Abstract
Abscisic acid (ABA) is a plant hormone found in all higher plants; it plays an important role in seed dormancy, embryo development, and adaptation to environmental stresses, most notably drought. The regulatory step in ABA synthesis is the cleavage reaction of a 9-cis-epoxy-carotenoid catalyzed by the 9-cis-epoxy-carotenoid dioxygenases (NCEDs). The parasitic angiosperm Cuscuta reflexa lacks neoxanthin, one of the common precursors of ABA in all higher plants. Thus, is C. reflexa capable of synthesizing ABA, or does it acquire ABA from its host plants? Stem tips of C. reflexa were cultured in vitro and found to accumulate ABA in the absence of host plants. This demonstrates that this parasitic plant is capable of synthesizing ABA. Dehydration of detached stem tips caused a big rise in ABA content. During dehydration, 18O was incorporated into ABA from 18O2, indicating that ABA was synthesized de novo in C. reflexa. Two NCED genes, CrNCED1 and CrNCED2, were cloned from C. reflexa. Expression of CrNCEDs was up-regulated significantly by dehydration. In vitro enzyme assays with recombinant CrNCED1 protein showed that the protein is able to cleave both 9-cis-violaxanthin and 9′-cis-neoxanthin to give xanthoxin. Thus, despite the absence of neoxanthin in C. reflexa, the biochemical activity of CrNCED1 is similar to that of NCEDs from other higher plants. These results provide evidence for conservation of the ABA biosynthesis pathway among members of the plant kingdom.
Abscisic acid (ABA) is found in all higher plants and algae and is also produced by some fungi (Oritani and Kiyota, 2003; Schwartz and Zeevaart, 2004; Nambara and Marion-Poll, 2005). In higher plants, ABA is involved in seed dormancy, embryo development, and adaptation to various abiotic stresses. ABA is a sesquiterpenoid (C15). In some fungi, there is a direct pathway from isopentenyl pyrophosphate (C5) via farnesyl pyrophosphate (C15; Oritani and Kiyota, 2003). Higher plants synthesize ABA via the C40 indirect pathway. A C40 carotenoid is oxidatively cleaved to form a C25 byproduct and the C15 precursor of ABA, xanthoxin. Biochemical and molecular evidence has shown that the cleavage reaction is the rate-limiting step in the ABA biosynthetic pathway.
Although the pathway of ABA biosynthesis in higher plants has been well established, there are still a few unresolved questions. One is the endogenous substrate of the cleavage reaction. The biochemical evidence has indicated that the C40 substrate for production of biologically active ABA is an epoxy-carotenoid in the 9-cis configuration in order for biologically active ABA to be produced. In higher plants, the major 9-cis-epoxy-carotenoids are 9′-cis-neoxanthin and 9-cis-violaxanthin. Because 9′-cis-neoxanthin is the most abundant 9-cis-epoxy-carotenoid in higher plants, it has been speculated that 9′-cis-neoxanthin is the main endogenous substrate of ABA. However, in vitro enzyme assays with recombinant 9-cis-epoxy-carotenoid dioxygenase (NCED) proteins from several plant species have shown that NCEDs are capable of cleaving both 9′-cis-neoxanthin and 9-cis-violaxanthin (Schwartz et al., 1997; Qin and Zeevaart, 1999; Chernys and Zeevaart, 2000), with a higher activity when 9-cis-violaxanthin is used as substrate (Schwartz et al., 2003b). This raised the question whether one or both are the endogenous substrates of ABA biosynthesis.
Cuscuta reflexa is of interest in addressing this question, because it lacks 9′-cis-neoxanthin, one of the major 9-cis-epoxy-carotenoids found in green plants. A study by Bungard et al. (1999) investigated the xanthophyll-carotenoid complement of the main light-harvesting complex involved in light capture of photosynthesis. They found that in most higher plants, the complex is highly conserved and includes, in addition to chlorophyll, the carotenoids neoxanthin, violaxanthin, and lutein. In C. reflexa, neoxanthin is replaced by another xanthophyll, lutein-5,6-epoxide. Therefore, C. reflexa lacks the step that converts violaxanthin to neoxanthin. There are two xanthophyll cycles in C. reflexa: under excess irradiance violaxanthin and lutein-5,6-epoxide are converted to zeaxanthin and lutein, respectively, by de-epoxidation; the cycle is reversed under low irradiance (Bungard et al., 1999).
C. reflexa is parasitic on the above-ground parts of other plants. The leafless stems coil around the host stems and petioles. C. reflexa contains only a small amount of chlorophyll and relies on obtaining photosynthetic assimilates from the phloem of its host. It forms specialized structures, called haustoria, which are used to acquire nutrients from its host plant (Hibberd et al., 1998).
The lack of 9′-cis-neoxanthin in C. reflexa may result in an inability of the plant to produce ABA, and the parasite may acquire it from its host, thereby eliminating the need to synthesize ABA on its own (Bungard et al., 1999). The objective of this study was to determine whether C. reflexa is able to synthesize ABA or acquires it from its host. Increased ABA in C. reflexa grown independently of a host would indicate that it is capable of ABA synthesis. We also searched for the presence of an NCED gene in the Cuscuta genome. Presence of an NCED gene would be evidence for the 9-cis-epoxy-carotenoid cleavage reaction, the regulatory step for ABA synthesis. Our results show that despite the lack of 9′-cis-neoxanthin, C. reflexa is capable of ABA synthesis in a manner similar to that in other higher plants.
RESULTS
Cultured Cuscuta Shoot Tips Accumulate ABA
ABA present in Cuscuta stems could be synthesized in situ, synthesized in the host plant and imported by the parasite, or acquired by a combination of import and endogenous synthesis. To determine whether Cuscuta is capable of producing ABA, it is necessary to grow the parasite independently of a host, as first reported by Baldev (1962).
When stem tips were cultured in vitro for approximately 20 d, the stems became thicker and much greener than material taken directly from host plants (Fig. 1). In our three experiments, the ABA content increased 4- to 8-fold over that initially present in the shoot tips (Table I). This result demonstrates that ABA was synthesized by the cultured shoot tips. In other words, Cuscuta is capable of producing ABA independently of a host plant.
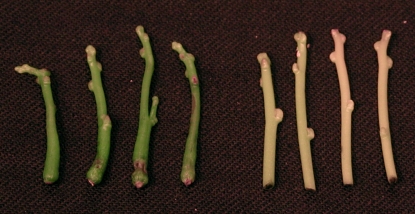
Figure 1.
Stem tips of C. reflexa. Left, Four stems cultured for 23 d with thickened stem at base and green color. Right, Four stems detached from plants parasitizing on Perilla have a pale yellow-orange color.
Table I.
Accumulation of ABA in Cuscuta shoot tips cultured in vitro
Cuscuta stem tips were grown in sterile culture under continuous light for approximately 20 d. Control represents ABA content of the tips prior to culture. Results of three separate experiments are presented.
| ABA Content | |||
|---|---|---|---|
| μg g−1DW | |||
| Experiment | 1 | 2 | 3 |
| Control tips | 5.2 | 12.5 | 3.9 |
| Cultured tips | 43.8 | 54.4 | 34.3 |
Dehydration of Cuscuta Shoot Tips Stimulates ABA Accumulation
Leaves of mesophytic plants can usually be dehydrated to 85% to 90% of their initial fresh weight (FW) by a stream of warm air from a hair dryer in a few minutes. The ABA content of such water-stressed leaves increases rapidly for approximately 4 h and then levels off (e.g. Qin and Zeevaart, 1999). C. reflexa produces only stems, which cannot be quickly dehydrated with a hair dryer. Instead, we dehydrated stem tips gradually by placing them in a desiccator in the presence of Drierite. Such water-stressed stems lost 20% to 30% of their FW over a 24-h period and then showed signs of loss of turgidity. The results in Table II show that dehydration caused a large increase in ABA content. Thus, water stress induces ABA accumulation in Cuscuta stems, as it does in leaves of nonparasitic plants. However, ABA content also increased in nonstressed stem tips, albeit much less than in dehydrated stems. This increase in ABA occurred even when the stems were stored in polythene bags in the presence of some water. This observation is in contrast to turgid detached leaves, which maintained turgor and never showed an increase in ABA when kept in polythene bags (Zeevaart, 1980). When Cuscuta stem tips are detached, a brown viscous fluid exudes from the cut surface, perhaps reducing turgor in the stems and inducing ABA biosynthesis (Pierce and Raschke, 1980).
Table II.
Dehydration induces ABA accumulation in Cuscuta stem tips
Detached stem tips of Cuscuta were kept in plastic bags (control) or in a desiccator in the presence of Drierite. Results of four separate experiments are presented. nd, Not determined.
| ABA Content | ||||
|---|---|---|---|---|
| μg g−1FW | ||||
| Experiment | 1 | 2 | 3 | 4 |
| Initial | 0.8 | 0.9 | 0.7 | 0.8 |
| 8 h control | nd | nd | 0.9 | 0.8 |
| 8 h dehydration | nd | nd | 3.8 | 3.1 |
| 24 h control | 4.4 | 2.9 | 6.1 | 2.9 |
| 24 h dehydration | 13.1 | 11.0 | 13.6 | 11.1 |
Incorporation of 18O into ABA during Dehydration
Previous work has established that ABA is a cleavage product of 9′-cis-epoxy-carotenoids (Zeevaart et al., 1989, 1991). Whether the carotenoid cleavage reactions are catalyzed by monooxygenases or dioxygenases has been debated (Leuenberger et al., 2001). For the biosynthesis of ABA and related compounds in plants, there is compelling evidence for a dioxygenase-catalyzed reaction (Schmidt et al., 2006). Consequently, when ABA synthesis takes place in an atmosphere enriched in 18O2, one 18O is incorporated into the carboxyl group of ABA. In long-term experiments, especially with tissues having a small xanthophyll pool, this pool is used up and has to be replenished, so that 18O is also incorporated into the ring positions of xanthophylls that become the 1′-hydroxyl and 4′-keto oxygens of ABA. Under these conditions, three atoms of 18O are incorporated into newly synthesized ABA (Zeevaart et al., 1989, 1991).
To demonstrate that ABA is synthesized de novo in Cuscuta and not released from a conjugated form, stem tips were incubated in an 18O2 atmosphere in the presence of Drierite for 24 h. ABA isolated from these shoots was analyzed as the methyl ester by chemical ionization mass spectrometry (MS). Instead of a single M− at mass-to-charge ratio (m/z) 278 as in unlabeled ABA, we observed a cluster of ions representing M−, M− +2, M− +4, and M− +6 (Table III). Thus, in addition to the carboxyl group of the side chain, the two O-atoms of the ring also became enriched in 18O. In the two experiments, 7% and 16%, respectively, of the ABA molecules had acquired three 18O atoms. These results indicate that dehydration resulted in extensive ABA biosynthesis. Furthermore, we can conclude that xanthophylls became depleted during the 24-h incubation period and had to be replenished by oxidation of carotenoid precursors.
Table III.
Incorporation of 18O into ABA in water-stressed shoot tips of Cuscuta
In two experiments, Cuscuta stem tips (25 and 20, respectively) were incubated in an atmosphere containing 20% 18O2 in the presence of Drierite for 24 h. The tips lost 30% of their FW during the 24-h incubation period. The m/z values 278 to 284 represent the relative abundances of the molecular ion cluster with zero, one, two, and three 18O atoms incorporated, respectively.
| Experiment | ABA Content | Relative Abundance | Labeled | |||
|---|---|---|---|---|---|---|
| μg | m/z | % | ||||
| 278 | 280 | 282 | 284 | |||
| 1 | 5.3 | 48 | 100 | 61 | 16 | 79 |
| 2 | 4.0 | 21 | 100 | 87 | 39 | 91 |
Isolation of C. reflexa NCED Genes
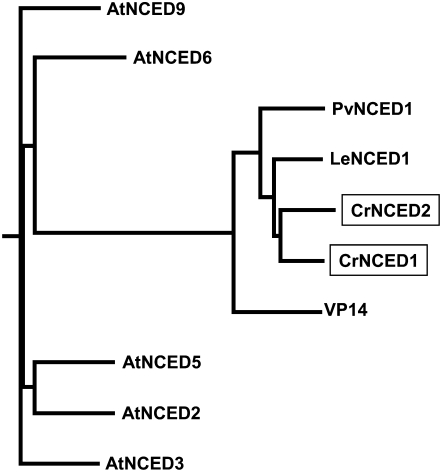
All higher plant NCED genes identified to date lack introns, thus providing an approach to isolate C. reflexa NCEDs from genomic DNA. The initial 625-bp partial sequence obtained by degenerate primers showed strong homology to other known NCEDs. A Southern blot hybridized with this partial fragment at high stringency showed two bands when SspI was used to restriction digest C. reflexa genomic DNA. Thus, we suspected that there may be two NCED sequences present in the C. reflexa genome and that they share significant sequence identity in this region. The inverse PCR identified two fragments, both NCED-related sequences. The final CrNCED clones were amplified from C. reflexa genomic DNA containing only the open reading frames. We assigned CrNCED1 to the clone that contained the exact 625-bp sequence obtained by the initial degenerate PCR and CrNCED2 to the second clone. Neither gene shows introns in its sequence. At the region where the degenerate primers were used to amplify the initial CrNCED1 fragment, CrNCED1 and CrNCED2 share 85.1% identity at the nucleic acid level. This explains why two bands were obtained when the Southern blot was probed with the CrNCED1 fragment. The CrNCED1 and CrNCED2 genes encode proteins of 586 and 636 amino acids, respectively (Supplemental Fig. S1). At the amino acid level, the sequence of CrNCED1 shows 76.1% identity with the tomato (Solanum lycopersicum, formerly Lycopersicon esculentum) LeNCED1, 68.7% identity with the bean (Phaseolus vulgaris) PvNCED1, and 61.0% identity with the maize (Zea mays) VP14. The CrNCED proteins are very similar to other NCEDs (Fig. 2). The presence of NCED genes in C. reflexa demonstrates that this parasitic plant possesses the key enzyme of the ABA biosynthetic pathway in higher plants. This suggests that C. reflexa is able to synthesize ABA via the same pathway as other higher plants, despite its parasitic habit.
Figure 2.
Phylogenetic tree of the NCED subfamily. Sequence data can be found in the GenBank/EMBL data libraries under the following accession numbers: C. reflexa (Cr), CrNCED1 (AY974807) and CrNCED2 (AY974808); Arabidopsis (At), AtNCED2 (NP-193569), AtNCED3 (NP-188062), AtNCED5 (NP-174302), AtNCED6 (NP-189064), and AtNCED9 (NP-177960); bean (Pv), PvNCED1 (AF190462); tomato (Le), LeNCED1 (AJ439079); and viviparous maize (VP), VP14 (U95953). Alignment was performed with the ClustalW method using the deduced full-length protein sequences. The two CrNCEDs are enclosed in boxes.
Northern-Blot Analysis of the Two CrNCED Genes
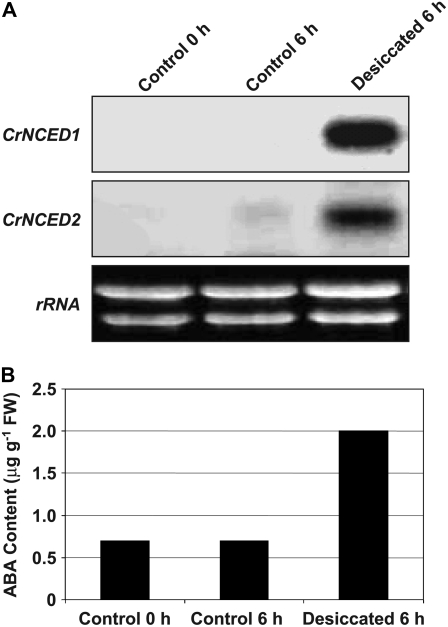
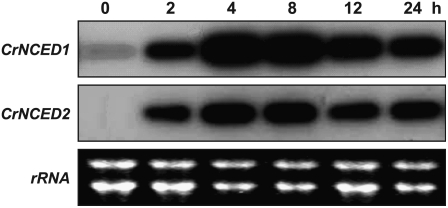
We studied expression of CrNCED1 and CrNCED2 in relation to dehydration by northern blotting of RNA isolated from C. reflexa stem tips. Because of the high identity of the two genes at the nucleotide level, we designed gene-specific probes (Supplemental Fig. S2). Figure 3 shows the expression of CrNCED1 and CrNCED2, as well as ABA accumulation, in detached C. reflexa stem tips prior to and during desiccation. Expression of both CrNCEDs was strongly induced after 6 h of dehydration. Weak expression of CrNCED2 in the control after 6 h supports the finding that nonstressed stems in long-term treatments accumulate ABA (Table II). The time course of CrNCED expression during dehydration is shown in Figure 4. Clearly, the level of transcripts was much increased after 2 h of dehydration, reached a maximum around 4 h, and then slowly declined. This pattern of expression is similar to that of PvNCED1 in water-stressed primary bean leaves (Qin and Zeevaart, 1999).
Figure 3.
Expression patterns of CrNCED1 and CrNCED2 and ABA content in detached C. reflexa shoot tips after 6 h of desiccation. Approximately 15 shoot tips per treatment were incubated in polythene bags or in the presence of Drierite for 6 h. Desiccated shoot tips had lost 10.7% of their FW after 6 h. A, Transcript levels of CrNCED1 and CrNCED2. The blot was successively probed with the 3′UTRs of CrNCED1 and CrNCED2, respectively. B, ABA content.
Figure 4.
Time course of changes in expression of CrNCEDs during dehydration of detached C. reflexa tips. Approximately 15 shoot tips per treatment were incubated in the presence of Drierite for various periods of time. Transcript levels of CrNCED1 and CrNCED2 are shown after 0, 2, 4, 8, 12, or 24 h. The blot was successively probed with the 3′UTRs of CrNCED1 and CrNCED2.
Biochemical Activity of CrNCED1
In higher plants, 9′-cis-neoxanthin is the most abundant epoxy-carotenoid in the cis configuration that can be used as substrate for ABA biosynthesis. However, in vitro assays with NCEDs from a variety of plants showed that recombinant protein has higher catalytic activity with 9-cis-violaxanthin than with 9′-cis-neoxanthin. The absence of neoxanthin in C. reflexa raised the question whether CrNCEDs have lost their ability to cleave neoxanthin. We produced recombinant protein of CrNCED1 and CrNCED2 in Escherichia coli strain BL21 cells. The induction levels were very high; however, both of the proteins remained mostly in the insoluble fraction. Purified CrNCED1 protein was concentrated with a Millipore Ultra-free 0.5 centrifugal filter and used for enzyme assays. The protein was able to cleave both 9-cis-violaxanthin and 9′-cis-neoxanthin. The cleavage product was identified as xanthoxin by gas chromatography (GC)-MS (Supplemental Fig. S3). This result indicates that the function of NCED is conserved in C. reflexa. As reported with other NCEDs, CrNCED1 also had higher activity with 9-cis-violaxanthin as substrate than with 9′-cis-neoxanthin.
DISCUSSION
Elucidation of the ABA biosynthetic pathway has been mainly achieved by characterization of ABA-deficient mutants and cloning of the corresponding genes (Schwartz et al., 2003a). So far, no gene has been isolated for the conversion of violaxanthin to neoxanthin. However, North et al. (2007) isolated a mutant, aba4, which lacks neoxanthin, accumulates violaxanthin, is deficient in ABA, and has a mild phenotype. In seeds and nonstressed vegetative tissue, sufficient ABA was synthesized (from 9-cis-violaxanthin) to give a normal phenotype. But upon dehydration, ABA accumulation was severely limited so that it was concluded that stress-induced ABA is predominantly derived from neoxanthin (North et al., 2007). However, it cannot be ruled out that under water stress, the substrate becomes limiting, considering that in these mutant plants 9-cis-violaxanthin will probably occupy the sites vacated by 9′-cis-neoxanthin in the major light-harvesting complex (Snyder et al., 2004).
The holoparasite C. reflexa also lacks neoxanthin (Bungard et al., 1999; J.A.D. Zeevaart, unpublished data). Despite the absence of neoxanthin in C. reflexa, this plant is capable of synthesizing ABA in the absence of a host, as shown by the following evidence. First, stem tips cultured in vitro separate from a host increase their ABA content (Table I). Second, dehydration of detached stem tips results in a large increase in ABA (Table II). Third, labeling stem tips with 18O2 during dehydration yields ABA that is enriched in one to three 18O atoms (Table III), thus conclusively demonstrating that ABA is synthesized de novo during water stress. The labeling of ABA with more than one 18O atom shows that the xanthophyll substrate was limiting and that precursor carotenoids were converted to 9-cis-epoxy-carotenoids during the 24-h incubation.
Considering the high level of 9-cis-violaxanthin in C. reflexa (Bungard et al., 1999; Snyder et al., 2005), it is likely that this xanthophyll is the C40 substrate for ABA. C. reflexa also contains lutein-5,6-epoxide, but because it is in the all-trans form (Bungard et al., 1999), it cannot serve as a precursor for ABA.
Bungard et al. (1999) speculated that host-acquired ABA might have eliminated the need for ABA biosynthesis in C. reflexa, which would have resulted in redundancy of neoxanthin. However, our results demonstrate that despite the lack of neoxanthin, C. reflexa does synthesize its own ABA like other higher plants.
Cuscuta stems are connected via haustoria with their host plants to obtain organic and inorganic nutrients, as well as water, from the host. Symplastic connections also permit transfer of proteins, viruses (Birschwilks et al., 2006, 2007), and even mRNA (Roney et al., 2007) from host to parasite. ABA is transported in both phloem and xylem (Zeevaart and Boyer, 1984), so that ABA produced in the host is undoubtedly transferred to Cuscuta. Stem tips from plants parasitizing on water-stressed Perilla contained 3 times more ABA than tips from well-watered plants (J.A.D. Zeevaart, unpublished data). However, in this case, ABA synthesized in the host and parasite cannot be distinguished. To study transfer of host ABA to the parasite, a Cuscuta species devoid of chlorophyll, such as C. odorata (Berg et al., 2003), and probably also lacking carotenoids would be suitable material.
Whereas ABA biosynthesis is preserved in C. reflexa, other aspects of ABA metabolism may differ from other plants. For example, rehydration of water-stressed stem tips did not cause a rapid decline in ABA levels. Furthermore, no phaseic acid (PA) or dihydrophaseic acid (DPA) could be detected in Cuscuta stem tips (J.A.D. Zeevaart, unpublished data). This is surprising, because even if Cuscuta does not convert ABA to PA and DPA, it would at least acquire these metabolites from the host plant. It is possible that Cuscuta rapidly conjugates or degrades both PA and DPA, so that the steady-state levels are below the level of detection. Thus, how ABA is catabolized by Cuscuta and the possible presence of genes encoding ABA 8′-hydroxylase (Kushiro et al., 2004; Yang and Zeevaart, 2006) remain for future studies.
MATERIALS AND METHODS
Plant Material
Seeds of Cuscuta reflexa were obtained from Dr. B. Baldev (Baldev, 1962). The seeds were germinated in a petri dish, and developing seedlings were clamped in cut stems of Ricinus communis plants. Later, Perilla ocymoides (green perilla) was used as host species for Cuscuta. The plants were grown in a greenhouse in which the natural daylength was extended with light from incandescent bulbs to prevent flowering in Cuscuta and Perilla, which are both short-day plants.
For in vitro culture, shoot tips (approximately 2 cm long) were cut from Cuscuta stems. The tips were surface sterilized in a 5% bleach solution for 5 min followed by rinsing in sterile distilled water. The tips were then placed in culture tubes containing 10 mL culture medium and with their basal ends inserted into the medium. The medium consisted of Murashige and Skoog basal medium with Gamborg vitamins, 5% Suc, and 0.8% agar. The cultures were maintained at 23°C for 3 weeks with continuous light from fluorescent lamps at a photo flux density of 40 μmol m−2 s−1. Upon harvest, the tips were frozen in liquid N2 and lyophilized.
For desiccation experiments, 3-cm stem tips (15–20 tips per treatment) were placed in a desiccator in the presence of Drierite (anhydrous calcium sulfate, 8 mesh). For labeling with 18O2, shoot tips were placed in the presence of Drierite in a 250-mL Erlenmeyer flask sealed with a serum stopper. The procedure for incubation with 18O2 (95%; Cambridge Isotope Labs) was as described by Creelman and Zeevaart (1984). The 18O2 in the flask was replenished after 12 h.
ABA Extraction, Purification, and Measurement
These procedures were as described by Tian et al. (2004). ABA was expressed in micrograms per gram dry weight (DW) or in micrograms per gram FW. The latter was used for desiccated shoot tips, which retained some moisture following lyophilization. DW was approximately 20% of FW.
GC-NCI-MS of Me-ABA was performed on a Waters GCT Premier Micromass mass spectrometer equipped with a 6890N HP gas chromatograph (Agilent). The column used was an SLB-5ms fused silica capillary column (10-m × 0.1-mm × 0.1-μm film thickness; Supelco) with helium as the carrier gas (flow rate 0.5 mL min−1). Gas-liquid chromatography conditions were as follows: the oven temperature was kept at 100°C for 1 min, then programmed from 100°C to 200°C at 25°C min−1, and finally from 200°C to 250°C at 12°C min−1. Methane was used as the reagent gas. The spectra were recorded at 10 scans per second at a collision energy of 70 eV in the mass range m/z 50 to 320.
Isolation of C. reflexa Genomic DNA
One gram of C. reflexa tips was pulverized in liquid N2 and extracted in 5 mL of extraction buffer (2.1 g urea, 0.5 m NaCl, 50 mm Tris-HCl at pH 8.0, 20 mm EDTA at pH 8.0, and 1% Sarkosyl detergent). An equal volume of phenol:chloroform (1:1, v/v) was added to the mixture and the sample was centrifuged for 5 min at 5,000g to separate the aqueous phase from the organic solvents. DNA in the supernatant was precipitated with 10 mL ethanol. The DNA pellet was then resuspended in water.
Cloning Fragments of NCED Genes
Based on the sequences of NCED genes from maize (Zea mays), bean (Phaseolus vulgaris), and Arabidopsis (Arabidopsis thaliana), two degenerate primers were designed to amplify a partial sequence of an NCED from C. reflexa. The primers were: JZ1114 (forward, 5′-GTNTTT/CCCNAAA/GGCNATA/C/TGG-3′) and JZ1142 (reverse, 5′-CCANGCG/AAACCANAG/AG/ATGG/AAA-3′; Supplemental Fig. S1). PCR yielded a fragment of 625 bp. Sequence analysis of this fragment and comparison with known NCEDs indicated that it was a putative partial NCED.
A Southern blot was performed with the partial CrNCED1 as probe to identify a suitable restriction enzyme that would generate a genomic fragment with the full-length CrNCED1 gene. Digestion of genomic DNA with SspI produced two bands at approximately 3.2 and 3.4 kb. This restriction enzyme was, therefore, selected for use in inverse PCR. One microgram of genomic DNA was digested with SspI for more than 6 h. The DNA fragments were ligated in a total volume of 600 μL at 4°C overnight in favor of self-ligation. The DNA was then precipitated with 2 volumes of ethanol and resuspended in 10 μL of distilled water. One microliter of this DNA was used in the first round PCR reaction with primers JZ1152 (forward, 5′-CGTCGTCGTCCCCGACCAGC-3′) and JZ1154 (reverse, 5′-CAGGTCGCCGGACGGTGTTAC-3′). The resulting product was diluted 100-fold with distilled water, and 1 μL of this diluted product was used as a template for a second round nested PCR amplification with primers JZ1151 (forward, 5′-CCACGGCGGCTCCCCGGTGG-3′) and JZ1153 (reverse, 5′-TAGACCAGACCGGCGTTGGC-3′). Two fragments were visible on agarose gel at sizes of 2.9 and 3.1 kb, respectively, which match the calculated size based on the Southern blot. The two fragments were cloned into the pGEM-T Easy vector (Promega) for sequencing at the MSU Research Technology Support Facility.
Cloning of CrNCED1 and CrNCED2 and the Construction of Bacterial Expression Vectors
The sequences of the two C. reflexa genomic fragments indicated that both are putative NCED sequences. The starting and stop codons were predicted according to other available NCED sequences. To amplify the CrNCED1 and CrNCED2 clones for construction of expression vectors, the primers used were: JZ1300 (CrNCED1 forward: 5′-ATGGCGAATTCTTTGTATAAACCC-3′) and JZ1301 (CrNCED1 reverse: 5′-CTAGACTTGGGTGGCCAAGTC-3′); and JZ1302 (CrNCED2 forward: 5′-TCCATGTTGCAACACGTTGG-3′) and JZ1303 (CrNCED2 reverse: 5′-TCACATGACTTCAGTTAATAAATC-3′). The PCR was carried out with high-fidelity Taq enzyme (Pfu Turbo polymerase; Stratagene) to reduce amplification errors. The amplification with C. reflexa genomic DNA as template yielded the full-length ORF sequences of CrNCED1 (1,761 bp) and CrNCED2 (1,911 bp), respectively. The products were cloned into the pGEM-T Easy vector (Promega), generating clones pGEM102 (CrNCED1) and pGEM103 (CrNCED2). To construct bacterial expression vectors, the NotI fragments of pGEM102 or pGEM103 were inserted into the GST-fusion vector pGEX-5X-3 digested with NotI. The sense-orientation clones were identified and named 5X102 and 5X103.
RNA Isolation and Northern Hybridization
Stem tips (2 cm) of Cuscuta were frozen in liquid N2 and ground with a mortar and pestle. Total RNA was isolated according to the CTAB (hexadecyltrimethylammonium bromide) method (Milligan, 1992) or by the acid AGPC (guanidinium thiocyanate-phenol-chloroform) method (Chomczynski and Sacchi, 1987). Total RNA (20 μg each) was separated by electrophoresis in formaldehyde-1.2% agarose gel and blotted onto a Hybond N+ membrane (Amersham-Pharmacia) in 20× SSC by capillary blotting. After crosslinking the RNA by UV irradiation, hybridization was carried out at 42°C for 16 h with gentle shaking in hybridization buffer containing a 32P-labeled cDNA probe (CrNCED1-3′UTR or CrNCED2-3′UTR; Supplemental Fig. S2), 50% (v/v) formamide, 10% dextran sulfate, 1× Denhardt's solution (0.02% [w/v] Ficoll 400, 0.02% [w/v] polyvinylpyrrolidone, 0.02% [w/v] bovine serum albumin, 0.5% [w/v] SDS, 3× SSC, and 50 mm Tris-HCl [pH 7.5]). The membrane was washed three times with gentle shaking in a solution containing 2× SSC/0.1% SDS at 65°C for 15 min and twice with 0.5× SSC/0.1% SDS at 50°C for 15 min. RNA was visualized with a PhosphorImager (Molecular Dynamics).
Gene-specific probes for CrNCED1 and CrNCED2 were designed as follows. 3′ RACE (Invitrogen) was performed following the manufacturer's protocol using poly(A+) RNA isolated from C. reflexa. Following RT with primers supplied by CLONTECH, the first-strand cDNA was used directly in 3′-RACE PCR reactions. Primary PCR amplification reactions were achieved using a high-fidelity enzyme (Pfu Turbo polymerase; Stratagene) and gene-specific primers JZ1167 (GTGAAGTTACCATCAAGAGTTC) and poly dT (CrNCED1), or JZ1169 (GGCATCTGTGAAATTTACCATC) and poly dT (CrNCED2) to generate the 3′-cDNA RACE fragments, respectively. A schematic representation of the CrNCED-3′UTRs structures is presented in Supplemental Figure S2. The PCR reaction consisted of the first denaturation for 3 min at 94°C, a series of 30 cycles (1 min at 94°C, 1 min at 52°C or 55°C, 1 min at 72°C), and a final extension for 7 min at 72°C. A 5-μL aliquot of the RACE reaction solution was analyzed by 1.2% agarose gel electrophoresis.
Recombinant Protein Purification
The plasmid 5X102 was transformed into Escherichia coli bacterial strain BL21 cells. An overnight culture of 5 mL was diluted to 100 mL with 2× YTA medium (16 g L−1 tryptone, 10 g L−1 yeast extract, 5 g L−1 NaCl, 100 mg L−1 ampicillin) and was grown at 28°C for 4 h. The culture was induced for protein expression for 2 h at 28°C with isopropyl β-d-1-thiogalactopyranoside at a final concentration of 0.1 mm. The cells were harvested, washed, and resuspended in 10 mL of PBS buffer containing 10 mg mL−1 lysozyme and frozen at −80°C. The frozen cells were thawed and sonicated in the presence of 0.5 mm dithiothreitol. Triton X-100 was added to the mixture to give a final concentration of 0.1%. The mixture was incubated in iced water for 30 min and then centrifuged at 10,000g for 10 min. The supernatant was applied to 1 mL of 50% slurry of glutathione Sepharose 4B (Amersham-Pharmacia) equilibrated in PBS buffer. After binding, the beads were washed with PBS buffer and were treated with 25 units of Factor Xa protease (Amersham-Pharmacia) for 4 h at 22°C. The purified CrNCED1 protein was eluted in the presence of 0.1% Triton X-100 and concentrated to 50 μL with a Millipore Ultrafree 0.5 centrifugal filter device equipped with BIOMAX 10 membrane.
Substrate Preparation and Enzyme Assay of CrNCED1
The C40-epoxycarotenoids all-trans-violaxanthin and 9′-cis-neoxanthin were isolated from spinach (Spinacia oleracea) leaves as described (Rock and Zeevaart, 1991). After isomerization with iodine, all-trans-violaxanthin, 9-cis-violaxanthin, all-trans-neoxanthin, and 9′-cis-neoxanthin were separated by normal phase HPLC with a 0.78- × 30-cm μPorasil semipreparative column (Waters) using a linear gradient from 10% to 100% ethyl acetate in hexane for 65 min at a flow rate of 2.5 mL min−1. The isomers of violaxanthin and neoxanthin were verified by their absorption spectra in ethanol and were quantified by spectrophotometry. The enzyme assay was performed as described by Schwartz et al. (1997). Appropriate amounts of protein and substrate were added in a total volume of 100 μL. Assays were incubated for 15 min at 22°C and then partitioned with ethyl acetate. The products were analyzed by HPLC with a 0.4- × 30-cm μPorasil column (Waters). The peaks eluting at the retention time of xanthoxin were collected and analyzed by GC-MS.
Sequence data for this article can be found in the GenBank/EMBL data libraries under accession numbers AY974807 and AY974808.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Amino acids sequence alignment of NCEDs.
Supplemental Figure S2. The CrNCED1- and CrNCED2-3′UTR gene regions.
Supplemental Figure S3. HPLC chromatograms of the cleavage reaction product xanthoxin.
Supplementary Material
Acknowledgments
We thank Bev Chamberlin of the Michigan State University Mass Spectrometry Facility for assistance with GC-MS.
This work was supported by the U.S. Department of Energy (grant no. DE–FG02–91ER20021).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Jan A.D. Zeevaart (zeevaart@msu.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Baldev B (1962) In vitro studies of floral induction on stem apices of Cuscuta reflexa Roxb.: a short-day plant. Ann Bot (Lond) 26 173–180 [Google Scholar]
- Berg S, Krupinska K, Krause K (2003) Plastids of three Cuscuta species differing in plastid coding capacity have a common parasite-specific RNA composition. Planta 218 135–142 [DOI] [PubMed] [Google Scholar]
- Birschwilks M, Haupt S, Hofius D, Neumann S (2006) Transfer of phloem-mobile substances from the host plants to the holoparasite Cuscuta sp. J Exp Bot 57 911–921 [DOI] [PubMed] [Google Scholar]
- Birschwilks M, Sauer N, Scheel D, Neumann S (2007) Arabidopsis thaliana is a susceptible host plant for the holoparasite Cuscuta spec. Planta 226 1231–1241 [DOI] [PubMed] [Google Scholar]
- Bungard RA, Ruban AV, Hibberd JM, Press MC, Horton P, Scholes JD (1999) Unusual carotenoid composition and a new type of xanthophyll cycle in plants. Proc Natl Acad Sci USA 96 1135–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernys JT, Zeevaart JAD (2000) Characterization of the 9-cis-epoxycarotenoid dioxygenase gene family and the regulation of abscisic acid biosynthesis in avocado. Plant Physiol 124 343–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction. Anal Biochem 162 156–159 [DOI] [PubMed] [Google Scholar]
- Creelman RA, Zeevaart JAD (1984) Incorporation of oxygen into abscisic acid and phaseic acid from molecular oxygen. Plant Physiol 75 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibberd JM, Bungard RA, Press MC, Jeschke WD, Scholes JD, Quick WP (1998) Localization of photosynthetic metabolism in the parasitic angiosperm Cuscuta reflexa. Planta 205 506–513 [Google Scholar]
- Kushiro T, Okamoto M, Nakabayashi K, Yamagishi K, Kitamura S, Asami T, Hirai N, Koshiba T, Kamiya Y, Nambara E (2004) The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: key enzymes in ABA catabolism. EMBO J 23 647–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuenberger MG, Engeloch-Jarret C, Woggon WD (2001) The reaction mechanism of the enzyme-catalyzed central cleavage of β-carotene to retinal. Angew Chem Int Ed Engl 40 2614–2617 [DOI] [PubMed] [Google Scholar]
- Milligan BG (1992) Plant DNA isolation. In AR Hoelzer, ed, Molecular Genetic Analysis of Populations. A Practical Approach. Oxford University, Oxford, pp 59–88
- Nambara E, Marion-Poll A (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56 165–185 [DOI] [PubMed] [Google Scholar]
- North HM, De Almeida A, Boutin JP, Frey A, To A, Botran L, Sotta B, Marion-Poll A (2007) The Arabidopsis ABA-deficient mutant aba4 demonstrates that the major route for stress-induced ABA accumulation is via neoxanthin isomers. Plant J 50 810–824 [DOI] [PubMed] [Google Scholar]
- Oritani T, Kiyota H (2003) Biosynthesis and metabolism of abscisic acid and related compounds. Nat Prod Rep 20 414–425 [DOI] [PubMed] [Google Scholar]
- Pierce M, Raschke K (1980) Correlation between loss of turgor and accumulation of abscisic acid in detached leaves. Planta 148 174–182 [DOI] [PubMed] [Google Scholar]
- Qin X, Zeevaart JAD (1999) The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proc Natl Acad Sci USA 96 15354–15361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock CD, Zeevaart JAD (1991) The aba mutant of Arabidopsis thaliana is impaired in epoxy-carotenoid biosynthesis. Proc Natl Acad Sci USA 88 7496–7499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roney JK, Khatibi PA, Westwood JH (2007) Cross-species translocation of mRNA from host plants into the parasitic plant dodder. Plant Physiol 143 1037–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H, Kurtzer R, Eisenreich W, Schwab W (2006) The carotenase AtCCD1 from Arabidopsis thaliana is a dioxygenase. J Biol Chem 281 9845–9851 [DOI] [PubMed] [Google Scholar]
- Schwartz SH, Zeevaart JAD (2004) Abscisic acid biosynthesis and metabolism. In PJ Davies, ed, Plant Hormones: Biosynthesis, Signal Transduction, Action! Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 137–155
- Schwartz SH, Qin X, Zeevaart JAD (2003. a) Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol 131 1591–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SH, Tan BC, Gage DA, Zeevaart JAD, McCarty DR (1997) Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276 1872–1874 [DOI] [PubMed] [Google Scholar]
- Schwartz SH, Tan BC, McCarty DR, Welch W, Zeevaart JAD (2003. b) Substrate specificity and kinetics for VP14, a carotenoid cleavage dioxygenase in the ABA biosynthetic pathway. Biochim Biophys Acta 1619 9–14 [DOI] [PubMed] [Google Scholar]
- Snyder AM, Clark BM, Bungard RA (2005) Light-dependent conversion of carotenoids in the parasitic angiosperm Cuscuta reflexa L. Plant Cell Environ 28 1326–1333 [Google Scholar]
- Snyder AM, Clark BM, Robert B, Ruban AV, Bungard RA (2004) Carotenoid specificity of light-harvesting complex II binding sites: occurrence of 9-cis-violaxanthin in the neoxanthin-binding site in the parasitic angiosperm Cuscuta reflexa. J Biol Chem 279 5162–5168 [DOI] [PubMed] [Google Scholar]
- Tian L, DellaPenna D, Zeevaart JAD (2004) Effect of hydroxylated carotenoid deficiency on ABA accumulation in Arabidopsis. Physiol Plant 122 314–320 [Google Scholar]
- Yang SH, Zeevaart JAD (2006) Expression of ABA 8′-hydroxylases in relation to leaf water relations and seed development in bean. Plant J 47 675–686 [DOI] [PubMed] [Google Scholar]
- Zeevaart JAD (1980) Changes in the levels of abscisic acid and its metabolites in excised leaf blades of Xanthium strumarium during and after water stress. Plant Physiol 66 672–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart JAD, Boyer GL (1984) Accumulation and transport of abscisic acid and its metabolites in Ricinus and Xanthium. Plant Physiol 74 934–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart JAD, Heath TG, Gage DA (1989) Evidence for a universal pathway of abscisic acid biosynthesis in higher plants from 18O incorporation patterns. Plant Physiol 91 1594–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart JAD, Rock CD, Fantauzzo F, Heath TG, Gage DA (1991) Metabolism of abscisic acid and its physiological implications. In WJ Davies, HG Jones, eds, Abscisic Acid: Physiology and Biochemistry. BIOS Scientific, Oxford, pp 39–52
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.