-type to transgenic plants (wt/399f and 399f/wt). The grafted plants were maintained under Pi-sufficient conditions to avoid the endogenous induction of miR399 expression. Scions and rootstocks from individual grafted plants were harvested for Pi assay and measurement of PHO2 mRNA, primary transcripts of miR399 and mature miR399 by quantitative reverse transcription (RT)-PCR. Specific mature miR399 species were measured by quantitative RT-PCR coupled with Taq-Man probe. Results from quantitative RT-PCR are presented as relative expression levels with 2−ΔCT × 102 or 2−ΔCT × 104 for easy observation (Livak and Schmittgen, 2001; Schmittgen et al., 2004) and comparison of the expression of the same transcript in different figures. Because of variations in shoot Pi concentration among individual plant, results from individual grafted plants were not averaged but, rather, displayed as a distribution of data collected from each plant (Fig. 1A). Like nongrafted plants, wt/wt self-grafted plants displayed normal shoot Pi concentration, but 399f/399f self-grafting plants showed a high level of Pi in the scions (Fig. 1A).
Figure 1.
Reciprocal grafting between wild-type and miR399f-overexpressing transgenic Arabidopsis plants. A, Pi concentration in the scions. Each spot indicates one data point from an independent grafted plant. B, Relative levels of PHO2 mRNA in the scions (black bars) and rootstocks (gray bars) of grafted plants. C, Relation between the Pi concentration in scions and the expression of PHO2 in rootstocks. D and E, Measurement of pri-miR399f (D) and mature miR399f (E) in scions (black bars) and rootstocks (gray bars), respectively. Amount of mature miR399f in wild-type self-grafting plants subjected to 1- or 2-d Pi starvation treatment were included in E. F, Relative expression of GFP:GUS, pri-miR399f, and mature miR399f in the rootstocks carrying miR399f promoter∷GFP:GUS transgene grafted with miR399f-overexpressing scions (399f/P399f) or self-grafted controls (P399f/P399f). Grafting combinations are shown as scion/rootstock. RNA was analyzed by quantitative RT-PCR and the levels could be compared with those shown in Figure 2. The error bars represent sd (n = 4–5 independent grafted plants). ND, Nondetectable. The difference between two dashed lines in E indicates the potential amount of moved miR399f.
Wild-type scions grafted onto miR399f-overexpressing rootstocks (wt/399f) showed a high level of Pi (Fig. 1A), which indicates that miR399f-overexpressing rootstocks alone are sufficient to cause high accumulation of Pi in wild-type scions. This observation is similar to that in pho2 rootstocks grafted with wild-type scions (Bari et al., 2006) because of the reduced or nonfunctional PHO2 protein in the rootstocks. Of note, miR399f-overexpressing scions also accumulated high concentrations of Pi when grafted onto wild-type rootstocks (399f/wt; Fig. 1A). PHO2 mRNA level in scions and rootstocks of all grafted plants was similar to that of the original genotypes, except in wild-type rootstocks of 399f/wt plants, which showed more than 10-fold lower PHO2 mRNA level than wt/wt controls (Fig. 1B). The increased Pi concentration in miR399-overexpressing scions is associated with reduced PHO2 mRNA in wild-type rootstocks.
Plotting Pi concentration in scions against the level of PHO2 mRNA in rootstocks gave a negative correlation, with three groupings (Fig. 1C): (1) wt/wt self-grafted plants accumulating normal Pi level in shoots and a high PHO2 mRNA level in roots; (2) 399f/399f and wt/399f plants with high Pi level in shoots but a low PHO2 mRNA level in roots; and (3) 399f/wt plants containing a high Pi level in shoots but a relatively low PHO2 mRNA level in roots. When the PHO2 mRNA in rootstocks drops to a critical level, as observed in 399f/wt, scions started to accumulate a high amount of Pi. High accumulation of Pi in scions coexists with a Pi toxic phenotype shown as chlorosis and necrosis in leaf tips (Chiou et al., 2006). In addition, the Pi concentration in rootstocks of all grafted plants was normal and similar to that in wt/wt controls (data not shown), as seen in roots of pho2 or miR399-overexpressing plants.
Reduced PHO2 expression in wild-type rootstocks of 399f/wt plants raises the possibility of movement of miR399f from miR399f-overexpressing scions to suppress the expression of PHO2 in rootstocks. Movement of miR399f was examined by detection of mature miR399f and its primary transcript (pri-miR399f) on quantitative RT-PCR (Fig. 1, D and E). As expected, a high level of mature miR399f and pri-miR399f was observed in scions and rootstocks from miR399f-overexpressing plants. Mature miR399f was at an extremely low level in wild-type scions or rootstocks except for a relatively small but substantial amount of mature miR399f in wild-type rootstocks of 399f/wt plants. Although the level of mature miR399f in wild-type rootstocks was only 0.35% of that in miR399f-overexpressing scions, it reached 18-fold of that in wt/wt controls (Fig. 1E). This level of miR399f did not result from de novo synthesis in the wild-type rootstocks, because pri-miR399f was barely detected and was as low as that in wt/wt control plants (Fig. 1D). Exclusion of de novo synthesis of miR399f was provided by grafts of rootstocks of transgenic plants expressing the GFP:GUS fusion gene driven by the miR399f promoter (Aung et al., 2006). The level of GFP:GUS in the rootstocks grafted with miR399f-overexpressing scions (399f/P399f) was as low as that in the rootstocks of self-grafted controls (P399f/P399f; Fig. 1F). From these observations, we conclude that the mature miR399f detected in the wild-type rootstocks should originate from the miR399f produced in the scions moving across graft unions to the rootstocks.
Association of miR399f movement with reduced PHO2 mRNA level in rootstocks suggests the cleavage of PHO2 mRNA by the scion-born miR399f. However, from our experience, the level of translocated miR399f seemed comparatively low, because it was undetectable by regular RNA gel-blot analysis. To determine whether such a small amount of miR399f is functional and sufficient to cleave PHO2 mRNA, we compared its level with that in Pi-starved wt/wt self-grafted plants. The wt/wt plants were treated under Pi-free medium for 1 or 2 d, and the expression of mature miR399f was analyzed. The level of miR399f (2−ΔCT × 102 = 11.50) in rootstocks of 399f/wt plants was higher than that in 1-d Pi-starved rootstocks (2−ΔCT × 102 = 1.09) and close to that in 2-d Pi-starved rootstocks of wt/wt plants (2−ΔCT × 102 = 13.0; Fig. 1E). Although the level was not as high as in long-term Pi-starved samples, in which the expression of miR399f (2−ΔCT × 102) can reach over 350 after 5-d starvation (Fig. 5D), constitutive accumulation of a low amount of miR399f should be effective in keeping PHO2 mRNA at the low level. Because the whole grafting process takes about 4 to 5 weeks, maintenance of a low level of miR399f in an extended period could be effective enough to reduce the PHO2 mRNA level. Furthermore, the miR399-guided cleavage products of PHO2 mRNAs in the rootstocks of 399f/wt plants were identified by analysis of RNA ligase-mediated 5′ rapid amplification of cDNA end (RLM-5′ RACE; red numbers in Supplemental Fig. S1).
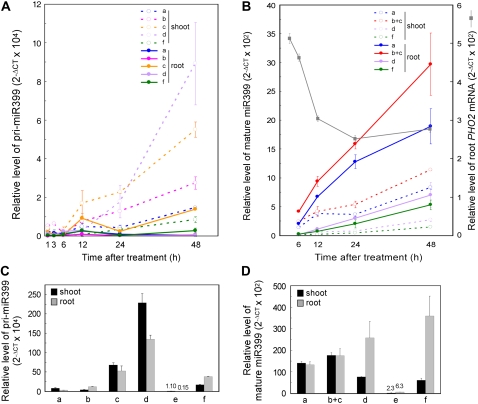
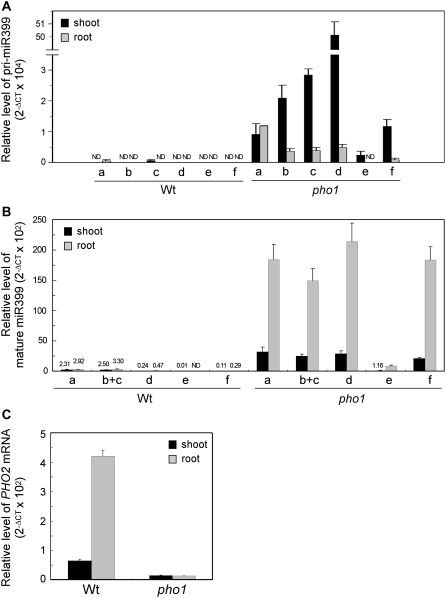
Figure 5.
A and B, Time-course analyses of different pri-miR399s (a, b, c, d, and f; A) and mature miR399 species (a, b+c, d, and f; B) during the initiation of Pi deprivation. The dashed and solid lines indicate the expression levels in shoots and roots, respectively. The level of PHO2 mRNA was included in B as a gray solid line. All miR399 species are shown except miR399e because of its low expression. The level of mature miR399b and c cannot be distinguished because of identical sequences. C and D, Relative expression levels of pri-miR399s (C) and mature miR399s (D) in shoots (black bars) and roots (gray bars) after 5 d of Pi starvation treatment. Please note that the mature miR399a and b/c could have been overestimated in B and D (see Supplemental Fig. S5). One of two biological replicates is presented, and the error bars indicate the sd of two technical replicates.
Differential Targeting of PHO2 mRNA by miR399b/c and miR399f
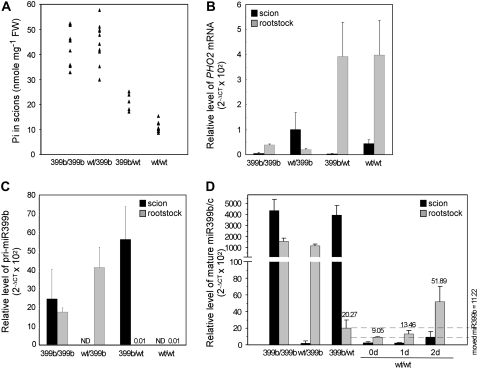
In addition to grafting with miR399f, we grafted miR399b-overexpressing plants, which have been shown to display a similar Pi toxic phenotype as miR399f-overexpressing plants (Chiou et al., 2006). To our surprise, unlike miR399f plants, transgenic scions of 399b/wt plants showed a Pi concentration not as high as that of wt/399b or 399b/399b plants (Fig. 2A). A low accumulation of Pi in transgenic scions was associated with a high expression of PHO2 in wild-type rootstocks of 399b/wt plants (Fig. 2B). The low efficiency in suppressing PHO2 in the rootstocks of 399b/wt plants was not due to lower expression of miR399b in the transgenic scions or less movement of miR399b compared to 399f/wt plants. Subtracting expression from the basal expression in wt/wt plants resulted in a comparable increased amount of mature miR399b (2−ΔCT × 102 = 11.22) or miR399f (2−ΔCT × 102 = 10.85) in the wild-type rootstocks of 399b/wt or 399f/wt grafting plants (Figs. 1E and 2D). This suggests that the movement of miR399b from scions to rootstocks could be as efficient as that of miR399f.
Figure 2.
Reciprocal grafting between wild-type and miR399b-overexpressing transgenic Arabidopsis plants. A, Pi concentration in the scions. Each spot indicates one data point from an independent grafted plant. B, Relative levels of PHO2 mRNA in the scions (black bars) and rootstocks (gray bars) of grafted plants. C and D, Measurement of pri-miR399b (C) and mature miR399b/c (D) in scions (black bars) and rootstocks (gray bars), respectively. The level of mature miR399b/c in wild-type self-grafted plants subjected to 1- or 2-d Pi starvation treatment were included in D. Because of the identical sequences, the level of mature miR399b and c cannot be distinguished. The error bars represent sd (n = 4–5 independent grafted plants). ND, Nondetectable. The difference between two dashed lines in D indicates the potential amount of moved miR399b.
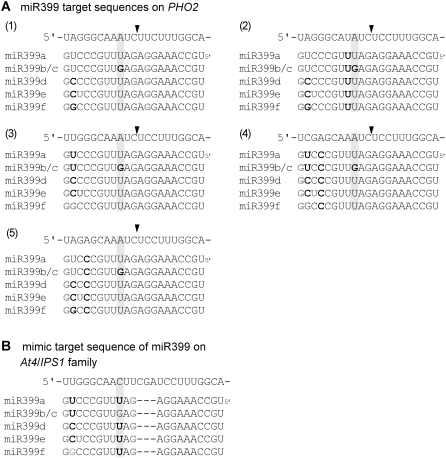
The differential efficiency on the suppression of PHO2 led us to inspect the sequence variation between miR399b and f. Sequences of different miR399 species (miR399a–f) aligned with the five potential target sites of the PHO2 transcript, in addition to a few mismatches toward the 3′ ends of the miR399s, revealed that the guanosine (G) at nucleotide 13 of miR399b/c is not base-paired to all of five target sequences (Fig. 3A). We postulated that the mismatch at this position may reduce the cleavage efficiency. This hypothesis is supported by a similar observation in transgenic plants with overexpressed miR399b replaced with miR399c as grafting materials (Supplemental Fig. S2) because of the identical sequences of mature miR399b and c.
Figure 3.
Sequence alignments among different miR399 species and five potential targets on PHO2 mRNA (A) and the mimicked target sequence on At4/IPS1 noncoding RNAs (B). Mismatched nucleotides are indicated in bold and GU base-pairings are marked in gray. The position of nucleotide 13 is highlighted. Solid triangles indicate the cleavage sites of target sequences by miR399s.
Movement of Arabidopsis miR399 in Tobacco Plants
We generated transgenic tobacco plants overexpressing Arabidopsis miR399b or miR399f, and, similar to Arabidopsis, transgenic tobacco plants overaccumulated Pi in shoots and displayed Pi toxicity (Supplemental Fig. S3, A and B). This finding suggests efficient targeting of the tobacco PHO2 homologue by Arabidopsis miR399. Targeting of tobacco PHO2 by heterologous Arabidopsis miR399s suggests a high degree of sequence homology between Arabidopsis and tobacco miR399s. These transgenic tobacco plants were used for reciprocal grafting materials as performed in Arabidopsis. The results from tobacco grafting were consistent with those from Arabidopsis. 399f/399f, wt/399f, and 399f/wt tobacco plants accumulated high Pi content in scions (Fig. 4A) as a consequence of suppression of tobacco PHO2 in rootstocks (Fig. 4C). Wild-type rootstocks of 399f/wt plants showed a significantly higher level of miR399f (2−ΔCT × 102 = 26.32) than wt/wt controls (2−ΔCT × 102 = 0.21; Fig. 4G). De novo synthesis of miR399f in the rootstocks was unlikely because of no detectable pri-miR399f (Fig. 4E). Tobacco plants also showed different performance of miR399f and b. In 399b/wt tobacco plants, the suppression of PHO2 in rootstocks was less efficient than in 399f/wt tobacco plants, which resulted in less Pi accumulation in scions (Fig. 4, B and D). Although the level of mature miR399b in the rootstocks of 399b/wt plants was lower than that of miR399f in the rootstocks of 399f/wt plants, possibly because of lower expression of miR399b in these transgenic lines, miR399b still moved from transgenic scions to wild-type rootstocks (Fig. 4H). Interestingly, sequence alignment also revealed the mismatch at nucleotide 13 of Arabidopsis miR399b/c to the four potential targets on the tobacco PHO2 transcript (Supplemental Fig. S3C). Taken together, similar results obtained from Arabidopsis and tobacco grafting experiments demonstrate the long-distance movement of miR399 from shoots to roots and suggest a common phenomena for the movement of miR399 in plants.
Figure 4.
Reciprocal grafting between wild-type and miR399-overexpressing transgenic tobacco plants. Pi concentration in the scions (A and B) and the relative level of PHO2 mRNA (C and D), pri-miR399 (E and F), and mature miR399 (G and H) in scions (black bars) and rootstocks (gray bars) of grafted plants were measured. A, C, E, and G represent data from grafts between wild-type and miR399f-overexpressing plants. Results from B, D, F, and H are from grafts between wild-type and miR399b-overexpressing plants. The error bars represent the sd (n = 4–5 independent grafted plants). ND, Nondetectable.
Long-Distance Movement of miR399 from Shoots to Roots as an Early Response to Pi Deficiency
Results from grafting experiments provide evidence of long-distance movement of miR399 from shoots to roots. However, both roots and shoots show a similar amount of mature miR399 (Chiou et al., 2006), and promoter∷reporter analysis revealed transcriptional activity of all six miR399 genes in vascular tissues of roots and shoots (Aung et al., 2006). Why do miR399s need to travel from shoots to roots if both tissues can synthesize miR399s and what is the biological significance of such movement? In previous study (Aung et al., 2006; Chiou et al., 2006), miR399s were examined after long Pi starvation (3–5 d), so we confined the treatment to a shorter period (1 h–2 d) and examined the level of all species of pri-miR399 and mature miR399 in roots and shoots, respectively.
We began to detect a considerable amount of pri-miR399s in shoots at 12 h after treatment, which continued to increase during treatment (Fig. 5A). In shoots, pri-miR399d showed the greatest accumulation, followed by pri-miR399c, b, a, and f after 2 d of treatment. The expression of miR399e was very low within 2 d of treatment and is therefore not shown. In contrast to pri-miR399 expression in shoots, that in roots was scarce prior to 1-d treatment. The accumulation of pri-miR399s in roots was much less abundant than that in shoots, which suggests a faster response of shoots than roots in up-regulating miR399 genes. When mature miR399s were examined, a high amount of pri-miR399s in the shoots did not result in high accumulation of mature miR399s. In fact, roots accumulated a higher amount of mature miR399s than shoots (Fig. 5B) despite a very low level of pri-miR399s (Fig. 5A). A high level of pri-miR399s was detected in shoots, but a high level of mature miR399s was noticed in roots. This finding was true for all different miR399 species. The accumulation of mature miR399s was associated with reduced PHO2 mRNA level in roots (Fig. 5B). The opposite accumulation of pri-miR399s and mature miR399s in shoots and roots implies the movement of mature miR399s from shoots to roots, which is important for the suppression of PHO2 in roots to activate the Pi acquisition from the rhizosphere and subsequent translocation during the onset of Pi starvation. After 5-d starvation, the expression of miR399 continued to increase, and the levels of pri-miR399s and mature miR399s were comparable in both shoots and roots except for a higher level of mature miR399d and f in roots (Fig. 5, C and D).
One may argue that the low level of pri-miR399s but high level of mature miR399s in roots is a consequence of higher processing efficiency from precursors to mature products. This scenario seems unlikely. From our calculations, the processing for miR399b and c in roots has to be 15-fold more efficient than that in shoots to explain the differences we observed. In fact, we compared the processing efficiency between shoots and roots by calculating the ratio of level of mature miR171b to that of its primary transcript and found similar processing efficiency in shoots and roots of wild-type plants grown under Pi-sufficient or -deficient conditions (Supplemental Fig. S4A). Moreover, on the basis of the expression of the endoplasmic reticulum-localized GFP reporter gene, the promoter activity of miR399a, b, c, or f was 2 to 20 times greater in shoots than in roots 2 d after Pi starvation (Supplemental Fig. S4B). Higher promoter activity is correlated with higher accumulations of pri-miR399s in shoots, which supports a faster induction of miR399 genes in shoots than in roots.
Systemic Regulation of PHO2 by miR399 in pho1
The pho1 mutant is defective in the loading of Pi into the xylem of roots and thus displays a Pi-deficient phenotype in shoots (Poirier et al., 1991). Because Pi level is low in shoots of pho1 but remains at normal levels in roots, we examined the expression and possible systemic regulation of miR399 and PHO2 in pho1. In response to an internal low Pi level, both pri-miR399s and mature miR399s accumulated in pho1 shoots (Fig. 6, A and B), along with an increased amount of pri-miR399s in pho1 roots, which suggests the existence of a shoot-derived signal to systemically up-regulate the expression of miR399 in roots (Fig. 6A). Similar to the observation during short-term Pi deficiency, a higher amount of pri-miR399s, except pri-miR399a, accumulated in shoots than in roots, whereas more mature miR399s were detected in roots (Fig. 6, A and B). The abundance of pri-miR399d in shoots was much higher than that of other miR399 species, but this did not result in a high accumulation of mature miR399d in shoots or roots. Whether this observation is caused by differential processing among different miR399 species requires further studies. The level of PHO2 mRNA in shoots and roots of the pho1 mutant was reduced as a result of increased miR399s (Fig. 6C). The miR399-guided cleavage of PHO2 mRNA was further revealed by RLM-5′ RACE (blue numbers in Supplemental Fig. S1). The analysis of pho1 offers additional lines of evidence for systemic suppression of PHO2 by the movement of miR399 from shoots to roots.
Figure 6.
Quantitative RT-PCR analyses of different pri-miR399s (a–f; A) and mature miR399 species (a, b+c, d, e, and f; B) and PHO2 mRNA (C) in the shoots (black bars) and roots (gray bars) of wild-type plants and pho1 mutant. Please note that the mature miR399a and b/c could have been overestimated in B (see Supplemental Fig. S5). The error bars represent the sd of two technical replicates. ND, Nondetectable.
Pi Starvation-Induced and miR399-Dependent PHO2 siRNAs
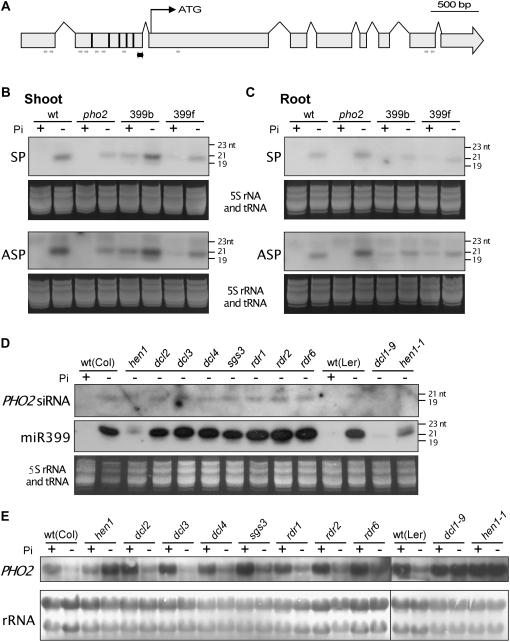
Two miRNA-directed cleavages may generate siRNAs (Axtell et al., 2006). Because the PHO2 transcript contains five potential miR399 target sites, examining whether targeting by miR399 can generate PHO2 siRNA is of interest. Several fragments corresponding to different regions of cleaved product were used as probes to detect the PHO2 small RNAs. The signal was observed when the downstream sequence at the fifth target site was used as a probe (Fig. 7A). Intriguingly, these PHO2 small RNAs were detected only in the Pi-starved samples of wild-type and pho2 plants, which indicates that their production is Pi-starvation dependent. Moreover, these PHO2 small RNAs were noticed in transgenic plants overexpressing miR399b or f, regardless of their Pi status (Fig. 7, B and C). The association between the expression of miR399s and PHO2 small RNAs suggests that the production of these small RNAs may rely on the cleavage of miR399s. These small RNAs are about 20 nucleotides long, are present in both roots and shoots, and are double-stranded, because they could be detected by both sense and antisense probes. They likely function as siRNAs to reinforce the repression of PHO2. Because the level of these siRNAs was enhanced in transgenic plants under Pi starvation, the enhancement may result from an additive effect by the overexpression of transgenes and induction of endogenous miR399.
Figure 7.
Detection of PHO2 siRNA. The location of probe (double-pointed black arrow) and sequenced small RNAs (single-pointed gray arrows indicating the direction of 5′ to 3′) corresponding to the PHO2 transcript are marked in A. The vertical bars within the second exon indicate the five miR399 target sites. B and C, RNA gel-blot analysis of PHO2 siRNAs in Pi-sufficient (+Pi) and 5-d Pi-starved (−Pi) shoots (B) and roots (C) of wild-type (wt), pho2, and miR399-overexpressing (399b and 399f) plants. The siRNA could be detected by both SP and ASP. D, Detection of PHO2 siRNA (ASP) and miR399 in the roots of small RNA biogenesis mutants subjected to 5-d Pi-starvation treatment. E, RNA gel-blot analysis of PHO2 mRNA in the roots of different mutants under Pi-sufficient or -deficient conditions. Staining of 5S ribosomal RNA and tRNA (B–D) and 25S and 18S ribosomal RNAs (E) is shown as the loading control.
To understand the biogenesis of PHO2 siRNAs, we analyzed mutants defective in small RNA biogenesis (Fig. 7D). PHO2 siRNAs could be detected in all Pi-starved mutants except dcl1 and hen1, two essential components in the biogenesis of miRNAs (Park et al., 2002; Chen, 2005). No signal of PHO2 siRNAs was detected in hen1-1 with a Landsberg erecta background, whereas a slightly larger band was observed in hen1 of the Columbia ecotype, likely resulting from the uridylation of a nonmethylated 3′ end (Li et al., 2005). An abnormal size or absence of PHO2 siRNAs is associated with a significant reduction in the level of miR399 in dcl1 and hen1 mutants (Fig. 7D). These results further support the involvement of miR399 in generating PHO2 siRNAs. Consistent with the low expression of miR399 in dcl1 and hen1 mutants, the level of PHO2 mRNA was not down-regulated in response to Pi deficiency (Fig. 7E).
DISCUSSION
From results of grafting in Arabidopsis and tobacco plants, we revealed the long-distance movement of miR399 from shoots to regulate the expression of PHO2 in roots. The observations of differential accumulation in expression of miR399 precursors and mature miR399 in shoots and roots of pho1 mutant plants and during Pi deficiency further supported this notion. Targeting of miR399b/c on the PHO2 transcript was less efficient than that of miR399f likely because of a mismatch at nucleotide 13. Furthermore, identification of Pi starvation-induced and miR399-mediated PHO2 siRNAs provides an additional layer of regulation of PHO2 expression.
Long-Distance Movement of miR399 via Phloem
Detection of mature miR399 in the wild-type rootstocks grafted with miR399-overexpressing scions provides evidence for the movement of miR399 from shoots to roots (Figs. 1, E and F, 2D, and 4, G and H). The increased level of miR399 did not result from de novo synthesis of miR399 in the rootstocks, because the level of miR399 precursors was undetectable or as low as that in wt/wt controls (Figs. 1D, 2C, and 4, E and F). Moreover, transcription of miR399f was not enhanced in the miR399f promoter∷reporter rootstocks grafted with miR399f-overexpressing scions (399f/P399f; Fig. 1F). Importantly, grafting results obtained from Arabidopsis could be reproduced in tobacco plants, which suggests that the movement and regulatory network of miR399 could be common and conserved. This suggestion is also supported by the identification of miR399 and PHO2 homologues in many plant species (Bari et al., 2006; Chiou et al., 2006; miRBase, release 10.1). Furthermore, downward but not upward movement of miR399 indicates the transportation of miR399 via the phloem stream, which is consistent with the observation of many miRNA species in phloem sap but no RNAs in xylem sap (Yoo et al., 2004; Buhtz et al., 2008). miR166 may function as a movable signal because of its progressively expanding expression pattern during leaf development and its accumulation in phloem (Juarez et al., 2004). However, synthetic miRNA targeting auxin response factors (miR-ARF) was not able to cross graft unions in tomato (Solanum lycopersicum) or tobacco plants in a study based on characterization of phenotype (Alvarez et al., 2006). Similar to miR399-overexpression plants used in this study, miR-ARF was driven by a 35S promoter, so the difference in mobility between miR399 and miR-ARF should not result from different expression patterns. Instead, the long-distance movement of the miRNA information network should be regulated tightly with selectivity and specificity.
Recently, Pant et al. (2008) reported the application of an Arabidopsis grafting approach similar to what we describe here. Although our two studies found a similar conclusion of the long-distance movement of miR399 from scion to suppress PHO2 expression in rootstocks, the detectable movement of miR399 differed. Pant et al. (2008) detected a very high level of mature miR399d in wild-type rootstocks of 399d/wt plants. The level was close to that in the original miR399d-overexpressing plants or the level of UBQ10, one of most abundant transcripts in plants. The similar level of miR399d in overexpressing scions and wild-type rootstocks suggests highly efficient movement of miR399d. However, we did not observe such highly efficient movement. In our system, the detectable miR399f or b level in wild-type rootstocks of 399/wt Arabidopsis or tobacco plants was relatively small (0.35%–5%) as compared with the original overexpressing scions. One explanation is the difference in transgenic lines expressing different miR399 species. Whether miR399d shows different mobility than miR399f would be interesting, because these two contain only one nucleotide difference at position 20 (Fig. 3). One nucleotide change in the potato (Solanum tuberosum) spindle tuber viroid was reported to interfere in its trafficking to phloem for systemic infection because of differences in tertiary structure (Zhong et al., 2007). Nevertheless, the size of miRNAs is much smaller than that of viroid RNAs, and the formation of a secondary or tertiary structure is not clear. Also, the discrepancy may result from different experimental systems, such as grafting procedures, growth conditions, or detection methods.
Under natural conditions, the movement of miR399s should occur when Pi is depleted because of the up-regulation of miR399s by Pi deprivation. However, the movement of miR399s was observed in grafted plants grown under Pi-sufficient conditions, which indicates that the movement of miR399s could be independent of Pi status and the machinery is ready and functional under both Pi-sufficient and -deficient conditions. Nevertheless, other components induced under Pi deficiency may be involved and facilitate the movement during deficiency.
One may wonder whether the miRNA precursors can move. If they can move, the precursors left from processes would have to move out of the nucleus, travel through the phloem, and be processed very quickly after arriving at roots, because we were unable to detect increased levels of primary transcript (pri-miR399) and stem-loop precursors (pre-miR399, data not shown) in the wild-type rootstocks of 399/wt plants. Moreover, the processing of precursors by dicer after long-distance movement is less possible, because they have to reenter the nucleus to be processed unless the process can take place in the cytosol. The lower abundance of miRNA precursors and nucleus localization of processing machinery (Chen, 2005; Fang and Spector, 2007) make them unlikely to be the mobile candidates.
At the beginning of this study, quantitative RT-PCR failed to detect mature miR399 in the wild-type rootstocks of 399/wt plants with a polyadenylation method (Shi and Chiang, 2005) used for cDNA synthesis. With a stem-loop priming method (Chen et al., 2005) coupled with the Taq-Man probe, we began to detect the mature miR399. The lower sensitivity of polyadenylation may be caused by the inhibition of tailing at the 3′ end of mature miRNA because of the methyl group on the 2′ OH of 3′ terminal nucleotide (Yang et al., 2006). One should be cautious about the detection of low abundance of miRNAs with this polyadenylation methodology.
Suppression of PHO2 by the Movement of miR399
In 399f/wt plants, the reduction of PHO2 mRNA was associated with the existence of miR399f in rootstocks (Fig. 1, B and E). Down-regulation of PHO2 was not due to the reduced transcriptional activity of PHO2, because the transcript level of the GFP:GUS fusion reporter driven by the PHO2 promoter in the rootstocks grafted with miR399f-overexpressing scions (399f/Ppho2) was not altered (data not shown). Down-regulation of PHO2 in wild-type rootstocks should result from the posttranscriptional cleavage by miR399f transported from scions to rootstocks. The level of miR399f in the rootstocks was close to that in 2-d Pi-starved wt/wt control plants (Fig. 1E). In fact, the level of PHO2 mRNA in roots was reduced 40% after 2-d starvation (Fig. 5B). One has to keep in mind that such reduction is a collective effect of all six miR399 species. Although the level of miR399f is not high, constitutive maintenance of such a low level could be sufficient to reduce PHO2 expression. Moreover, PHO2 siRNAs derived from the targeting of miR399s (Fig. 7) may enhance the suppression effect.
Even though transgenic plants overexpressing miR399b, c, or f all display Pi toxicity due to the inhibition of PHO2 (Chiou et al., 2006), our grafting experiments showed that miR399b and c were not as efficient as miR399f in suppressing PHO2 in rootstocks and accumulating Pi in scions (Fig. 2; Supplemental Fig. S2). These observations are not likely due to the different mobility of these miR399s (Figs. 1E and 2D). The difference might reside in the variation at nucleotide 13 of the miR399s. The G at nucleotide 13 of miR399b/c is not complementary to any of five potential target sequences on PHO2 mRNA, but interestingly, makes better base-pairing with the consensus sequence of At4/IPS1 noncoding RNAs (Fig. 3). The function of noncoding RNAs from the At4/IPS1 family was recently described to inhibit the cleavage of miR399 by mimicking target sequences of PHO2 (Franco-Zorrilla et al., 2007). The degree of complementary to the target sequences of PHO2 or consensus sequence of At4/IPS1 noncoding RNAs may explain the difference in cleavage efficiency between miR399b/c and miR399f. This differential targeting may be observed only when the amount of miR399 is limited, as in grafted plants; however, the low-targeting efficiency by miR399b/c can be compensated by a high level of expression, as with miR399b- or c-overexpressing transgenic plants (Chiou et al., 2006). In addition to finding this variation in nucleotide 13 in Arabidopsis, we also observed the variation in other miR399s from rice, Medicago, and Populus. Interestingly, like the G at position 13 in miR399b/c in Arabidopsis, that in the other miR399s results in mismatches to all their target sequences but better base-pairing to the homologues of At4/IPS1 noncoding RNAs in the species. We hypothesize that this feature may be involved in a mechanism to control the abundance of PHO2 mRNA through the regulation of targeting efficiency.
Previously, the miR399-guided cleavage of PHO2 mRNA was validated by 5′ RACE with Pi-replete tissues, and the target sites 2 and 3 appeared to be the predominant cleavage sites (Allen et al., 2005). In our assay, most of the 5′ end of PHO2 mRNA fragments was mapped to target sites 3, 4, and 5 on analyses of rootstocks of 399f/wt plants and roots of pho1 and Pi-starved wild-type plants (Supplemental Fig. S1). Because five miR399 target sites exist on PHO2 mRNA, mapping the 5′ end of RNA fragments on site 5 does not exclude the cleavage on the other upstream sites. The discrepancy among these assay results may be caused by differences in the abundance of miR399s in these samples.
Refining PHO2 by miR399-Mediated PHO2 siRNA
Production of transacting siRNAs (tasiRNAs) by miRNA-direct cleavage on noncoding TAS genes has been well documented (Vazquez et al., 2004; Allen et al., 2005; Williams et al., 2005; Yoshikawa et al., 2005). Like miRNAs, tasiRNAs function in trans to inhibit the expression of other genes. In addition, detection of siRNAs corresponding to several miRNA target genes in wild-type plants, but not the ago1 or dcl1 mutant, suggests that miRNAs can trigger the production of secondary siRNAs (Ronemus et al., 2006). Here, we revealed a population of PHO2 siRNAs that is accumulated in response to Pi deficiency and associated with the expression of miR399 (Fig. 7). These siRNAs may function to reinforce or fine-tune the regulation of PHO2 by miR399s. Detection of PHO2 siRNAs in the mutants dcl2, dcl3, dcl4, sgs3, rdr1, rdr2, and rdr6 suggests that the biogenesis of these siRNAs is different from that of tasiRNAs or may not be mediated on a single dicer or RNA-dependent RNA polymerase.
From current small-RNA databases, ASRP (http://asrp.cgrb.oregonstate.edu/db/download.html), MPSS (http://mpss.udel.edu/at/), and 454 pyrosequencing (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE5228), several small RNAs corresponding to the PHO2 gene were identified and shown in Figure 7A. The significance of these small RNAs is not clear because of their very low abundance in the databases, usually only 1 or 2 reads. None were located in the region of the probe giving the signals. This finding is not surprising, because the current small-RNA databases are generated from plants grown under Pi-sufficient conditions.
Because transgene-derived coat protein siRNAs move in the long-distance phloem across graft unions (Yoo et al., 2004), we wondered whether the PHO2 siRNAs can also move like miR399s. From RNA gel-blot results, we were unable to detect their movement. More sensitive detection methods are needed to validate their mobility. Because no specific sequence is available for these siRNAs, quantitative RT-PCR cannot be employed. Deep sequencing of small RNAs from phloem sap isolated from Pi-deficient plants will provide sequence information for these siRNAs.
Biological Relevance of the Movement of miR399
Time course analyses of Pi starvation treatment indicated the early induction of miR399s in shoots (before 12 h), whereas the induction is much slower in roots (after 1–2 d; Fig. 5A). However, the high accumulation of mature miR399s was noticed initially in roots rather than shoots despite the high accumulation of pri-miR399s in shoots (Fig. 5, A and B). This inconsistency between the accumulation of pri-miR399s and mature miR399s in shoots and roots offers indirect evidence for the movement of mature miR399s from shoots to roots. Earlier and greater induction of miR399s in shoots provides a reasonable explanation for the need for miR399 movement to roots. Shoot-derived miR399s are responsible for early clean-up of PHO2 mRNA in roots, where miR399s are not readily expressed. Of note, the suppression of PHO2 by miR399s has to take place in roots to activate Pi uptake from the rhizosphere and Pi translocation from roots to shoots. Such communication between shoots and roots could be crucial as one of the early defense machineries in response to Pi deficiency. For extended starvation, roots will express sufficient miR399s to suppress the PHO2 expression (Fig. 5, C and D). The role for the movement of miR399s during long-term Pi deficiency requires further study.
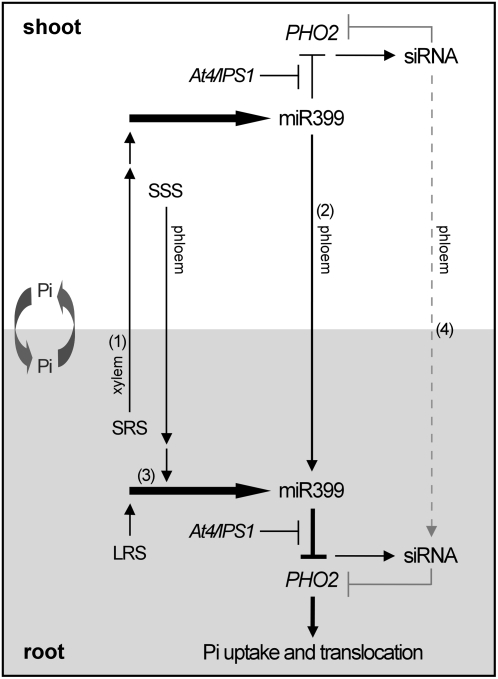
In Figure 8, we propose a hypothesis for signal communication between roots and shoots to regulate the expression of miR399s and PHO2. Upon initiation of Pi deficiency, roots first sense the changes and emit two types of response: one is the local response restricted to the roots directly exposed to the nutrient; the other is a systemic response of sending a signal to shoots (Forde, 2002; Atkins and Smith, 2007; Schachtman and Shin, 2007). A systemic root-born signal is delivered through the xylem to trigger the initial expression of miR399s in shoots (Fig. 8, 1). Mature miR399 produced in shoots move down to roots via a phloem stream (Fig. 8, 2). Soon after the induction of miR399s in shoots, a systemic shoot-born signal, alone or together with a local root-born signal, then activates the expression of miR399s in roots (Fig. 8, 3). Detection of pri-miR399s in pho1 roots (Fig. 6A) suggests that the activation of miR399s in pho1 roots is triggered by a signal derived from Pi-starved shoots. The existence of a local root-born signal is supported by the expression of miR399s in the detached Pi-starved roots (K. Aung, Y.-N. Chen, and T.-J. Chiou, unpublished data). The miR399s accumulated in roots, derived from shoots or de novo synthesis in roots, inhibit PHO2 expression and result in enhanced Pi uptake and translocation. Generation of PHO2 siRNAs by the targeting of miR399 may reinforce the suppression of PHO2 at a later stage (Fig. 8, 4). Movement of PHO2 siRNAs is not known and remains to be uncovered.
Figure 8.
A proposed model for the regulation of miR399 and PHO2 through signal communication between roots and shoots in response to Pi deficiency. SRS, Systemic root-born signal; SSS, systemic shoot-born signal; LRS, local root-born signal. Movement of PHO2 siRNAs is not known and is indicated as a dashed line. Allocation and recycling of Pi between shoots and roots may be involved in this long-distance signaling network and are indicated as circular arrows. See text for a detailed description.
The root-shoot communication is necessary for plants to survive through unfavorable nutrient conditions, because roots are responsible for the nutrient uptake and transportation that has to coordinate with the demand of shoots. As proposed previously, Pi allocation and recycling between shoots and roots is considered to provide the systemic signals (Drew and Saker, 1984; Jeschke et al., 1997; Burleigh and Harrison, 1999). Thus, Pi allocation and recycling may reflect the Pi status in different tissues. Reduced transport of Pi from roots to shoots during the onset of Pi deficiency may trigger the induction of miR399s in shoots. The miR399s produced in shoots serve as long-distance signals to activate Pi transport systems in roots by the inhibition of PHO2 expression. In the future, identification of the associated partners of miR399s during the long-distance transportation will provide further insight into the regulation of miRNA trafficking. Whether the phloem SMALL RNA BINDING PROTEIN1 (CmPSRP1) involved in small RNA trafficking in pumpkin (Yoo et al., 2004) also participates in the movement of miR399 awaits investigation. In addition to miR399, miR395 and miR398 were also detected in the phloem sap of Brassica napus under sulfate and copper starvation, respectively (Buhtz et al., 2008). The progress of these innovative studies has opened a new prospect for the systemic role of miRNAs in the regulation of nutrient homeostasis.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia and tobacco (Nicotiana benthamiana) wild-type and transgenic plants overexpressing miR399 were used for grafting experiments. pho1-2 (CS8507) was obtained from the Arabidopsis Biological Research Center. Seeds of small RNA biogenesis mutants were gifts from Drs. Xuemei Chen and Shu-Hsing Wu: dcl1-9 (CS3828), dcl2-1 (SALK_064627), dcl3-1 (SALK_005512), dcl4-2 (GABI_160G05), rdr1-1, rdr2-1 (Xie et al., 2004), rdr6-11 (CS24285), sgs3-11 (CS24289), hen1 (SALK_049197), and hen1-1 (Chen et al., 2002). For time-course analysis, wild-type seeds were surface sterilized and germinated on agar plates with one-half Murashige and Skoog medium for 9 d. The seedlings were then transferred to hydroponic culture with one-half Hoagland solution containing 250 μm KH2PO4 for an additional 8 d. Pi starvation was initiated by replacing 250 μm KH2PO4 with 10 μm KH2PO4. Seedlings were harvested at 1, 3, 6, 12, 24, and 48 h and 5 d after treatment. Nutrient solution was replenished every 5 d. Root and shoot samples were collected separately for Pi assay or RNA isolation. All growth conditions were at 22°C under a 16-hr photoperiod with cool fluorescent light at 100 to 150 μE m−2 s−1. The pho1 mutant was also grown hydroponically under Pi-sufficient conditions.
Grafting of Arabidopsis and Tobacco Plants
Hypocotyl reciprocal grafting was performed with wild-type plants and miR399-overexpressing transgenic (Chiou et al., 2006) and miR399f promoter∷reporter lines (Aung et al., 2006). For Arabidopsis, micrografting was performed as previously described (Turnbull et al., 2002) with minor modifications. Seeds were germinated on one-half Hoagland medium for 4 d, and seedlings were transferred to vertical plates and incubated in the dark for 2 d. Scions and rootstocks were obtained by use of a no. 15 scalpel blade, and grafting involved use of a dissecting microscope. Grafted plants were incubated vertically in the dark for 1 d prior to being transferred to the culture room. To observe and remove adventitious roots generated from scions, grafting was carried out without collars. Two weeks after grafting, plants were transferred to hydroponic culture and grew for another 10 d. Grafted plants were checked without adventitious roots before harvest. Scions and rootstocks were collected separately for Pi assay and RNA isolation. Lack of contamination of adventitious roots in the rootstocks of 399/wt grafted plants was confirmed by the absent expression of hygromycin-resistant gene located within the transgene by PCR. Tobacco grafting was conducted similar to that in Arabidopsis, except that 2-week-old plants were used for grafting and the samples were harvested after 1 month of hydroponic culture. All grafting experiments were repeated at least three times independently, and similar results were obtained. Data from one of them was presented.
Phosphate Content Analysis
Pi content was determined as described (Ames, 1966) with minor modifications (Chiou et al., 2006).
RNA Isolation and Quantitative RT-PCR
RNA isolation was performed as described previously (Lin et al., 2005) using TRIzol reagent (Invitrogen). Total RNA was treated with DNase I (Ambion) prior to quantitative RT-PCR analyses to eliminate genomic DNA contamination. cDNA was synthesized from 0.5 μg total RNA by Moloney murine leukemia virus reverse transcriptase (Promega) using oligo(dT) primer. Quantitative RT-PCR of pri-miR399 and PHO2 transcript involved the Power SYBR Green PCR Master Mix kit (Applied Biosystems) on the 7300 Real Time PCR system (Applied Biosystems) following the manufacturer's instructions. For comparable quantitation among different sets of primers, PCR efficiency was determined as described (Schmittgen et al., 2004) by a series of 2-fold dilutions of cDNAs. The calculated efficiency of all primers was 1.96 to 2.17. Relative expression levels were normalized to that of an internal control (UBQ10 for Arabidopsis and actin for tobacco) and presented as 2−ΔCT × 102 or 104 to simply the presentation of data (Livak and Schmittgen, 2001; Schmittgen et al., 2004). The primer sequences are listed in Supplemental Table S1. The primers for quantitation of PHO2 mRNA were designed across the second miRNA399 target site.
cDNAs used for quantitation RT-PCR of mature miRNA were synthesized from 10 ng of total RNA by use of the Taq-Man MicroRNA Reverse Transcription kit (Applied Biosystems). Content of specific mature miR399 species (miR399a, b/c, e, and f) was quantified by use of Taq-Man Gene Expression Master Mix combined with Taq-Man MicroRNA Assay (Applied Biosystems) following the manufacturer's instructions. Taq-Man MicroRNA Assays quantitate only mature miRNA, not the precursors. Relative expression levels were normalized to the amount of mature miR171b, whose expression was compatible in shoots and roots and was not altered in miR399-overexpressing lines or by Pi deficiency. Because a commercial detection kit for miR399d was unavailable, the specific primers and Taq-Man probe were designed in-house (see Supplemental Table S1). Two experimental repeats were performed for all PCR reactions.
According to the manufacturer's declaration, the Taq-Man MicroRNA assay is able to discriminate between highly homologous miRNA sequences differing by only a single nucleotide (Chen et al., 2005). We found that the detection is specific to each miR399 species, except for 19% to 28% of cross detection between miR399a and b/c (Supplemental Fig. S5). Thus, our assays possibly entailed overestimation of miR399a and b/c in Figures 5, B and D, and 6B.
RNA Gel-Blot Analysis
RNA gel-blot analysis was performed as described (Chiou et al., 2006). For the detection of PHO2 siRNAs, PCR fragments (sense probe [SP] and antisense probe [ASP]; see Supplemental Table S1 for primer sequences) containing the RNA polymerase promoter sequence (T7 or SP6) were used as templates for in vitro transcription reaction with the use of MAXIscript kit (Ambion) and [α-32P]UTP (800 Ci mmol−1; Perkin Elmer). Unincorporated 32P was removed by a ProbeQuant G25 microcolumn (Amersham Biosciences) before hybridization.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number EU375892 (N. benthamiana PHO2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Identification of miR399-guided cleavage products.
Supplemental Figure S2. Reciprocal grafts between wild-type and miR399c-overexpressing Arabidopsis plants.
Supplemental Figure S3. Overexpressing of Arabidopsis miR399b or miR399f in tobacco plants.
Supplemental Figure S4. Processing efficiency of miR171b and promoter activities of miR399a, b, c, or f in shoots versus in roots.
Supplemental Figure S5. Specificity of Taq-Man miRNA assays among different miR399 species.
Supplemental Table S1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Drs. Shu-Hsing Wu and Chun-lin Su for critically reading the manuscript and Drs. Wolf-Rüdiger Scheible and William Lucas for discussion of the results. We also thank Ho-Ming Chen for analysis of PHO2 small RNAs and Ya-Shiuan Lai and Ya-Ni Chen for technical assistance.
This work was supported by the Academia Sinica and by the National Science Council of the Republic of China (grant nos. 95–2311–B–001–044–MY3 and 96–2321–B–001–018 to T.-J.C.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Tzyy-Jen Chiou (tjchiou@gate.sinica.edu.tw).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Allen E, Xie Z, Gustafson AM, Carrington JC (2005) microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121 207–221 [DOI] [PubMed] [Google Scholar]
- Alvarez JP, Pekker I, Goldshmidt A, Blum E, Amsellem Z, Eshed Y (2006) Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell 18 1134–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames BN (1966) Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol 8 115–118 [Google Scholar]
- Atkins CA, Smith PMC (2007) Translocation in legumes: assimilates, nutrients, and signaling molecules. Plant Physiol 144 550–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung K, Lin SI, Wu CC, Huang YT, Su Cl, Chiou TJ (2006) pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol 141 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Jan C, Rajagopalan R, Bartel DP (2006) A two-hit trigger for siRNA biogenesis in plants. Cell 127 565–577 [DOI] [PubMed] [Google Scholar]
- Bari R, Datt Pant B, Stitt M, Scheible WR (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates TR, Lynch JP (1996) Stimulation of root hair elongation in Arabidopsis thaliana by low phosphorus availability. Plant Cell Environ 19 529–538 [Google Scholar]
- Bonnet E, Van de Peer Y, Rouze P (2006) The small RNA world of plants. New Phytol 171 451–468 [DOI] [PubMed] [Google Scholar]
- Buhtz A, Springer F, Chappell L, Baulcombe DC, Kehr J (2008) Identification and characterization of small RNAs from the phloem of Brassica napus. Plant J 53 739–749 [DOI] [PubMed] [Google Scholar]
- Burleigh SH, Harrison MJ (1999) The down-regulation of Mt4-like genes by phosphate fertilization occurs systemically and involves phosphate translocation to the shoots. Plant Physiol 119 241–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al (2005) Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33 e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X (2005) microRNA biogenesis and function in plants. FEBS Lett 579 5923–5931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Liu J, Cheng Y, Jia D (2002) HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development 129 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ (2007) The role of microRNAs in sensing nutrient stress. Plant Cell Environ 30 323–332 [DOI] [PubMed] [Google Scholar]
- Chiou TJ, Aung K, Lin SI, Wu CC, Chiang SF, Su Cl (2006) Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 18 412–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Randall PJ (1995) Characterization of a phosphate-accumulator mutant of Arabidopsis thaliana. Plant Physiol 107 207–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B, Rengel Z, Delhaize E (1998) Uptake and translocation of phosphate by pho2 mutant and wild-type seedlings of Arabidopsis thaliana. Planta 205 251–256 [DOI] [PubMed] [Google Scholar]
- Drew MC, Saker LR (1984) Uptake and long-distance transport of phosphate, potassium and chloride in relation to internal ion concentrations in barley: evidence of non-allosteric regulation. Planta 160 500–507 [DOI] [PubMed] [Google Scholar]
- Fang Y, Spector DL (2007) Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr Biol 17 818–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde BG (2002) The role of long-distance signalling in plant responses to nitrate and other nutrients. J Exp Bot 53 39–43 [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, Garcia JA, Paz-Ares J (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39 1033–1037 [DOI] [PubMed] [Google Scholar]
- Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK (2005) A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol 15 2038–2043 [DOI] [PubMed] [Google Scholar]
- Hamburger D, Rezzonico E, MacDonald-Comber Petetot J, Somerville C, Poirier Y (2002) Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 14 889–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeschke WD, Kirkby EA, Peuke AD, Pate JS, Hartung W (1997) Effects of P deficiency on assimilation and transport of nitrate and phosphate in intact plants of castor bean (Ricinus communis L.). J Exp Bot 48 75–91 [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol 57 19–53 [DOI] [PubMed] [Google Scholar]
- Juarez MT, Kui JS, Thomas J, Heller BA, Timmermans MCP (2004) microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428 84–88 [DOI] [PubMed] [Google Scholar]
- Li J, Yang Z, Yu B, Liu J, Chen X (2005) Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol 15 1501–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SI, Wang JG, Poon SY, Su Cl, Wang SS, Chiou TJ (2005) Differential regulation of FLOWERING LOCUS C expression by vernalization in cabbage and Arabidopsis. Plant Physiol 137 1037–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-[delta][delta]CT method. Methods 25 402–408 [DOI] [PubMed] [Google Scholar]
- Lough TJ, Lucas WJ (2006) Integrative plant biology: role of phloem long-distance macromolecular trafficking. Annu Rev Plant Biol 57 203–232 [DOI] [PubMed] [Google Scholar]
- Mallory AC, Vaucheret H (2006) Functions of microRNAs and related small RNAs in plants. Nat Genet 38 S31–S36 [DOI] [PubMed] [Google Scholar]
- Marschner H (1995) Mineral Nutrition of Higher Plants. Academic Press, London
- Martin AC, del Pozo JC, Iglesias J, Rubio V, Solano R, de la Pena A, Leyva A, Paz-Ares J (2000) Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. Plant J 24 559–567 [DOI] [PubMed] [Google Scholar]
- Pant BD, Buhtz A, Kehr J, Scheible WR (2008) MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J 53 731–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X (2002) CARPEL FACTORY, a dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12 1484–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier Y, Bucher M (2002) Phosphate transport and homeostasis in Arabidopsis. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. The American Society of Plant Biologists, Rockville, MD, doi: 10.1199/tab.0024, www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed]
- Poirier Y, Thoma S, Somerville C, Schiefelbein J (1991) A mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol 97 1087–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama KG (1999) Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol 50 665–693 [DOI] [PubMed] [Google Scholar]
- Robinson D (1994) The responses of plants to non-uniform supplies of nutrients. New Phytol 127 635–674 [DOI] [PubMed] [Google Scholar]
- Ronemus M, Vaughn MW, Martienssen RA (2006) MicroRNA-targeted and small interfering RNA-mediated mRNA degradation is regulated by Argonaute, Dicer, and RNA-dependent RNA polymerase in Arabidopsis. Plant Cell 18 1559–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martin AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtman DP, Shin R (2007) Nutrient sensing and signaling: NPKS. Annu Rev Plant Biol 58 47–69 [DOI] [PubMed] [Google Scholar]
- Schikora A, Schmidt W (2001) Iron stress-induced changes in root epidermal cell fate are regulated independently from physiological responses to low iron availability. Plant Physiol 125 1679–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Jiang J, Liu Q, Yang L (2004) A high-throughput method to monitor the expression of microRNA precursors. Nucleic Acids Res 32 e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R, Chiang VL (2005) Facile means for quantifying microRNA expression by real-time PCR. Biotechniques 39 519–525 [DOI] [PubMed] [Google Scholar]
- Sunkar R, Chinnusamy V, Zhu J, Zhu JK (2007) Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci 12 301–309 [DOI] [PubMed] [Google Scholar]
- Ticconi CA, Delatorre CA, Lahner B, Salt DE, Abel S (2004) Arabidopsis pdr2 reveals a phosphate-sensitive checkpoint in root development. Plant J 37 801–814 [DOI] [PubMed] [Google Scholar]
- Turnbull CGN, Booker JP, Leyser HMO (2002) Micrografting techniques for testing long-distance signalling in Arabidopsis. Plant J 32 255–262 [DOI] [PubMed] [Google Scholar]
- Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crete P (2004) Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell 16 69–79 [DOI] [PubMed] [Google Scholar]
- Williams L, Carles CC, Osmont KS, Fletcher JC (2005) A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proc Natl Acad Sci USA 102 9703–9708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC (2004) Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2 E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Ebright YW, Yu B, Chen X (2006) HEN1 recognizes 21-24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res 34 667–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo BC, Kragler F, Varkonyi-Gasic E, Haywood V, Archer-Evans S, Lee YM, Lough TJ, Lucas WJ (2004) A systemic small RNA signaling system in plants. Plant Cell 16 1979–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M, Peragine A, Park MY, Poethig RS (2005) A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev 19 2164–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Tao X, Stombaugh J, Leontis N, Ding B (2007) Tertiary structure and function of an RNA motif required for plant vascular entry to initiate systemic trafficking. EMBO J 26 3836–3846 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.