Abstract
Expression of AtPHO1;H10, a member of the Arabidopsis (Arabidopsis thaliana) PHO1 gene family, is strongly induced following numerous abiotic and biotic stresses, including wounding, dehydration, cold, salt, and pathogen attack. AtPHO1;H10 expression by wounding was localized to the cells in the close vicinity of the wound site. AtPHO1;H10 expression was increased by application of the jasmonic acid (JA) precursor 12-oxo-phytodienoic acid (OPDA), but not by JA or coronatine. Surprisingly, induction of AtPHO1;H10 by OPDA was dependent on the presence of CORONATINE INSENSITIVE1 (COI1). The induction of AtPHO1;H10 expression by wounding and dehydration was dependent on COI1 and was comparable in both the wild type and the OPDA reductase 3-deficient (opr3) mutant. In contrast, induction of AtPHO1;H10 expression by exogenous abscisic acid (ABA) was independent of the presence of either OPDA or COI1, but was strongly decreased in the ABA-insensitive mutant abi1-1. The involvement of the ABA pathway in regulating AtPHO1;H10 was distinct between wounding and dehydration, with induction of AtPHO1;H10 by wounding being comparable to wild type in the ABA-deficient mutant aba1-3 and abi1-1, whereas a strong reduction in AtPHO1;H10 expression occurred in aba1-3 and abi1-1 following dehydration. Together, these results reveal that OPDA can modulate gene expression via COI1 in a manner distinct from JA, and independently from ABA. Furthermore, the implication of the ABA pathway in coregulating AtPHO1;H10 expression is dependent on the abiotic stress applied, being weak under wounding but strong upon dehydration.
Jasmonates, which include jasmonic acid (JA) and its methyl ester (MeJA), act as plant growth regulators and signal molecules involved in numerous development processes and responses to the environment (Farmer et al., 2003; Devoto and Turner, 2005; Schilmiller and Howe, 2005; Wasternack, 2007; Browse and Howe, 2008). A prominent role of jasmonates has been described in the response of plants to wounding, pathogen attack as well as water stress and UV damage. JA is also essential in anther development (Feys et al., 1994; Stinzi and Browse, 2000; He et al., 2002).
JA biosynthesis occurs through the octadecanoic acid pathway and is initiated by the oxidation of α-linolenic acid to 13-hydroperoxylinolenic acid (Schaller et al., 2005). This hydroperoxide is then dehydrated by allene oxide synthase (AOS) and cyclized by allene oxide cyclase to form the cyclopentenone (9S,13S)-12-oxo-phytodienoic acid (OPDA). The enzyme OPDA reductase 3 (OPR3) then reduces the pentacyclic ring double bond to form 3-oxo-2(2′(Z)-pentenyl)-cyclopentane-1-octanoic acid (OPC:8). OPC:8 then goes through three cycles of β-oxidation in the peroxisome to yield 3R,7S-JA, which can be converted to biologically active derivatives, such as jasmonyl-l-Ile (Staswick and Tiryaki, 2004).
Several components of the JA-mediated signal transduction cascade have been characterized. One key component is CORONATINE INSENSITIVE1 (COI1). COI1 contains an F-box domain and associates with Skp-like proteins, cullin, and AtRbx1, a RING-box 1 protein to form an active SCFCOI1 complex that is thought to function as an E3-type ubiquitin ligase (Xu et al., 2001; Devoto et al., 2002). COI1 is required for numerous JA-dependent responses, including resistance to several pathogens and insects, and anther development (Feys et al., 1994; Xie et al., 1998). Recently, the JASMONATE ZIM (JAZ) domain transcriptional repressors have been identified as targets of the SCFCOI1 complex and jasmonate treatment induces their degradation by proteasomes (Chini et al., 2007; Thines et al., 2007). Members of the JAZ protein family interact and negatively regulate MYC2, a key transcriptional activator of jasmonate responses (Chini et al., 2007). Mutations and natural truncations in JAZ family members lead to reduced sensitivity to JA and impairs resistance to insect herbivory (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007; Chung et al., 2008).
Apart from being a precursor to JA, OPDA has been found to be an active signal molecule. Transient increase of both OPDA and JA has been described in response to wounding and osmotic stress (Kramell et al., 2000; Reymond et al., 2000; Stinzi et al., 2001). Treatment of barley (Hordeum vulgare) leaves with OPDA led to the activation of genes that were not activated by endogenous JA (Kramell et al., 2000). Furthermore, tendril coiling of Bryonia dioica was found to be more responsive to OPDA then JA (Stelmach et al., 1998; Blechert et al., 1999). The Arabidopsis (Arabidopsis thaliana) opr3 mutant accumulates OPDA following wounding but is deficient in JA biosynthesis (Stinzi et al., 2001). This mutant exhibits delayed anther dehiscence, resulting in male sterility (Sanders et al., 2000; Stinzi and Browse, 2000). Application of JA but not of OPDA can restore fertility, indicating that JA and not OPDA is the active molecule that regulates anther development. However, opr3 plants were shown to be as resistant as wild type to both the dipteran Bradysia impatiens and the fungal pathogen Alternaria brassicicola (Stinzi et al., 2001). Through the analysis of a set of 150 defense-related genes, OPDA was found to not only up-regulate COI1-dependent genes that are also regulated by JA, but also induced several genes in a COI1-independent fashion that are not induced by JA (Stinzi et al., 2001). More recently, from an oligonucleotide array containing 21,500 Arabidopsis genes, a set of approximately 150 genes were identified that were induced by exogenous OPDA but not by exogenous JA or MeJA (Taki et al., 2005). Approximately half of these OPDA-specific response genes were induced by wounding, and analysis of a subset of six revealed that all were induced by OPDA in a COI1-independent fashion. Thus, no gene induced by OPDA in a COI1-dependant pathway, and not by JA, has been identified from these studies.
Numerous responses of plants that are mediated by jasmonates, such as wounding or defense against pathogens, are also influenced by other hormones that may interact in an antagonistic or synergistic fashion depending on the stress or developmental process (Lorenzo and Solano, 2005). Significant synergistic interactions between the ethylene and jasmonate pathways have been described for wounding, with the ETHYLENE RESPONSE FACTOR1 (ERF1) being a convergence point between these pathways (Lorenzo et al., 2003). The salicylic acid (SA)-mediated pathway can be either synergistic or antagonist to the JA pathway, depending on the concentration of SA and JA (Lorenzo and Solano, 2005; Mur et al., 2006). The abscisic acid (ABA) signal transduction pathway has been shown to contribute to the induction of PIN2 (proteinase inhibitor II) and of other genes in tomato (Solanum lycopersicum) and potato (Solanum tuberosum) leaves following wounding, as well as to affect the feeding of insects on Arabidopsis (Pena-Cortes et al., 1995; Herde et al., 1996; Carrera and Prat, 1998; Bodenhausen and Reymond, 2007). However, the activation of several genes by JA were shown to be independent of ABA in barley and potato (Lee et al., 1996; Dammann et al., 1997). ABA interacts both in an antagonistic and synergistic fashion with JA in the response to distinct pathogens (Anderson et al., 2004; Mauch-Mani and Mauch, 2005; Adie et al., 2007).
This study describes the response of one member of the PHO1 gene family in Arabidopsis to biotic and abiotic stresses. PHO1 is involved in the transfer of phosphate to the xylem in the root, and the Arabidopsis genome contains 10 additional genes showing homology to PHO1 (named AtPHO1;H1 to AtPHO;H10; Poirier et al., 1991; Hamburger et al., 2002; Wang et al., 2004). The gene AtPHO1;H10 was found to be strongly induced by numerous stresses, including in the local response to wounding. The signal molecules and transduction pathways involved in the regulation of AtPHO1;H10 following wounding and dehydration was thus investigated. AtPHO1;H10 expression was induced by OPDA but not by JA or by coronatine, a bacterial polyketide metabolite that mimics jasmonates. Surprisingly, induction of AtPHO1;H10 by both external OPDA application and wounding occurred via a COI1-dependent pathway. AtPHO1;H10 was also induced by ABA in a COI1-independent manner, and both the ABA- and OPDA-mediated signal transduction pathways were involved in the induction of AtPHO1;H10 following dehydration, while OPDA was the main signal involved in wounding.
RESULTS
AtPHO1;H10 Is Regulated by Numerous Biotic and Abiotic Stresses
Analysis by RNA gel-blot analysis of the expression of the AtPHO1;H10 gene in soil-grown plants revealed that the gene was well expressed in roots and flowers but weakly in leaves and the inflorescence stems (Fig. 1A). A similar stronger expression in roots compared to leaves was observed for plants grown in agar-solidified medium (Fig. 1A). In transgenic plants expressing the GUS reporter gene under the control of 1.0 kb of the AtPHO1;H10 promoter sequence (pH10∷GUS line), strong GUS expression was evident in the epidermal and cortical cells of the roots, particularly in the primary roots and more weakly in the emerging secondary roots (Fig. 1, B and C). In the leaves, expression was primarily limited to the hydathodes and more weakly to trichomes (Fig. 1D).
Figure 1.
Regulation of AtPHO1;H10 expression by various biotic and abiotic stresses. A, Expression of AtPHO;H10 in roots (R), leaves (L), stems (S), and flowers (F) from soil-grown plants, or from leaves (L) and roots (R) from plants grown on agar. B to D, Pattern of expression of the GUS reporter gene in roots (B and C) and leaves (D) of ppH10∷GUS plants. E, Comparison of the AtPHO1;H10 mRNA levels between control untreated plants (ctl) and leaves 2 h after wounding with razor blade (wo), leaves 2 h after dehydration (de), leaves 12 h after a cold treatment at 4°C (co), senescent leaves (se), seedlings 2 h after transfer to medium containing100 mm NaCl (na), leaves 4 h after spraying of 10 μm paraquat (pa), seedlings 4 h after transfer to medium containing 250 mm mannitol (ma), and leaves 18 h after infection with P. syringae pv. tomato DC3000 (Pst) or Pst with the avirulence gene avrRpm1. All stresses were performed on the leaves of 6-week-old plants grown in soil at 20°C under short-day photoperiod (10-h day/14-h night), except for the NaCl and mannitol treatments, which were realized on the whole 10-d-old plants grown under continuous light at 20°C in agar-solidified medium containing half-strength MS and 1% Suc. Probes for marker genes induced by the various stresses were used as control. The AtRD29A gene was chosen to confirm stress induced by dehydration, cold, NaCl, and mannitol treatments. Similarly, AtJR3 was used for the wounding stress, AtSAG12 for senescence, AtGST6 for the paraquat treatment, and AtPR1 for the infection with P. syringae. F, Transgenic ppH10∷GUS plants were treated by wounding (wo), dehydration (de), cold (co), addition of NaCl (na), infection by P. syringae pv. tomato DC3000 (Pst) or Pst with the avirulence gene avrRpm1, as indicated above. Senescing (se) cotyledons and first leaves from a 6-week-old rosette are indicated by the arrow. G, AtPHO1;H10 mRNA levels in leaves after a heat stress at 37°C for 1 h. Probe for the heat shock gene AtHSP18.2 was used as control for heat stress and RNA extracted from wounded leaves (wo) was used as a positive control for hybridization with the AtPHO;H10 probe. For all RNA gel blots, 25 μg of total RNA was used and hybridization with an Arabidopsis α-tubulin probe was used as loading control.
Following an initial observation of strong GUS expression at the cutting edge of a petiole in pH10∷GUS plants (data not shown), expression of AtPHO1;H10 in wild-type plants treated with a variety of abiotic and biotic stresses was examined by RNA gel-blot analysis. In soil-grown plants, a strong accumulation of AtPHO1;H10 mRNA was observed in leaves 2 h after mechanical wounding, 2 h after initiation of dehydration, 12 h after cold treatment at 4°C, 4 h after paraquat treatment, 18 h after infection of leaves with Pseudomonas syringae pv. tomato DC3000 expressing or not the avirulence gene avrRpm1, and in senescing leaves (Fig. 1E). Similarly, a strong expression of AtPHO1;H10 in plants grown in agar-solidified medium was observed after 2 h of transfer to medium containing 100 mm NaCl or 4 h after transfer to 250 mm mannitol (Fig. 1E). Treatment of transgenic pH10∷GUS plants with a subset of the stresses described above also showed strong GUS expression in either the roots (NaCl stress) or leaves (wounding, dehydration, senescence, and P. syringae infection [Fig. 1F]). However, no induction of AtPHO1;H10 expression could be detected by RNA gel-blot analysis in leaves following a heat shock at 37°C for 1 h (Fig. 1G).
In pH10∷GUS transgenic plants, GUS expression was restricted to the wound site for either leaves wounded mechanically with a razor blade, or challenged with the chewing caterpillar Pieris rapae (Fig. 2, B and D). RNA gel-blot analysis confirmed that the up-regulation of AtPHO1;H10 expression in wounded or eaten leaves was restricted to the zone surrounding the wound site and not at a distal region in the same wounded leaves (Fig. 2, A and C).
Figure 2.
Local accumulation of AtPHO1;H10 mRNA in leaves near the wound site and in areas infiltrated with OPDA. Wounding was performed on leaves either with a razor blade (A and B) or by feeding with the caterpillar P. rapae (C and D). RNA gel-blot anaylsis experiments (A and C) with RNA extracted from unwounded leaves (ctl), from tissues 3 mm around the wound site (lo, local), and from the remaining part of the wounded leaves (di, distal). B and D, Localization of GUS activity in leaves of transgenic ppH10∷GUS plants following wounding with a razor blade (B) or by P. rapae feeding (D). Samples for northern or GUS staining were harvested 3 and 5 h after wounding by a razor blade or P. rapae feeding, respectively. E, The right side of the abaxial side of ppH10∷GUS plants was infiltrated with either water (left) or 3 nmol of OPDA (right) and stained for GUS activity after 3 h.
AtPHO1;H10 Is Induced by ABA and OPDA But Not by JA or Coronatine
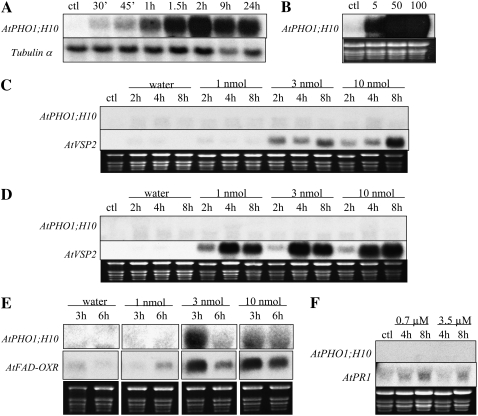
The induction of AtPHO1;H10 following treatment of plants with various hormones and signal molecules involved in the response of plants to different biotic and abiotic stresses was examined by RNA gel-blot analysis. Treatment of plants with 100 μm ABA led to strong expression of AtPHO1;H10, with increased expression being detectable as early as 30 min after ABA addition and maximal expression observed at 2 h (Fig. 3A). Strong expression of AtPHO1;H10 was also observed after 2 h of treatment with ABA concentrations ranging from 5 to 100 μm (Fig. 3B). Analysis of leaves 2, 4, and 8 h after infiltration with 1, 3, and 10 nmol of either JA or coronatine did not lead to the induction of AtPHO1;H10, whereas induction of the marker gene AtVSP2 was observed for JA and coronatine, respectively (Fig. 3, C and D). In contrast, infiltration of leaves with OPDA led to overexpression of AtPHO1;H10, with maximal expression observed after 3 h of infiltration with 3 nmol of OPDA (Fig. 3E). Infiltration of leaves of pH10∷GUS plants with 3 nmol OPDA led to a local induction of GUS in the infiltrated area (Fig. 2E). No induction of AtPHO1;H10 expression was observed after 3 h of infiltration of leaves with 10 nmol oleic acid or linolenic acid (data not shown). Similarly, no induction of AtPHO1;H10 expression was observed in plants treated with methyl salicylate (Fig. 3F).
Figure 3.
Regulation of AtPHO1;H10 transcript levels by exogenous addition of phytohormones. A, Plants grown on agar-solidified medium for 7 d were transferred to the same medium containing 100 μm ABA for times ranging from 30 min up to 24 h before being harvested. Control (ctl) plants were transferred to agar-solidified medium without ABA for 24 h. B, Plants grown on agar-solidified medium for 7 d were transferred to the same medium containing no ABA (ctl) or 5, 50, or 100 μm ABA for 2 h before being harvested. Leaves of plants grown in soil were infiltrated with either 10 μL of water or water containing 5% ethanol, or 10 μL of solutions containing JA (C), coronatine (D), or OPDA (E), representing the infiltration of 1, 3, or 10 nmol of jasmonates. Infiltrated leaves were harvested between 2 and 8 h postinfiltration. Control leaves (ctl) were not infiltrated. F, Plants were placed in boxes containing the volatile methyl salicylate at a final concentration in air of 0.7 or 3.5 μm and leaves harvested after 4 to 8 h. Control plants (ctl) were put in boxes without methyl salicylate for 8 h. Probes used for hybridization are indicated on the left of every panel.
Induction of AtPHO1;H10 by Wounding Is Mediated by OPDA via a COI1-Dependent Pathway
Induction of AtPHO1;H10 by wounding was analyzed in various mutants affected in the jasmonate biosynthetic or perception pathway. The aos mutant is defective in the AOS gene, and is deficient in both OPDA and JA synthesis (Park et al., 2002). In contrast, the opr3 mutant is defective in the gene encoding the peroxisomal OPDA reductase 3 and is able to accumulate OPDA but not JA (Stinzi and Browse, 2000; Stinzi et al., 2001). Finally, the coi1-1 mutant is deficient in numerous JA-mediated responses (Feys et al., 1994; Xie et al., 1998).
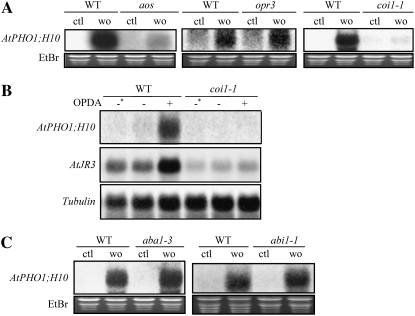
Overexpression of AtPHO1;H10 following mechanical wounding was nearly abolished in both the aos and coi1-1 mutant while it remained unaffected in the opr3 mutant (Fig. 4A), indicating that endogenous OPDA synthesized following wounding was capable of inducing AtPHO1;H10 expression via a COI1-dependent pathway. Furthermore, induction of AtPHO1;H10 expression by infiltration of OPDA was also abolished in the coi1-1 mutant (Fig. 4B).
Figure 4.
Regulation of the AtPHO1;H10 expression following wounding and OPDA treatment in JA and ABA mutants. A and C, RNA was extracted from unwounded leaves (ctl) or leaves 2 h after wounding with a razor blade (wo). B, RNA was extracted from uninfiltrated leaves (−*), leaves 3 h following infiltration with water (−), or 3 h following infiltration with 10 nmol OPDA (+). Wild-type plants used as control were of the same accession genotype as the corresponding mutants, namely Col for the aos and coi1-1 mutants, Ws for opr3, and Ler for aba1-3 and abi1-1.
Wounding involves dehydration at the wound site and may involve an ABA-mediated response (Reymond et al., 2000). Induction of AtPHO;H10 following mechanical wounding was thus analyzed in the aba1-3 mutant deficient in ABA synthesis (Koornneef et al., 1982), and in the ABA-insensitive mutant abi1-1, deficient in multiple responses of plants to ABA (Koornneef et al., 1984). For both aba1-3 and abi1-1 mutants, the overexpression of AtPHO1;H10 following wounding was unchanged compared to the control (Fig. 4C).
AtPHO1;H10 Expression under Dehydration Involves Both ABA and OPDA-Mediated Signaling
The contribution of both the jasmonate- and ABA-mediated signaling pathway to the induction of AtPHO1;H10 expression following dehydration of rosette was assessed. Induction of AtPHO1;H10 was strongly reduced in both the aos and coi1-1 mutants, whereas only a slight reduction was observed in the opr3 mutant (Fig. 5A). A strong reduction in AtPHO1;H10 expression following dehydration was also observed in the aba1-3 and abi1-1 mutant (Fig. 5B). Together, these results indicate a contribution of both OPDA and ABA to the response of AtPHO1;H10 to dehydration.
Figure 5.
Regulation of the AtPHO1;H10 expression following dehydration in JA and ABA mutants. RNA was extracted from leaves of well-watered plants (ctl) or leaves 2 h after dehydration was induced by cutting a rosette at the hypocotyl and placing it on a filter paper. Wild-type plants used as control were of the same accession genotype as the corresponding mutants, namely Col for the aos and coi1-1 mutants, Ws for opr3, and Ler for aba1-3 and abi1-1.
AtPHO1;H10 Expression by ABA Involves the ABI1 and ROP10 Signaling Pathway and Is Independent of the Jasmonate Pathway
Induction of AtPHO1;H10 expression by ABA in seedlings was strongly suppressed in abi1-1 (Fig. 6A). ROP10 is a member of the ROP subfamily of Rho GTPases located in the plasma membrane and was shown to negatively regulate ABA-mediated responses (Zheng et al., 2002). The rop10 mutant was previously shown to exhibit enhanced responses to ABA and the induction of AtPHO1;H10 by ABA was enhanced in the rop10 mutant compared to wild-type plants (Fig. 6B; Zheng et al., 2002).
Figure 6.
Regulation of the AtPHO1;H10 expression by exogenous treatment with ABA in mutants or transgenic plants affected in JA or ABA responses. Plants were grown for 10 d in agar-solidified medium containing half-strength MS and 1% Suc before being transferred to the same medium supplemented with 10 μm ABA for 2 h. RNA was extracted from the whole plants. Wild-type plants used as control were of the same accession genotype as the corresponding mutants, namely Ler for the abi1-1 mutant (A) and the transgenic lines 35S-ABF3, 35S-ABF4, and 35S-ABI5 (C), Ws for rop10 (B), and Col for the aos and coi1-1 mutants (D). Probes used for hybridization are indicated on the leaf of every panel.
Analysis of 1 kb of promoter sequence of AtPHO1;H10 revealed the sequences CACGTGTC and CACGTGGC 619 and 508 bp upstream of the start codon, respectively, which conforms to the cis-acting ABA responsive elements (ABREs). Furthermore, the sequence CACGCGT, which conforms to the CE3 coupling element, is found 23 bp upstream of the CACGTGGC ABRE (Rock, 2000). These sequences are recognized by members of the bZIP (basic-domain Leu zipper) transcription factors, which include ABF3, ABF4, and ABI5, three proteins involved in ABA-mediated responses (Rock, 2000; Yamaguchi-Shinozaki and Shinozaki, 2004). Expression of AtPHO1;H10 was analyzed in transgenic lines overexpressing the ABF3, ABF4, and ABI5 transcription factors under the control of the CaMV35S promoter (Kang et al., 2002; Lopez-Molina et al., 2002). In plants treated or untreated with ABA, overexpression of AtPHO1;H10 relative to wild-type plants was only observed in the ABI5-overexpressing lines (Fig. 6C). For the ABF3-overexpressing line, AtPHO1;H10 expression following ABA treatment was reduced compared to control (Fig. 6C). Finally, the overexpression of AtPHO1;H10 following ABA treatment was found to be unaffected in the aos and coi1-1 mutant (Fig. 6D).
DISCUSSION
The observed pattern of induction of AtPHO1;H10 expression by multiple stresses is largely in agreement with its induction by either exogenous ABA or OPDA applications. Changes in the endogenous level of ABA and the implication of the ABA signaling pathway has been well described for stresses mediated by wounding, dehydration, cold, senescence, salt, and osmotic stress (Pena-Cortes et al., 1989, 1995; Birkenmeier and Ryan, 1998; Zhu, 2002; Gusta et al., 2005; de Torres-Zabala et al., 2007). ABA has also been implicated in the response of plants to pathogen infection, including infection with P. syringae (Anderson et al., 2004; Mauch-Mani and Mauch, 2005; Adie et al., 2007; de Torres-Zabala et al., 2007). Similarly, increase in endogenous level of OPDA and/or JA, as well as the implication of the JA signaling pathway has been described for wounding, dehydration, senescence, salt, osmotic, and oxidative stresses, as well as for infection with pathogens (Harms et al., 1995; Lehmann et al., 1995; Pena-Cortes et al., 1995; Herde et al., 1996; Moons et al., 1997; Reymond et al., 2000; Stinzi and Browse, 2000; He et al., 2002; Sasaki-Sekimoto et al., 2005; Truman et al., 2007). Thus, although it is not excluded that other molecules or signaling pathways could be involved in modulating the expression of AtPHO1;H10 in other stresses, these results reveal that both ABA and OPDA are important regulators for this gene.
Previous studies have shown that OPDA can have a biological activity that can either overlap with JA or be distinct from JA. Overlap in the activity of OPDA and JA has been shown by the maintenance of resistance to the dipteran B. impatiens and fungus A. brassicicola in the Arabidopsis mutant opr3, synthesizing OPDA but not JA (Stinzi et al., 2001). In contrast, the defect in pollen maturation observed in the opr3 mutant can be complemented only by exogenous application of JA, indicating that JA and not OPDA is the active molecule that regulates anther development (Stinzi and Browse, 2000). Similarly, the ozone-treated opr3 mutant failed to activate several JA-regulated genes that are involved in antioxidant metabolism, and the opr3 mutant was more sensitive to ozone exposure compared to wild type (Sasaki-Sekimoto et al., 2005). At the genetic level, the combination of gene expression studies between the wild type and opr3 mutants following wounding or infiltration with either OPDA or JA has revealed genes, such as JR3 (At1g51760) and HPL (At4g15440), which can be activated either by OPDA or JA, both in a COI1-dependent manner (Stinzi et al., 2001; Taki et al., 2005). However, some genes, such as VSP2 (At5g24770) and MBP (At3g16460), were activated by JA and not OPDA, also in a COI1-dependent manner (Stinzi et al., 2001; Taki et al., 2005). Approximately 150 OPDA-regulated genes (ORGs) in Arabidopsis were found to be activated by exogenous OPDA but not JA (Stinzi et al., 2001; Taki et al., 2005). Expression analysis of a subset of ORGs, including ERF5 (At5g47230) and FAD-OXR (At4g20860), revealed that all were activated via a COI1-independent pathway, clearly indicating that ORGs are activated via a mechanism distinct from the JA-COI1 pathway (Stinzi et al., 2001; Taki et al., 2005). In contrast, this work reveals that AtPHO1;H10 can be activated by OPDA in a COI1-dependent manner, but not by JA. Together, these studies reveal that the output of the COI1-dependent pathway on gene activation can be quite distinct depending on whether OPDA or JA is involved (Fig. 7).
Figure 7.
Model showing the distinct regulation of gene expression mediated by OPDA and JA via COI1-dependent and -independent pathways. A selection of genes activated by each pathway is indicated and is based on this work and the studies of Taki et al. (2005) and Stinzi et al. (2001).
In accordance with the participation of JAZ proteins in the jasmonate signaling network, altered expression of some JAZ proteins lead to reduced sensitivity to JA (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007). Physical interaction between COI1 and JAZ3/JAI3 or JAZ1 has been demonstrated in cell-free protein extracts, while interaction between JAZ1 and COI1 was further demonstrated in a yeast two-hybrid system (Chini et al., 2007; Thines et al., 2007). Remarkably, interaction between JAZ1 and COI1 was found to be dependent on the presence of JA-Ile, whereas OPDA, JA, or MeJA did not promote interactions between these proteins (Thines et al., 2007). Furthermore, proteasome degradation of JAZ3/JAI3 was impaired in the jar1 mutant deficient in the enzyme responsible for the synthesis of JA-Ile conjugate (Chini et al., 2007). Together, these results reveal that JA-Ile is an active hormone promoting the interaction between COI1 and either JAZ1 or JAZ3/JAI3. Furthermore, although OPDA was previously regarded as a close structural analog of coronatine (Weiler et al., 1994), the current data rather suggest that coronatine is a structural analog of JA-Ile (Staswick, 2008). In this context, what then is the significance of the activation of AtPHO1;H10 by OPDA and not JA or coronatine, in a COI1-dependent fashion? One hypothesis would be that OPDA promotes the interaction of COI1 with a distinct member of the JAZ protein family, which comprises 12 members in Arabidopsis. Alternatively, OPDA may activate AtPHO1;H10 through the interaction of COI1 with other unidentified proteins. The sets of genes regulated by COI1 would thus depend on both the nature of the jasmonate (e.g. OPDA or JA-Ile) and the presence of distinct effector molecules interacting with COI1. The distinct role of jasmonates other than JA-Ile has recently been supported by findings that wound-induced expression of COI1-dependent genes in the Arabidopsis JA-Ile-deficient jar1-1 mutant is not significantly impaired, and by the distinct role of JA and JA-Ile in herbivore resistance of Nicotiana attenuata (Wang et al., 2007; Chung et al., 2008).
Several physiological and developmental processes are influenced by either a synergistic or antagonistic cross talk between ABA and jasmonates, including seed germination (Wilen et al., 1991; Staswick et al., 1992; Ellis and Turner, 2002) and defense against pathogens (Anderson et al., 2004; Mauch-Mani and Mauch, 2005; Adie et al., 2007). Positive interactions between ABA and JA have been reported for wounding, mainly in potato and tomato. ABA synthesis or perception via ABI1 was required for the local increase in JA and the induction of PIN2 expression following wounding in both potato and tomato (Pena-Cortes et al., 1989, 1995, 1996; Hildmann et al., 1992; Herde et al., 1996; Carrera and Prat, 1998). Increases in ABA were also measured following wounding in both local and systemic leaves, and the expression of several JA-regulated genes, including PIN2, in the wounded ABA-deficient mutant can be restored either via ABA or JA application (Pena-Cortes et al., 1989, 1995; Creelman and Mullet, 1995; Herde et al., 1996; Birkenmeier and Ryan, 1998). Together, these studies indicated that in potato and tomato, ABA synthesis and perception was necessary for activation of the JA biosynthetic and signaling pathway following wounding. In Arabidopsis, ABA was found to activate AtMYC2 in a COI1-dependent manner, indicating that ABA precedes JA in the activation of AtMYC2-mediated wound responses (Lorenzo et al., 2004). It was, however, noted that distinct genes can be induced by either ABA or JA in various tissues in potato and barley (Lee et al., 1996; Dammann et al., 1997), and that a higher level of ABA can be maintained in dehydrated tissues without activation of JA synthesis or JA-regulated genes in soybean (Glycine max) and tomato (Creelman and Mullet, 1995; Birkenmeier and Ryan, 1998).
Activation of AtPHO1;H10 expression by external ABA was independent of the presence of both AOS and COI1, but was strongly influenced by the presence of ABI1 and ROP10, indicating the lack of involvement of the jasmonate pathway and the main participation of the ABA signaling pathway in this activation. The presence of ABRE and C3 cis-acting elements in the promoter of AtPHO1;H10 and the activation of AtPHO1;H10 expression in the transgenic line overexpressing the ABI5 transcription factor further highlight the main contribution of the ABA signaling pathway via ABI5 in AtPHO1;H10 regulation. The lack of response of the AtPHO1;H10 gene to overexpression of ABF3 and ABF4 is in contrast to the strong activation of AtPHO1;H10 by ABI5 (Fig. 6B). These data likely reflect differential strength of interaction of these three transcription factors to the cis-promoter elements present in AtPHO1;H10.
The contribution of the ABA signaling pathway to AtPHO1;H10 expression following wounding and dehydration was distinct. Thus, AtPHO1;H10 induction by wounding was found to be unaffected in the aba1-3 and abi1-1 mutants, indicating that in contrast to the ABA-dependent activation of PIN2 in wounded potato and tomato leaves, induction of ATPHO1;H10 expression by wounding mainly involved the OPDA-COI1 pathway and was independent of the ABA pathway. In contrast, upon dehydration, both the ABA and OPDA-COI1 pathway needed to be present to achieve maximal AtPHO1;H10 expression. The implication of ABA upon dehydration could perhaps be linked to a higher increase in ABA level in dehydrated tissues compared to wounding (Creelman and Mullet, 1995; Birkenmeier and Ryan, 1998). Interestingly, a role for ABA in defense against insect herbivory has been demonstrated, with the weight of Spodoptera littoralis larvae feeding on the ABA-deficient mutant aba2-1 being higher than larvae feeding on wild-type plants (Bodenhausen and Reymond, 2007). The same study revealed that a number of ABA-regulated genes are induced upon insect feeding either in a COI1-dependent or COI1-independent manner. Together, these results highlight that the extent of the interaction between the ABA and jasmonate pathways in biotic and abiotic stress is highly dependent on the nature of the stress involved. Similar conclusions are also emerging from the study of ABA and jasmonate signaling pathways in plant-pathogen interactions, whereby ABA can be either play a synergistic or antagonistic role with jasmonates depending on the pathogen (Anderson et al., 2004; Mauch-Mani and Mauch, 2005; Adie et al., 2007).
At present, the role of AtPHO1;H10 in the plant's response to wounding or water stress remains unclear. Although both AtPHO1 and its closest homolog AtPHO1;H1 are involved in the loading of inorganic phosphate to the xylem (Stefanovic et al., 2007), the distinct pattern of expression of AtPHO1;H10 makes it unlikely that it could have a similar role in Pi export. Interestingly, the ATPHO1;H4 gene has been implicated in the response of the hypocotyl to blue light, indicating that AtPHO1 family members could be involved in signal transduction cascades associated with different environmental stimuli (Ni and Kang, 2006). However, comparison of gene expression using complementary DNA (cDNA) microarray between the wild type and T-DNA null mutant for AtPHO1;H10 in response to abiotic wounding (3 h after wounding) or exogenous ABA treatment (1 and 12 h after addition of 100 μm ABA) has failed to reveal differences that could implicate AtPHO1;H10 in early events in ABA or OPDA signal transduction cascade (data not shown). Nevertheless, the strong and localized expression of AtPHO1;H10 at the wound site provides a novel reporter for the study of cell signaling in the immediate vicinity of the wound. This zone, restricted to only a few cells, is particularly susceptible to dehydration and has a direct contact to the atmosphere, distinguishing it from leaf zones distal to the wound. The unique regulation of AtPHO1;H10 provides a valuable marker to study the factors that enable distinct signaling cascades to emerge from the SCFCOI1 complex depending on the presence of either OPDA or JA.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Seeds of wild-type Arabidopsis (Arabidopsis thaliana) accession Columbia (Col), Landsberg erecta (Ler), or Wassilewskija (Ws), as well as of mutants (ecotypes of the mutants are indicated in parenthesis) aos (Col; Park et al., 2002), opr3 (Ws; Stinzi and Browse, 2000), coi1-1 (Col; Feys et al., 1994), aba1-3 (Ler; Koornneef et al., 1982), abi1-1 (Ler; Koornneef et al., 1984), 35S-ABF3 (Ler; Kang et al., 2002), 35S-ABF4 (Ler; Kang et al., 2002), 35S-ABI5 (Ler; Lopez-Molina et al., 2002), and rop10 (Ws; Zheng et al., 2002) were sown in 7-cm-diameter pots containing potting compost and vernalized for 4 d at 4°C. Plants were then grown for 6 to 7 weeks in a growth room (20°C at 70% relative humidity and with 10-h/14-h light/dark cycle at 100 μEm−2 s−1). For some experiments, plants were grown in agar-solidified half-strength Murashige and Skoog (MS) medium under constant illumination.
Wounding, Dehydration, Pathogen, and Chemical Treatments
Wounding was done with incisions made with a razor blade across the whole surface at intervals of approximately 2 mm. The main vein and the edges of the leaf were left undamaged. Following wounding, plants were kept in the same growth room for 3 h before being harvested and frozen in liquid nitrogen. For biotic wounding experiments, Pieris rapae caterpillars were placed on plants and were allowed to feed under light for 5 h at 20°C until approximately 40% of the leaf surface was removed. Larvae were then removed, and all plant leaves were immediately frozen in liquid nitrogen or used for GUS assay. Dehydration was performed by excising the whole rosettes from their roots with a razor blade and then placing them on a paper in the same growth chamber at 20°C and 70% humidity for various times before freezing in liquid nitrogen. Stock solutions of 10 mm (±)-cis, trans-ABA (Sigma) was prepared in water with a few microliters of NaOH 1 n to help the dissolution. Solutions of 9S,13S-OPDA (Larodan), coronatine (Sigma), and the (±)-JA (Sigma) were infiltrated into the abaxial surface of the leaf with a syringe without a needle, at 10 μL per leaf. Before infiltration of OPDA, the ethanol was evaporated and the residues were dissolved in a volume of ethanol adjusted so that the final concentration of ethanol after water addition was 5% (v/v). To improve solubilization, OPDA solution was sonicated in a water bath. JA and coronatine were directly dissolved into water. Control leaves were infiltrated with water or water containing the same concentration of ethanol. Volatile methyl-salicylate (Sigma) treatments were done in hermetic plexiglass boxes (11.4 L) by applying either 7.9 μmol or 40 μmol of MeJA on a Q-tip (final concentration of 0.7 μmol or 3.5 μmol to 1 L of air volume, respectively). For infection of leaves with Pseudomonas syringae pv. tomato DC3000 with or without the avirulence gene avrRpm1, the abaxial surface of leaves was infiltrated with 2.5 × 105 colony-forming units in 10 μL of 10 mm MgCl2 using a syringe without a needle.
RNA Isolation and Northern Hybridization Analysis
Total RNA was extracted from plants tissues by phenol:chloroform separation and lithium chloride precipitation followed by washes with sodium acetate and ethanol as previously described (Reymond et al., 2000). Northern analysis was performed by separating 25 μg of total RNA on agarose gels containing formaldehyde, transferring to nylon membranes (Hybond N+; Amersham Biosciences), and hybridizing with P32-radiolabeled specific probes according to standard procedures under high stringency conditions. Specific probes corresponded to the gene-specific tags (designed in the microarray CATMA project; Crowe et al., 2003) amplified by PCR with a specific set of primers, or to full-length cDNA.
GUS Staining
Transgenic plants expressing the uidA reporter gene under the control of a 1-kb fragment of the AtPHO1;H10 promoter (Wang et al., 2004) were selected in agar-solidified medium containing 50 μg/mL of kanamycin. The resistant plants were then transferred in pots containing fertilized soil for 5 weeks. Rosettes were stained for GUS activity as previously described (Wang et al., 2004).
Acknowledgments
The authors thank Louis Lopez-Molina, Soo Young Kim, and Zhenbiao Yang for providing us with seeds for some of the mutants and transgenic lines used in this study, Caroline Darimont for her work on cDNA microarrays, and Pasqualina Magliano and Syndie Delessert for help with plant care.
This work was supported the Fonds National Suisse (grant no. 3100A0–105874 to Y.P.) and the Etat de Vaud.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Yves Poirier (yves.poirier@unil.ch).
References
- Adie BAT, Pérez-Pérez J, Pérez-Pérez MM, Godoy M, Sanchez-Serrano J, Schmelz EA, Solano R (2007) ABA is an essential signal for plant resistance of pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19 1665–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JP, Badruzsaufari E, Schenk PM, Manners JM, Desmond OJ, Ehlert C, Maclean DJ, Ebert PR, Kazan K (2004) Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16 3460–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenmeier GF, Ryan CA (1998) Wound signaling in tomato plants. Evidence that ABA is not a primary signal for defense gene action. Plant Physiol 117 687–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blechert S, Bockelmann C, Füsslein M, Schrader TV, Stelmach B, Nielsen U, Weiler EW (1999) Structure-activity analyses reveal the existence of two separate groups of active octadecanoids in elicitation of the tendril-coiling response of Bryonia dioica Jacq. Planta 207 470–479 [Google Scholar]
- Bodenhausen N, Reymond P (2007) Signaling pathways controlling induced resistance to insect herbivores in Arabidopsis. Mol Plant Microbe Interact 11 1406–1420 [DOI] [PubMed] [Google Scholar]
- Browse J, Howe GA (2008) New weapons and a rapid response against insect attack. Plant Physiol 146 832–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera E, Prat S (1998) Expression of the Arabidopsis abi1-1 mutant allele inhibits proteinase inhibitor wound-induction in tomato. Plant J 15 765–771 [DOI] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernandez G, Adie B, Chico JM, Lorenzo O, Garcia-Casado G, Lopez-Vidriero I, Lozano FM, Ponce MR, et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448 666–671 [DOI] [PubMed] [Google Scholar]
- Chung HS, Koo AJK, Gao X, Jayanty S, Thines B, Jones AD, Howe GA (2008) Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol 146 952–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creelman RA, Mullet JE (1995) Jasmonic acid distribution and action in plants: regulation during development and response to biotic and abiotic stress. Proc Natl Acad Sci USA 92 4114–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe ML, Serizet C, Thareau V, Aubourg S, Rouze P, Hilson P, Beynon J, Weisbeek P, van Hummelen P, Reymond P, et al. (2003) CATMA: a complete Arabidopsis GST database. Nucleic Acids Res 31 156–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammann C, Rojo E, Sanchez-Serrano J (1997) Abscisic acid and jasmonic acid activate wound-inducible genes in potato through separate, organ-specific signal transduction pathways. Plant J 11 773–782 [DOI] [PubMed] [Google Scholar]
- de Torres-Zabala M, Truman W, Bennett MH, Lafforgue G, Mansfiled JW, Egea PR, Bögre L, Grant M (2007) Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J 26 1434–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto A, Nieto-Rostro M, Xie D, Ellis C, Harmston R, Patrick E, Davis J, Sherratt L, Coleman M, Turner JG (2002) COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J 32 457–466 [DOI] [PubMed] [Google Scholar]
- Devoto A, Turner JG (2005) Jasmonate-regulated Arabidopsis stress signalling network. Physiol Plant 123 161–172 [Google Scholar]
- Ellis C, Turner JG (2002) A conditionally fertile coi1 allele indicates cross-talk between plant hormone signalling pathways in Arabidopsis thaliana seeds and young seedlings. Planta 215 549–556 [DOI] [PubMed] [Google Scholar]
- Farmer EE, Alméras E, Krishnamurthy V (2003) Jasmonates and related oxylipins in plant responses to pathogenesis and herbivory. Curr Opin Plant Biol 6 372–378 [DOI] [PubMed] [Google Scholar]
- Feys BF, Benedetti CE, Penfold CN, Turner JG (1994) Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male-sterile, insensitive to methyl jasmonate, and are resistant to a bacterial pathogen. Plant Cell 6 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusta LV, Trischuk R, Weiser CJ (2005) Plant cold acclimation: the role of abscisic acid. J Plant Growth Regul 24 308–318 [Google Scholar]
- Hamburger D, Rezzonico E, MacDonald-Comber Petetot J, Somerville C, Poirier Y (2002) Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 14 889–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms K, Atzorn R, Brash A, Kühn H, Wasternack C, Willmitzer L, Pena-Cortes H (1995) Expression of a flax allene oxide synthase cDNA leads to increased endogenous jasmonic acid (JA) levels in transgenic plants but not to a corresponding activation of JA-responding genes. Plant Cell 7 1645–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Fukushige H, Hildebrand DF, Gan S (2002) Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol 128 876–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herde O, Atzorn R, Fisahn J, Wasternack C, Willmitzer L, Pena-Cortes H (1996) Localized wounding by heat initiates the accumulation of proteinase inhibitor II in abscisic acid-deficient plants by triggering jasmonic acid biosynthesis. Plant Physiol 112 853–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildmann T, Ebneth M, Pena-Cortes H, Sanchez-Serrano J, Willmitzer L, Prat S (1992) General roles of abscisic and jasmonic acids in gene activation as a result of mechanical wounding. Plant Cell 4 1157–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J-Y, Choi H-I, Inn M-Y, Kim SY (2002) Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Jorna ML, Brinkhorst-van der Swan DLC, Karssen CM (1982) The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L.) Heynh. Theor Appl Genet 61 385–393 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Reuling G, Karssen CM (1984) The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol Plant 61 377–383 [Google Scholar]
- Kramell R, Miersch O, Atzorn R, Parthier B, Wasternack C (2000) Octadecanoid-derived alteration of gene expression and the “oxylipin signature” in stressed barley leaves. Implications for different signaling pathways. Plant Physiol 123 177–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Parthier B, Löbler M (1996) Jasmonate signalling can be uncoupled from abscisic acid signalling in barley: identification of jasmonate-regulated transcripts which are not induced by abscisic acid. Planta 199 625–632 [DOI] [PubMed] [Google Scholar]
- Lehmann J, Atzorn R, Brückner C, Reinbothe S, Leopold J, Wasternack C, Parthier B (1995) Accumulation of jasmonate, abscisic acid, specific transcripts and proteins in osmotically stressed barley leaf segments. Planta 197 156–162 [Google Scholar]
- Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH (2002) ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J 32 317–328 [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sanchez-Serrano J, Solano R (2004) JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Piqueras R, Sanchez-Serrano J, Solano R (2003) ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Solano R (2005) Molecular players regulating the jasmonate signalling network. Curr Opin Plant Biol 8 532–540 [DOI] [PubMed] [Google Scholar]
- Mauch-Mani B, Mauch F (2005) The role of abscisic acid in plant-pathogen interactions. Curr Opin Plant Biol 8 409–414 [DOI] [PubMed] [Google Scholar]
- Moons A, Prinsen E, Bauw G, Van Montagu M (1997) Antagonistic effects of abscisic acid and jasmonates on salt stress-inducible transcripts in rice roots. Plant Cell 9 2243–2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur LAJ, Kenton P, Atzorn R, Miersch O, Wasternack C (2006) The outcome of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol 140 249–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Kang X (2006) Arabidopsis SHORT HYPOCOTYL UNDER BLUE1 contains SPX and EXS domains and acts in cryptochrome signaling. Plant Cell 18 921–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J-H, Halitschke R, Kim HB, Baldwin IT, Feldmann KA, Feyereisen R (2002) A knock-out mutation in allele oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J 31 1–12 [DOI] [PubMed] [Google Scholar]
- Pena-Cortes H, Fisahn J, Willmitzer L (1995) Signals involved in wound-induced proteinase inhibitor II gene expression in tomato and potato plants. Proc Natl Acad Sci USA 92 4106–4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena-Cortes H, Prat S, Atzorn R, Wasternack C, Willmitzer L (1996) Abscisic acid-deficient plants do not accumulate proteinase inhibitor II following systemin treatment. Planta 198 447–451 [Google Scholar]
- Pena-Cortes H, Sanchez-Serrano J, Mertens R, Willmitzer L, Prat S (1989) Abscisic acid is involved in the wound-induced expression of the proteinase inhibitor II gene in potato and tomato. Proc Natl Acad Sci USA 86 9851–9855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier Y, Thoma S, Somerville C, Schiefelbein J (1991) A mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol 97 1087–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE (2000) Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock CD (2000) Pathways to abscisic acid-regulated gene expression. New Phytol 148 357–396 [DOI] [PubMed] [Google Scholar]
- Sanders PM, Lee PY, Biesgen C, Boone JD, Beals TP, Weiler EW, Goldberg RB (2000) The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12 1041–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki-Sekimoto Y, Taki N, Obayashi T, Aono M, Matsumoto F, Sakurai N, Suzuki H, Hirai MY, Noji M, Saito K, et al. (2005) Coordinated activation of metabolic pathways for antioxidants and defence compounds by jasmonates and their roles in stress tolerance in Arabidopsis. Plant J 44 653–668 [DOI] [PubMed] [Google Scholar]
- Schaller F, Schaller A, Stinzi A (2005) Biosynthesis and metabolism of jasmonates. J Plant Growth Regul 23 179–199 [Google Scholar]
- Schilmiller AL, Howe GA (2005) Systemic signalling in the wound response. Curr Opin Plant Biol 8 369–377 [DOI] [PubMed] [Google Scholar]
- Staswick PE (2008) JAZing up jasmonate signaling. Trends Plant Sci 13 66–71 [DOI] [PubMed] [Google Scholar]
- Staswick PE, Su W, Howell SH (1992) Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc Natl Acad Sci USA 89 6837–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I (2004) The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16 2117–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic A, Ribot C, Rouached H, Wang Y, Chong J, Belbahri L, Delessert S, Poirier Y (2007) Members of the PHO1 gene family show limited functional redundancy in phosphate transfer to the shoot, and are regulated by phosphate deficiency via distinct pathways. Plant J 50 982–994 [DOI] [PubMed] [Google Scholar]
- Stelmach BA, Müller A, Hennig P, Laudert D, Andert L, Weiler EW (1998) Quantitation of the octadecanoid 12-oxo-phytodienoic acid, a signalling compound in mechanotransduction. Phytochemistry 47 539–589 [DOI] [PubMed] [Google Scholar]
- Stinzi A, Browse J (2000) The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci USA 97 10625–10630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinzi A, Weber H, Reymond P, Browse J, Farmer EE (2001) Plant defense in the absence of jasmonic acid: the role of cyclopentenones. Proc Natl Acad Sci USA 98 12837–12842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taki N, Sasaki-Sekimoto Y, Obayashi T, Kikuta A, Kobayashi K, Ainai T, Yagi K, Sakurai N, Suzuki H, Masuda T, et al (2005) 12-Oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiol 139 1268–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J (2007) JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signaling. Nature 448 661–665 [DOI] [PubMed] [Google Scholar]
- Truman W, Bennett MH, Kubigsteltig I, Turnbull C, Grant M (2007) Arabidopsis systemic immunity uses conserved defense signaling pathways and is mediated by jasmonates. Proc Natl Acad Sci USA 104 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Allmann S, Wu J, Baldwin IT (2007) Comparisons of LIPOXYGENASE3- and JASMONATE RESISTANT4/6-silenced plants reveal that jasmonic acid and jasmonic acid-amino acid conjugates play different roles in herbivore resistance of Nicotiana attenuata. Plant Physiol 146 904–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ribot C, Rezzonico E, Poirier Y (2004) Structure and expression profile of the Arabidopsis PHO1 gene family indicates a broad role in inorganic phosphate homeostasis. Plant Physiol 135 400–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C (2007) Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann Bot (Lond) 100 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler EW, Kutchan TM, Gorba T, Brodschelm W, Niesel U, Bublitz F (1994) The Pseudomonas phytotoxin coronatine mimics octadecanoid signalling molecules of higher plants. FEBS Lett 345 9–13 [DOI] [PubMed] [Google Scholar]
- Wilen RW, van Rooijen GJH, Pearce DW, Pharis RP, Holbrook LA, Moloney MM (1991) Effects of jasmonic acid on embryo-specific processes in Brassica and Linum oilseed. Plant Physiol 95 399–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280 1091–1094 [DOI] [PubMed] [Google Scholar]
- Xu L, Liu F, Wang Z, Peng W, Huang R, Huang D, Xie D (2001) The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14 1919–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (2004) Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci 10 88–94 [DOI] [PubMed] [Google Scholar]
- Yan Y, Stolz S, Chételat A, Reymond P, Pagni P, Dubugnon L, Farmer EE (2007) A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng ZL, Nafisi M, Tam A, Li H, Crowell DN, Chary SN, Schroeder JI, Shen J, Yang Z (2002) Plasma membrane-associated ROP10 small GTPase is a specific negative regulator of abscisic acid responses in Arabidopsis. Plant Cell 14 2787–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J-K (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Physiol Plant Mol Biol 53 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]