RNA silencing has become a major focus of molecular biology and biomedical research around the world. This is highlighted by a simple PubMed search for “RNA silencing,” which retrieves almost 9,000 articles. Interest in gene silencing-related mechanisms stemmed from the early 1990s, when this phenomenon was first noted as a surprise observation by plant scientists during the course of plant transformation experiments, in which the introduction of a transgene into the genome led to the silencing of both the transgene and homologous endogenes. From these initial studies, plant biologists have continued to generate a wealth of information into not only gene silencing mechanisms but also the complexity of these biological pathways as well as revealing their multilevel interactions with one another. The plant biology community has also made significant advancements in exploiting RNA silencing as a powerful tool for gene function studies and crop improvements.
In this article, we (1) review the rich history of gene silencing research and the knowledge it has generated into our understanding of this fundamental mechanism of gene regulation in plants; (2) describe examples of the current applications of RNA silencing in crop plants; and (3) discuss improvements in RNA silencing technology and its potential application in plant science.
YESTERDAY'S RNA SILENCING IN PLANTS—SO MANY CURIOUS FINDINGS!
Transfer DNA (T-DNA) vectors, modified from the tumor-inducing plasmid of Agrobacterium tumefaciens, have been used extensively for plant transformation to study gene expression. In an early study by Matzke et al. (1989), two T-DNA vectors encoding different selectable markers were sequentially introduced into the tobacco (Nicotiana tabacum) genome by Agrobacterium-mediated transformation. The authors reported that the selectable marker encoded by the first T-DNA became inactive in a subset of their double-transformant population following the introduction of the second vector. The observed transgene inactivation was correlated with the methylation of the promoter sequences driving the expression of the selectable marker gene, delivered by the initial transformation event, and the initiation of DNA methylation and gene inactivation was dependent on the genomic integration of the second T-DNA. The authors suggested that the substantial homology shared by the two T-DNA vectors, including two copies of the nopaline synthase promoter per T-DNA insert, may have initiated the methylation of the first vector. The following year, petunia (Petunia hybrida) plants, engineered to harbor additional transgene copies of the flower pigmentation gene, chalcone synthase (CHS), provided additional insights into homology-dependent gene silencing mechanisms in plants. These plants were modified to overexpress CHS with the aim of intensifying the purple coloration of flowers. However, the flowers of these modified plants expressed a dramatic range of pigmentation, including intense purple, patterns of purple and white, and flowers that were completely white. Molecular dissection of these transformed populations revealed that in some plant lines both the introduced and endogenous forms of the CHS gene were “turned off” or silenced to differing degrees, in a phenomenon the authors referred to as “cosuppression” (Napoli et al., 1990; van der Krol et al., 1990).
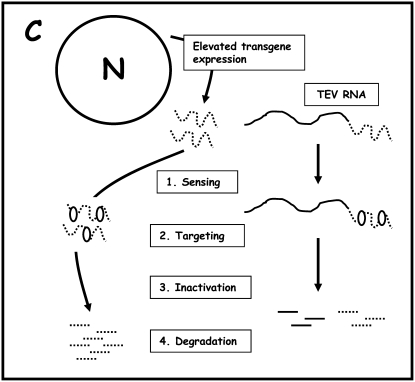
The underlying mechanisms responsible for these initial curious observations of gene silencing in plants remained unknown for many years, especially how it could be so sequence specific. Around the same time, cosuppression-like observations were being made in plants engineered to express virus-encoded sequences, namely the viral coat protein (CP) or a segment of viral replicase. Plants expressing one of these viral proteins generally conferred resistance to the virus from which the protein sequences were derived or to closely related viral strains. In one such study, Lindbo and colleagues (1993) transformed tobacco plants with the gene sequence of the tobacco etch virus (TEV) CP to provide TEV resistance. They noted that TEV could initiate replication in transformed plants, producing the typical systematic symptoms of infection, but these plants were able to outgrow TEV infection approximately 3 to 5 weeks after the initial inoculation event, returning to a “recovered” healthy noninfected state. Recovered leaves did not support subsequent inoculations with TEV, but they did support replication of the unrelated virus Potato virus X (PVX). Molecular analyses of the recovered tissue, using nuclear run-off assays and northern blotting, showed that introduced TEV sequences were still actively transcribed, but corresponding mRNA failed to accumulate. These observations led the authors to speculate that the gene silencing or cosuppression initiated by the transgene and viral trigger was localized to the cytoplasm and occurred at the posttranscriptional level. Among other models to account for the sequence specificity of transgene-mediated TEV resistance, the authors proposed that this process is initiated by high levels of RNA, above a certain threshold, in the cytoplasm and that a plant-encoded RNA-dependent RNA polymerase could be involved to generate a complementary RNA strand, which identified and hybridized with the invading viral RNA to disrupt its function and cause degradation. Figure 1 is a simplified reproduction of the prescient model proposed by Lindbo et al. (1993) to explain the gene silencing mechanisms observed using their viral system.
Figure 1.
Simplified reproduction of the model proposed by Lindbo et al. (1993) to explain the RNA degradation and antiviral state observed in their study. The plant cell is able to detect elevated levels or aberrant forms of RNA in the cytoplasm (C), leading to the targeting and inactivation of the RNA by a protein or nucleic acid cellular factor that degrades the targeted RNA.
To avoid the homology-dependent gene silencing observed in plant lines harboring multiple T-DNA insertions at either the transcriptional or posttranscriptional level, Angell and Baulcombe (1997) developed an alternative strategy to introduce transgenes into the plant genome. Amplicons or transgene vectors were developed, consisting of a cDNA of replicating PVX RNA into which a foreign gene could be inserted. Viral replication was predicted to result in a steady-state level of introduced transgene RNA that could mask the variations in gene expression associated with the use of existing T-DNA-based vectors, resulting in a reproducibly high level of transgene expression throughout the entire transformant population. However, instead of observing high expression levels of the introduced transgene (GUS) initiated by PVX amplicon replication, the authors noted that the amplicon-based transgene system induced a high level of posttranscriptional gene silencing (PTGS) in all plant lines analyzed. Tobacco plants inoculated with the PVX:GUS:CP amplicon accumulated virus-derived sequences at a much lower than expected level and showed no symptoms of PVX infection. The unexpected phenotype of PVX:GUS:CP infection was similar to those reported for other homology-dependent resistance studies of plants expressing viral genome sequences (Swaney et al., 1995).
An additional study from the Baulcombe group (Ratcliff et al., 1997) added extra weight to Lindbo's original hypothesis that a cytoplasmic RNA trigger was responsible for both virus resistance and homology-dependent gene silencing. Leaves of Nicotiana clevelandii plants initially inoculated with the Tomato black ring nepovirus (strain W22) were subsequently inoculated with additional virus strains that were progressively less related to W22 once the leaves had returned to a recovered state. The authors observed increased virus-associated symptoms in plants inoculated with viral strains less closely related to W22. PVX, an unrelated virus, was shown to be able to infect recovered N. clevelandii leaves, but when the PVX virus was modified to carry W22 sequences, the modified viral mRNA failed to accumulate to detectable levels. On the basis of these results, the authors suggested that gene silencing occurs when the plant erroneously perceives an introduced transgene or its RNA product as part of an invading virus, silencing the invading nucleic acids as part of a natural defense mechanism.
Continuing work focusing on the expression of CHS in petunia flowers also suggested that transgene-derived RNA was acting as the trigger to induce the sequence specificity of cosuppression. Purple-colored flowers were shown to express the CHS gene at very high levels, whereas the expression of CHS mRNA was suppressed in white flowers actively undergoing cosuppression. Molecular analyses of cosuppressed flowers revealed that not only was the expression of full-length CHS transcript reduced, but additional truncated transcripts also accumulated in plants with white flowers, accounting for the majority of CHS-specific transcripts detected in cosuppressed plant lines. A percentage of the truncated transcripts were predicted to form extensive secondary structure. This led the authors (Metzlaff et al., 1997) to suggest that overexpression of the CHS transgene resulted in the formation of aberrant RNA species through endonucleatic cleavage of secondary structures and that the aberrant RNA induced cosuppression by pairing with complementary regions of endogene mRNA, rendering the mRNA a target of endonucleatic cleavage. In the same year that the Baulcombe and Metzlaff groups were favoring the high level of expression of aberrant RNA species as the trigger for these processes, members of the Kooter group, based on their analysis of integration patterns of T-DNA insertions in CHS-silenced petunia, were advocating the idea of ectopic DNA-DNA pairing as the initiator (Stam et al., 1997). They found a correlation between the initiation of CHS silencing and the presence of multiple T-DNAs integrated into the plant genome at the same locus in an inverted-repeat (IR) orientation, even if the transgene lacked a promoter, but they had difficultly reconciling their IR findings with the previous descriptions of gene silencing induced by monomeric loci (Dorlhac de Borne et al., 1994; Palauqui and Vaucheret, 1995; Jorgensen et al., 1996; Thierry and Vaucheret, 1996).
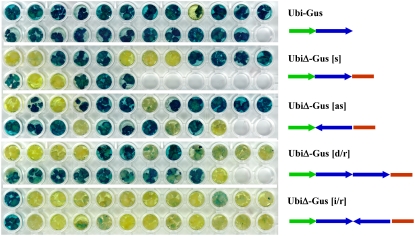
With the seemingly conflicting evidence about the initiation of gene silencing, our group took the approach of directly testing double-stranded RNA (dsRNA) as the initiator (Waterhouse et al., 1998). We showed that when sense and antisense transgenes containing potato virus Y (PVY) sequences were brought together by crossing, the plants exhibited silencing that protected them against the virus. However, the parental plant lines that contained either sense or antisense transgenes alone were not protected against viral infection. Also, the expression of a hairpin RNA (hpRNA) from an IR transgene was a much more efficient initiator of silencing than the expression of a sense or an antisense transgene alone. The IR construct encoding hpRNA of a truncated version of the GUS reporter gene silenced the expression of GUS in 90% of lines when this construct was superimposed on endogenous GUS activity in rice (Oryza sativa) callus via Agrobacterium-mediated transformation (Waterhouse et al., 1998; Wang and Waterhouse, 2000). When the GUS sense or antisense transgenes were used to supertransform the same rice material, the silencing efficiencies were significantly lower than that of the IR construct (Fig. 2). The GUS and PVY experiments provided the first solid evidence that the formation of a dsRNA molecule was a crucial step in the initiation of gene silencing in plants and provided the design of hpRNA constructs (Smith et al., 2000; Wang and Waterhouse 2000; Wesley et al., 2001) that are widely used in both plants and animals today.
Figure 2.
RNA silencing of GUS in rice callus using a sense, antisense, or IR vector. Waterhouse et al. (1998) clearly showed the significantly increased silencing efficiency offered by their IR GUS vector, which produces hpRNA to direct GUS silencing, compared with the silencing efficiencies of existing plant expression vectors, which only expressed unidirectional sequences in either the sense or the antisense orientation.
Following the discovery that dsRNA induces RNA interference (RNAi) in nematodes (Fire et al., 1998), plants (Waterhouse et al., 1998), protozoa (Ngo et al., 1998), and insects (Kennerdell and Carthew, 1998), the second major breakthrough in RNA silencing was made the following year: the identification and association of small RNA (sRNA) molecules in plants actively undergoing PTGS. Hamilton and Baulcombe (1999) screened for sRNA species in four different silencing backgrounds: (1) tomato (Solanum lycopersicum) plants undergoing cosuppression of an endogenous gene following homologous transgene insertion; (2) tobacco plants undergoing PTGS of a GUS transgene; (3) Nicotiana benthamiana plants in which the GFP gene was systemically silenced following inoculation with Agrobacterium harboring a GFP construct; and (4) N. benthamiana plants inoculated with PVX. sRNAs approximately 25 nucleotides in length and specific to the nucleic acid sequence undergoing silencing were identified in all four silencing backgrounds, and interestingly, the authors also showed correlation between the level of sRNA accumulation and the silencing efficiency conferred by each of these systems.
A detailed picture of the RNA silencing pathway in plants was starting to take shape: the introduction of foreign nucleic acid, be it transgene or virus derived, into the plant cell results in the production of a dsRNA molecule that is subsequently processed into the sRNA species that direct the sequence specificity of the observed silencing. These species of sRNA, typically 21 to 24 nucleotides in length, are now referred to as small-interfering RNAs (siRNAs). In vitro experiments in fly (Drosophila melanogaster) embryos demonstrated that the ribonuclease (RNase) III-like endonuclease termed Dicer was the endonuclease class responsible for processing siRNA species from dsRNA (Bernstein et al., 2001). An Arabidopsis (Arabidopsis thaliana) mutant line, carpel factory (caf), characterized by floral meristem determinacy defects as well as other whole-plant organ morphogenesis abnormalities was initially identified in a developmental screen by Steve Jacobsen. Molecular characterization of the caf mutant revealed that the CAF gene encodes a protein with similarities to both DExH/DEAD-box type RNA helicases and RNase III proteins (Jacobsen et al., 1999). We now know that CAF is one of four DICER-LIKE genes (DCL1–DCL4) encoded by the Arabidopsis genome (Finnegan et al., 2003; Xie et al., 2004; Margis et al., 2006) and that the floral defects associated with the caf phenotype result from the mutant's inability to process a class of endogenous sRNAs from their precursor dsRNA molecules (Golden et al., 2002; Park et al., 2002). CAF is now referred to as DCL1, the founding member of the plant-specific RNase III-like endonuclease family of proteins that cleave endogenous dsRNAs to produce sRNAs.
AGRONAUTE1 (AGO1) was isolated in an earlier developmental mutant screen, with ago1 Arabidopsis plants characterized by unexpanded pointed cotyledons, very narrow rosette leaves, and a single thickened and partially fasciated inflorescence (Bohmert et al., 1998). Homology searches not only revealed the existence of nine other AGO proteins encoded by the Arabidopsis genome but also identified substantial homology between AGO1 and the Caenorhabditis elegans protein RNAi-deficient 1 (RDE-1). RDE-1, like AGO1, was identified in a mutant screen for proteins required for RNAi in C. elegans, and rde-1 mutant worms were shown to be unable to mount an RNAi response when exposed to dsRNA (Tabara et al., 1999). AGO1 has since been shown to be a crucial component for RNA silencing in plants, with AGO1 using the guide strand of sRNA duplexes to target complementary mRNAs for cleavage in Arabidopsis (Baumberger and Baulcombe, 2005). The characterization of AGO1 was the last piece of the simplest form of the RNA silencing puzzle in plants; that is, following the formation of dsRNA, a DCL protein recognizes the dsRNA and processes it into shorter (approximately 21–25 nucleotides) sRNA duplexes from which one strand—the guide strand—is used by an AGO protein to target complementary mRNA to repress gene expression.
TODAY'S RNA SILENCING IN PLANTS—STILL A LONG WAY TO GO!
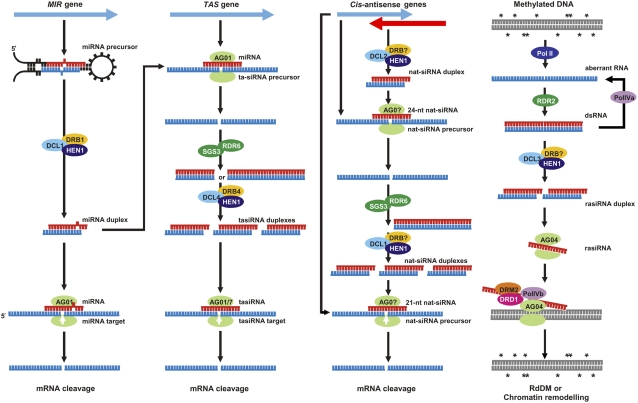
We now have a much greater understanding of the endogenous gene silencing pathways in Arabidopsis and their crucial involvement in controlling the expression of developmentally regulated genes, repressing the activity of the vast array of repetitive elements in the plant genome, and providing resistance against invading viral nucleic acids. In addition to the four DCL and 10 AGO protein family members encoded by the Arabidopsis genome, two other gene families have been shown to work in concert with the DCL and AGO proteins, namely the RNA-directed RNA polymerase (RDR) and double-stranded RNA-binding domain (dsRBP) gene families, of which there are six and five members, respectively, in Arabidopsis. Various members of the DCL, AGO, RDR, and dsRBP gene families play central roles in the parallel gene silencing pathways in Arabidopsis, including the microRNA (miRNA), trans-acting siRNA (tasiRNA), natural-antisense siRNA (natsiRNA), and repeat-associated siRNA (rasiRNA)/RNA-directed DNA methylation (RdDM) pathways. Figure 3 gives a schematic representation of the sequential steps involved in the parallel pathways of Arabidopsis gene silencing.
Figure 3.
The parallel RNA silencing pathways of Arabidopsis. Schematic representation of the parallel DCL/sRNA-directed RNA silencing pathways in the model dicotyledonous species Arabidopsis, outlining the specific step or steps in each pathway for the individual RNA silencing-associated proteins mentioned in the text of this review.
The miRNA Pathway
sRNAs are classed into two categories based on their mode of biogenesis: siRNAs are processed from long, perfectly double-stranded RNA, and miRNAs from single-stranded RNA transcripts (transcribed from MIR genes) that have the ability to fold back onto themselves to produce imperfectly double-stranded stem loop precursor structures. The first miRNA, lin-4, was discovered in C. elegans in 1993 by Victor Ambros (Lee et al., 1993), and today hundreds of miRNAs have been identified in plants and animals, including several hundred unique miRNAs in Arabidopsis alone (Millar and Waterhouse, 2005). In Arabidopsis, the primary-miRNA transcript is cleaved by DCL1 in the nucleus with the help of the dsRBP, HYPONASTIC LEAVES1 (HYL1), to produce the shorter precursor-miRNA (pre-miRNA) dsRNA molecule. The first DCL1-catalyzed cleavage step in the miRNA biogenesis pathway is made just below the miRNA duplex region of the dsRNA stem loop (Lu and Fedoroff, 2000). The miRNA duplex is then released from the pre-miRNA stem loop structure by the second cleavage step of the miRNA pathway, which is again directed by the combined action of DCL1 and HYL1 (Vazquez et al., 2004). The two-nucleotide 3′ overhangs of the liberated miRNA duplex are methylated by the sRNA-specific methyltransferase HUA ENHANCER1 (HEN1). The duplexes of siRNAs are also methylated by HEN1, a process that appears to be plant specific and is assumed to protect all sRNA species from polyuridylation and degradation (Chen et al., 2002; Yu et al., 2005). The miRNA duplex is then transported to the cytoplasm, with several classes of miRNA relying on the action of the Drosophila Exportin-5 ortholog HASTY (HST) for this nuclear exportation step. However, the exact role of HST in miRNA biogenesis remains unclear, as other families of miRNA appear to be transported to the cytoplasm via a HST-independent mechanism (Park et al., 2002). Similarly, the specific function of another factor that is required for miRNA biogenesis, SERRATE, remains unclear (Yang et al., 2006). In the cytoplasm, the mature single-stranded miRNA is loaded onto AGO1, the catalytic center of plant RNA-induced silencing complexes (RISC), to guide the Slicer activity of AGO1 to repress the expression of complementary mRNAs, and in plants, miRNA-directed repression of gene expression is predominantly mediated by transcript cleavage. This differs from the miRNA-mediated repression of gene expression in animals and insects, in which the predominant mode of action is translational repression mediated by the binding of the miRNA to its target mRNA, with mammalian miRNA target sequences primarily located in the 3′ untranslated region (UTR) of mammalian transcripts (Mallory and Vaucheret, 2006).
The tasiRNA Pathway
Two miRNAs, miR173 and miR390, have been shown to induce an addition level of complexity to the control of gene expression for normal development in plants (Axtell et al., 2007). These miRNAs bind to their target tasiRNA transcripts (TAS), directing cleavage of the TAS transcript in a miRNA/DCL1/HYL1-mediated manner. However, instead of becoming silenced, these cleaved noncoding RNA transcripts are used as templates for dsRNA synthesis by the RDR, RDR6, with the help of the coiled-coil protein, SUPPRESSOR OF GENE SILENCING3 (SGS3). The dsRNA is then processed into phased 21-nucleotide tasiRNAs by DCL4 in a sequential process initiated at the miRNA cleavage site. Similar to DCL1/HYL1 processing of MIR-derived hpRNA, DCL4 functions in tandem with the dsRBP, DRB4, to generate the phased tasiRNA from RDR6/SGS3-generated TAS dsRNA. The tasiRNAs then target their own specific cognate mRNAs for degradation, and a number of the developmentally important auxin response factors have been shown to be tasiRNA targets (Howell et al., 2007).
The natsiRNA Pathway
The Arabidopsis genome encodes more than 2,000 natural-antisense gene pairs, and these endogenous cis-antisense genes are transcribed from different DNA strands to produce dsRNA transcripts that harbor regions of complementarity at their 3′ ends (Borsani et al., 2005). The dsRNA molecule formed by these complementary end sequences provides a substrate for DCL2 cleavage and the generation of a single 24-nucleotide natsiRNA. This single 24-nucleotide natsiRNA subsequently targets one of the cis-antisense gene pair transcripts for cleavage, and the cleaved RNA molecule is converted to dsRNA by RDR6 and SGS3. The RDR6/SGS3-synthesized dsRNA molecule is then processed into phased 21-nucleotide natsiRNAs by the action of DCL1. The phased 21-nucleotide natsiRNAs, like the tasiRNA class of endogenous sRNAs, are in turn used as guides to direct sequence-specific silencing of homologous mRNAs.
The rasiRNA and RdDM Pathway
Another RNA silencing-related pathway in Arabidopsis that is regulated at the sRNA level is transcriptional gene silencing (TGS), which is an epigenetic mechanism resulting in the silencing of a transgene or an endogenous gene through the inactivation of their promoter sequences. DNA methylation is essential for normal plant and animal development and is also a hallmark of TGS (Mette et al., 2000). In fact, the majority of methylation in plants is associated with repeat sequences, such as transposons, and methylation of these sequences is thought to occur as a natural suppressor to control their expression (Wassenegger, 2005). In Arabidopsis, repeat sequences have been shown to be an extremely rich source of a unique class of siRNAs, termed rasiRNAs, which are of the 24-nucleotide size class, and rasiRNAs have been suggested to direct DNA methylation and hence to transcriptionally silence repetitive DNA sequences in the plant genome (Chan et al., 2005). Wassenegger and colleagues (1994) were the first to demonstrate that homologous transgenes could be methylated following the replication of introduced RNA viroid sequences, suggesting that an RdDM mechanism was responsible. Jones et al. (1998) went on to show that nuclear DNA could be methylated by introducing a homologous cytoplasmically replicating RNA virus. Both groups speculated that a sequence-specific RNA signal was able to enter the nucleus to direct DNA methylation. Since these initial findings, several studies have concentrated on producing RdDM mutants in Arabidopsis to identify the gene silencing machinery involved (Mette et al., 2000; Kanno et al., 2004, 2005). In brief, methylated DNA is thought to act as a template for the transcription of aberrant RNA by a protein with RNA polymerase activity, either RNA polymerase II, RDR2, or the plant-specific PolIVa. This aberrant RNA is then converted to dsRNA by RDR2 or PolIVa, or alternatively, PolIVa uses RDR2-transcribed dsRNA to produce additional aberrant RNA molecules in a self-perpetuating loop (Vaucheret, 2005). The dsRNA is processed by DCL3 into 24-nucleotide siRNAs that are methylated by HEN1 and used by AGO4 to direct the actual sequence-specific DNA methylation step of RdDM, mediated by the combined actions of the SNF2-like chromatin-remodeling protein, DEFECTIVE IN RNA-DIRECTED DNA METHYLATION1 (DRD1), the alternative form of the plant-specific PolIV, PolIVb, and the primary de novo DNA methyltransferase, DOMAINS REARRANGED METHYLASE2 (DRM2; Cao et al., 2003; Zilberman et al., 2003; Kanno et al., 2004, 2005). Once established, the DNA methyltransferases, METHYLTRANSFERASE1 (MET1) and CHROMOMETHYLASE3 (CMT3), are primarily responsible for maintaining DNA methylation, depending on the sequence context of the methylated cytosine residue. Table I summarizes the proteins discussed above that have been identified as functioning in the parallel gene silencing pathways of Arabidopsis.
Table I.
Proteins involved in the Arabidopsis sRNA-regulated gene silencing pathways
| Protein | Locus | Protein Class/Function |
|---|---|---|
| AGO1 | At1g48410 | RNA slicer/core component of plant RISC |
| AGO4 | At2g27040 | RNA slicer/involved in the establishment phase of RdDM |
| AGO6 | At2g32940 | RNA slicer/rasiRNA-directed heterochromatin formation |
| AGO7 | At1g69440 | RNA slicer/tasiRNA biogenesis and juvenile-to-adult transition |
| CMT3 | At1g69770 | Methyltransferase/maintenance phase of RdDM |
| DCL1 | At1g01040 | RNase III/miRNA, natsiRNA, and tasiRNA biogenesis |
| DCL2 | At3g03300 | RNase III/natsiRNA biogenesis and viral defense |
| DCL3 | At3g43920 | RNase III/rasiRNA biogenesis and establishment phase of RdDM |
| DCL4 | At5g20320 | RNase III/tasiRNA biogenesis and viral defense |
| DRM2 | At3g17310 | Methyltransferase/establishment phase of RdDM |
| HYL1 | At1g09700 | dsRBP/miRNA and tasiRNA biogenesis |
| DRB4 | At3g62800 | dsRBP/miRNA and tasiRNA biogenesis |
| DRD1 | At2g16390 | SNF2-like chromatin-remodeling factor/establishment phase of RdDM |
| HEN1 | At4g29160 | sRNA-specific methyltransferase/sRNA biogenesis |
| HST | At3g05040 | Exportin-5 ortholog/miRNA exportation from nucleus |
| MET1 | At5g49160 | Methyltransferase/maintenance phase of RdDM methylation |
| NRPD1a | At1g63020 | DNA-dependent RNA polymerase/establishment phase of RdDM |
| NRPD1b | At2g40030 | DNA-dependent RNA polymerase/establishment phase of RdDM |
| NRPD2 | At3g23780 | DNA-dependent RNA polymerase/establishment phase of RdDM |
| RDR1 | At1g14790 | RNA-dependent RNA polymerase/viral defense |
| RDR2 | At4g11130 | RNA-dependent RNA polymerase/rasiRNA biogenesis |
| RDR6 | At3g49500 | RNA-dependent RNA polymerase/tasiRNA and natsiRNA biogenesis |
| SGS3 | At5g23570 | Coiled-coil protein/tasiRNA and natsiRNA biogenesis |
Examples of Current Applications of Gene Silencing in Plants
For almost a decade, RNAi has been used as a research tool to discover or validate the functions of genes, and we are now starting to see the use of this technology for commercially focused applications in plants. The applications cover a wide spectrum, from designer flower colors to plant-produced medical therapeutics. They fall into two types of approach: protection of the plant against attack and fine-tuning of metabolic pathways.
Unsurprisingly, given the history of the discovery of PTGS, protection of plants from viral infection has been one of the first commercial outcomes resulting from the application of a gene silencing technique. Transgenic papaya (Carica papaya) with resistance to Papaya ringspot virus (PRSV; Fuchs and Gonsalves, 2007) and the Monsanto-produced NewLeaf Plus and NewLeaf Y potatoes (Solanum tuberosum) with resistance to Potato leafroll virus and PVY (www.research.cip.cgiar.org/) were among the first commercial releases. When released in the late 1990s, the mechanisms responsible for providing protection against these invading viruses remained unknown, but the PRSV-resistant papaya has proven to be a great success over the past decade, helping to save the papaya industry in Hawaii. Since then, there have been many different crop plants protected against a whole range of viruses using constructs that initiate a PTGS response (Fuchs and Gonsalves, 2007). Wang et al. (2000) applied the first deliberate use of RNA silencing for virus protection in the important cereal crop species barley (Hordeum vulgare). Barley yellow dwarf virus (BYDV) is a virus of global importance, as it infects and reduces yields of several cereal species worldwide. In this study, a hpRNA-encoding construct driven by the maize (Zea mays) ubiquitin promoter and targeting the 5′ end of the virus was transformed into barley to produce lines with complete immunity to BYDV. As most plant viruses have single-stranded RNA genomes, they have been the obvious targets for RNAi technology; indeed, we now know that a natural role of the RNA silencing/PTGS pathway in plants is for virus defense (Fusaro et al., 2006).
Controlling viruses by destroying their RNA within a plant cell is a relatively straightforward process and can also be achieved using artificial miRNAs (amiRNAs). Niu and colleagues (2006) used a 273-bp sequence of the Arabidopsis miR159a pre-miRNA transcript expressing amiRNAs against the viral suppressor genes P69 and HC-Pro to provide resistance against turnip yellow mosaic virus and turnip mosaic virus infection, respectively. In addition, a dimeric construct harboring two unique amiRNAs against both viral suppressors conferred resistance against these two viruses in inoculated Arabidopsis plants. Using a different amiRNA vector to target the 2b viral suppressor of the Cucumber mosaic virus (CMV), a suppressor that interacts with and blocks the Slicer activity of AGO1 was also shown to confer resistance to CMV infection in transgenic tobacco. A strong correlation between virus resistance and the expression level of the 2b-specific amiRNA was also shown for individual plant lines (Qu et al., 2007).
A more recently asked question is, can RNAi be used to protect a plant against invading or attacking organisms other than viruses? Agrobacterium is a soil-borne bacterium that causes crown gall disease, which imposes significant economic losses in perennial crops worldwide. It has a horizontal gene transfer system for a suite of oncogenes that, when integrated into the plant genome, generates tumor formation (the tumor-inducing plasmid mentioned earlier in the review). Escobar and colleagues (2001) tested whether RNAi could be used to control this plant parasitic pathogen. They transformed tomatoes with hpRNA constructs against the iaaM and ipt oncogenes, which are required for tumor formation. The authors demonstrated that the target mRNAs were silenced to produce transformant lines that are highly resistant to crown gall disease across a range of biovars, demonstrating the feasibility of such an approach. As the mechanism of resistance is based on hpRNA sequence homology to the mRNAs of the invading genes, it may be more durable than the highly specific receptor-ligand interactions characteristic of traditional plant resistance genes and, as such, might find broad application in agriculture and horticulture.
Plant parasitic nematodes, such as the root-knot (Meloidogyne spp.) and cyst (Heterodera and Globodera spp.) nematodes, cause significant damage to important crops such as legumes, vegetables, and cereals in most parts of the world. When this is coupled with the history of RNA silencing discovery from studies using C. elegans, it was also almost inevitable that the possibility of protecting plants from nematode damage by RNA silencing would be explored. Two approaches have been taken. One relies on targeting plant genes that are involved with the infection process, and the second approach targets essential genes within the nematode. Heterodera schachtii induces syncytial feeding structures in the roots of host plants, and this requires the up-regulation of Suc transporter genes to facilitate increased nutrient flow to the developing structure. Targeting these genes and down-regulating them with RNA silencing resulted in a significant reduction of female nematode development (Hoffman et al., 2008). RNA silencing can be induced in C. elegans by feeding it dsRNA, so it was reasoned that expressing hpRNAs containing sequences of vital nematode genes in the host plant might deliver dsRNA to a feeding nematode to incapacitate or kill it. Indeed, tobacco plants transformed with hpRNA constructs against two such root-knot nematode genes have shown such an effect: the target mRNAs in the plant parasitic nematodes were dramatically reduced, and the plants showed effective resistance against the parasite (Fairbairn et al., 2007). Although neither of these approaches have reached the commercial product stage in agronomic crops, the promise of an inexpensive and environmentally clean way of controlling plant parasitic nematodes, which are estimated to cause annual crop losses of over $125 billion worldwide (Chitwood, 2003), suggests that as long as the resistance holds true in the field, the application of such technology will only be a matter of time.
That hpRNA encoded in a plant can induce RNA silencing in a nematode that feeds upon it may rely on the intimate interactions between the plant and the feeding nematode. However, very recent work describes how this approach can be taken even further, as a protection method against herbivorous insect pests with much less intimate feeding associations. Baum et al. (2007) fed western corn rootworm larvae on artificial diets supplemented with specific dsRNAs, to screen a large number of genes for effective targets, and identified 14 whose knockdown by dsRNA killed the larvae. Transforming maize with a hpRNA against one of these genes, a subunit of the midgut enzyme vacuolar ATPase, gave protection against western corn rootworm infestation at a level that was comparable to that provided by the Bacillus thuringiensis (Bt) toxin transgene. Indeed, the hope that this approach might provide a backup for Bt protection in crops like cotton (Gossypium hirsutum) and maize, in which insects are continuing to develop resistance to Bt, is an attractive alternative strategy.
The examples described above are all RNA silencing-based strategies that protect the plant from pest or pathogen attack, but another widely embraced use of RNA silencing technology has been for reshaping metabolic pathways. For example, RNA silencing has been used to improve the human health attributes of cottonseed oil. Cotton is the world's sixth largest source of vegetable oil, but the oil profile has relatively high levels of palmitic acid, which, although providing stability at the high temperatures used in deep frying, also gives it low-density lipoprotein cholesterol-raising properties in humans. Oils that are low in palmitic acid and rich in either oleic acid or stearic acid have thermostability without the associated low-density lipoprotein cholesterol-raising properties. Liu et al. (2002) have used hpRNA constructs to silence the Δ9 and Δ12 desaturases, which catalyze the biosynthesis of these fatty acids, and have obtained plants that produce seed oil that is much more suitable for human consumption. In a similar vein, modification of the starch composition of wheat (Triticum aestivum) destined for human consumption in affluent countries, by altering its amylose-amylopectin ratio, has the potential to reduce the incidence of cardiovascular disease and colon cancers. Regina and colleagues (2006) have used hpRNA constructs to silence an isoform of a starch-branching enzyme to produce a high-amylose transgenic wheat line, which if widely adopted in western countries could have significant public health benefits.
RNA silencing technology also has many important nonfood applications, such as altering photosynthetic pathways in algae to give increased bioreactor performance (Mussgnug et al., 2007) and reshaping the morphine pathway in poppies (Papaver somniferum) to increase the yield of pharmaceutically significant compounds (Allen et al., 2004). An interesting application in the medical therapeutic arena has been in engineering plant-produced antibodies. Monoclonal antibodies are widely used in the therapeutic treatment of cancer, autoimmune, and inflammatory diseases, and plant-based production of these antibodies is becoming increasingly popular. However, plant-directed glycosylation of the Fc region of an antibody may compromise its ability to mediate effector functions and may also be immunogenic. To combat these potential problems, RNA silencing has been used in the algal Lemna production system to silence two endogenous glycan-transferase activities, resulting in the production of therapeutic antibodies with glycosylation homogeneity, which improves not only the antibody's safety but also its functionality (Cox et al., 2006).
Ironically, the plant RNAi application most likely to be the next commercial reality is one that delivers aesthetic rather than nutritional, medical, or environmental benefits to humankind. It has been a long quest to produce a blue rose, but it has now been achieved with the help of RNA silencing. Roses lack an enzyme for the biosynthesis of dihydromyricetin, an intermediate compound required for the production of delphinidin-based anthocyanins, the major constituents of violet and blue flowers. When the gene encoding this enzyme in Viola was transferred to roses, its expression resulted in the generation of transformed plant lines with purple petals, because one of the rose enzymes involved with the conversion of dihydromyricetin into delphinidin also converts other intermediate compounds into red and yellow pigments. However, silencing this gene using RNAi and introducing the homologous gene from Iris gave transformed rose plants that bore flowers with pure blue hues never seen before (Katsumoto et al., 2007).
FUTURE OF RNA SILENCING TECHNOLOGIES AND ITS CHALLENGES IN PLANTS
Can miRNAs Be Silenced Themselves?
An alternative amiRNA-like strategy was recently employed in plants, not to artificially overexpress a particular miRNA but to antagonize an endogenous miRNA's ability to cleave its specific target(s), providing a new functional analysis tool to study plant miRNAs. This strategy, termed “target mimicry,” relies on the expression of a small non-protein-coding mRNA that contains a complementary miRNA binding site within its sequence. A 23-nucleotide motif is engineered into the noncoding mRNA to contain critical mismatches to the miRNA under study, most notably a mismatched bulge opposite the miRNA cleavage site at positions 9 to 11 of the miRNA. Franco-Zorrilla et al. (2007) have used this approach to study the effects of knocking out the expression of the Arabidopsis miRNAs miR156 and miR319. They produced transformant lines with marked developmental phenotypes, suggesting that target mimicry, consisting of a noncleavable mRNA that forms a nonproductive interaction with the corresponding miRNA, could become an effective approach for studying miRNA activity in plants for altering plant architecture through miRNA repression for agricultural applications. The exact mode of action by target mimicry remains unclear: it not only suppresses the function of, but also reduces the abundance of, the target miRNA under analysis (Franco-Zorrilla et al., 2007).
Can Promoter-Induced Silencing Be Used to Achieve Efficient Gene Silencing in Plants?
TGS accompanied by de novo methylation of a target promoter in plants can be triggered by recombinant viruses or long hpRNA constructs containing promoter sequences (Jones et al., 1998; Mette et al., 2000). Such dsRNA-induced promoter silencing has long been proposed as a potential technology for achieving potent and heritable gene silencing in plants. It has been expected that transcriptional inactivation of promoters could lead to the complete silencing of target genes, a desirable scenario in many applications but often difficult to achieve through silencing of coding sequences. In addition, DNA methylation triggered by dsRNA transgenes could be maintained by DNA methyltransferases to allow stable inheritance of gene silencing in subsequent generations even in the absence of the inducer transgenes. Indeed, consistent and/or heritable silencing has been achieved for promoters of transgenes using both virus-induced gene silencing vectors and hpRNA constructs (Table II; Mette et al., 2000; Jones et al., 2001; Sijen et al., 2001; Kanno et al., 2004; Okano et al., 2008).
Table II.
A summary of promoter silencing in plants
| Target Promoter | Transgene or Endogene | Inducer/Plant | Level of Silencing | Reference |
|---|---|---|---|---|
| NOS | Transgene | hpRNA/Arabidopsis | Strong | Mette et al. (2000) |
| 35S | Transgene | Virus-induced gene silencing/N. benthamiana | Strong and inheritable | Jones et al. (2001) |
| hpRNA/petunia | Strong | Sijen et al. (2001) | ||
| hpRNA/rice | Strong | Okano et al. (2008) | ||
| α′ | Transgene | hpRNA/Arabidopsis | Strong | Kanno et al. (2004) |
| dfrA | Endogenous | hpRNA/petunia | Moderate | Sijen et al. (2001) |
| MS45 | Endogenous | hpRNA/maize | Strong | Cigan et al. (2005) |
| bs7 | Endogenous | hpRNA/maize | Strong | Cigan et al. (2005) |
| GBSS1 | Endogenous | hpRNA/potato | Moderate silencing induced only with a construct containing a 194-bp 5′ transcribed region | Heilersig et al. (2006) |
| se5 | Endogenous | hpRNA/rice | Moderate | Okano et al. (2008) |
| OsRac1 | Endogenous | hpRNA/rice | No silencing | Okano et al. (2008) |
| OsRac3 | Endogenous | hpRNA/rice | No silencing | Okano et al. (2008) |
| OsRac4 | Endogenous | hpRNA/rice | No silencing | Okano et al. (2008) |
| Cen8-9 | Endogenous | hpRNA/rice | No silencing | Okano et al. (2008) |
| Cen8-11 | Endogenous | hpRNA/rice | No silencing | Okano et al. (2008) |
| Cen8-18 | Endogenous | hpRNA/rice | No silencing | Okano et al. (2008) |
| Phytoene desaturase | Endogenous | hpRNA/Arabidopsis | No silencing | M.-B. Wang (unpublished data) |
| Chalcone synthase | Endogenous | hpRNA/Arabidopsis | Weak silencing in small proportions of lines | M.-B. Wang (unpublished data) |
| EIN2 | Endogenous | hpRNA/Arabidopsis | Moderate silencing, but construct contains 410-bp 5′ transcribed sequence | M.-B. Wang (unpublished data) |
Silencing of endogenous genes using promoter hpRNA constructs has also been reported (Table II). Sijen et al. (2001) was the first to show that an endogenous gene can be transcriptionally silenced in petunia using a promoter-specific hpRNA construct, although the silencing appeared to be less efficient than that induced by an hpRNA construct targeting the coding region. Using constitutively expressed promoter hpRNA constructs, Cigan et al. (2005) successfully silenced two anther-expressed genes in maize. However, recent studies have indicated that endogenous promoters are not as amenable to silencing as transgene promoters using hpRNA constructs. Okano et al. (2008) showed that hpRNA transgenes induced de novo DNA methylation in all seven endogenous promoters tested in rice, but only one of the targeted genes was significantly silenced (Table II). Heilersig and colleagues (2006) tried several versions of promoter hpRNA constructs to silence the granule-bound starch synthase I (GBSSI) gene in potato, but efficient silencing was achieved only with a construct that contains part of the 5′ transcribed region of the GBSSI gene, suggesting that the silencing was in fact at the posttranscriptional level. In addition, we have tried to silence three different endogenous genes in Arabidopsis using promoter hpRNA constructs, but we only obtained efficient silencing with a construct that contains a 5′ transcribed sequence of the target gene (Table II). It is unclear why differences exist among endogenous promoters or between endogenous and transgene promoters in their susceptibility to dsRNA-induced TGS in plants. Cytosine content and local DNA features have been proposed as important factors affecting RNA-directed TGS in plants (Fischer et al., 2007; Okano et al., 2008). Interestingly, the small numbers of endogenous promoters silenced efficiently in plants to date are all derived from tissue-specifically expressed genes, which may suggest that tissue- or organ-specific promoters are more susceptible to TGS than a constitutively expressed promoter.
Can Systemic and Transitive Silencing Be Exploited for Silencing Endogenous Genes?
An interesting feature of dsRNA-mediated transgene silencing in plants is its systemic nature; silencing can spread from cell to cell and over long distances via vascular-mediated transport (Voinnet, 2005). Transgenes containing both endogenous and exogenous sequences, such as the nitrite reductase (Palauqui et al., 1997) and GFP (Voinnet et al., 2000) genes, can be silenced systemically through grafting. Systemic silencing requires DCL4 and RDR6, suggesting an involvement of 21-nucleotide siRNAs in the signaling process and RDR6 in the amplification of signals (Dunoyer et al., 2005; Voinnet, 2005). Surprisingly, factors involved in RdDM are required for graft transmission of transgene silencing, indicating that chromatin modification plays a role in the perception and perpetuation of long-distance silencing signals (Brosnan et al., 2007).
Systemic spread of silencing could have both advantageous and detrimental consequences with respect to the application of gene silencing technologies in plants. Long-distance gene silencing induced by localized introduction of dsRNA or by grafting would be particularly useful in horticultural crops such as grapevine (Vitis vinifera) and fruit trees because of the difficulty in generating transgenic plants from these species, plus the normally heterozygous state of their genomes. On the other hand, cell-to-cell and long-distance spread of silencing would make it difficult to achieve tissue- or organ-specific gene silencing, which might be necessary in certain applications. Interestingly, systemic silencing in plants has only been observed when transgenes are used as both the inducer and the target of silencing. To date, no systemic silencing has been associated with the use of endogenous genes as a target (Wang and Metzlaff, 2005), although short-distance cell-to-cell spread of silencing has been reported for one endogenous gene (Dunoyer et al., 2005). However, organ-specific silencing has been achieved in plants against several endogenous genes (Liu et al., 2002), suggesting that localized silencing may not be difficult to achieve in plants, but the application of a broad systemic silencing mechanism for efficient silencing of endogenous genes may be problematic.
Silencing of a transgene can spread from a dsRNA-targeted region to adjacent nontargeted sequences. This phenomenon, known as transitivity, has only been observed when transgenes are used as the target (Garcia-Perez et al., 2004). How silencing spreads in a transitive manner remains unclear, but the process may resemble the biogenesis pathway of tasiRNAs, in which the primary TAS transcript is first cleaved by a miRNA and the cleavage fragment is used by RDR6 to synthesize dsRNA, giving rise to siRNAs in a phased manner. This transitive nature of transgene silencing may have been inadvertently incorporated into two previously reported gene silencing technologies in plants. A 1-aminocyclopropane-carboxylate oxidase 1 (ACO1) transgene containing two upstream IRs of a 79-bp segment of the 5′ UTR ACO1 sequence induces strong silencing in tomato against both the target ACO1 gene and the closely related gene ACO2. ACO2 shares significant sequence homology with ACO1 only within its coding region, and a subsequent study showed that the strong silencing is associated with the accumulation of siRNAs from both the region targeted by the 5′ UTR IRs and the coding sequences immediately downstream (Han and Grierson, 2002). This suggests that silencing has spread from the 5′ UTR targeted by the IR hpRNA to downstream ACO1 coding sequences of the transgene. This transitive spread of silencing could then lead to cross-silencing of ACO2 due to its sequence homology to ACO1 in the coding region. Similar to the ACO1 construct, sense constructs carrying an IR at the 3′ UTR have been shown to induce consistent silencing to a number of endogenous genes in tomato and Arabidopsis (Brummell et al., 2003). The 3′ IR shares no sequence homology with the target gene sequences, so the silencing must be induced by siRNAs corresponding to upstream sense sequences of the transgene. Thus, in both cases, silencing spreads transitively from the IR-targeted regions to downstream or upstream untargeted regions. It is possible that primary siRNAs derived from the IR region cause the cleavage of transgene transcripts, giving rise to TAS-like RNA fragments that are then converted to dsRNA by RDR6 to produce secondary siRNAs that direct the spread of silencing into surrounding nontargeted sequences.
The Role of RNA Silencing in Plant Defense against Nonviral Pathogens: Can This Be Exploited to Generate Resistance against a Broad Range of Pathogens in Plants?
As discussed earlier, viruses are a direct target of RNA silencing mechanisms, and hpRNA-based constructs targeting viral RNAs have proven superior to previous transgenic approaches for generating resistance in plants against viruses. With the exception of Agrobacterium, whose T-DNA-encoded genes have been shown to be targeted by PTGS, there has been no evidence that genes of nonviral pathogens are a direct target of RNA silencing in plants. Despite the aforementioned resistance to Agrobacterium, nematode, or insects that has been achieved using hpRNA constructs in plants, it remains to be seen whether direct targeting of pathogen-encoded genes will become a practical approach for controlling a broad range of plant diseases.
Recent studies have provided evidence that RNA silencing pathways also play a role in plant defense against nonviral pathogens and insects, which could provide an alternative platform for developing disease and insect control strategies in plants. For instance, the natsiRNA nat-siRNAATGB2 is strongly induced in Arabidopsis upon infection by Pseudomonas syringae pv tomato and down-regulates a PPRL gene that encodes a negative regulator of the RPS2 disease resistance pathway. As a result, the induction of nat-siRNAATGB2 increases the RPS2-mediated race-specific resistance against P. syringae pv tomato in Arabidopsis (Katiyar-Agarwal et al., 2006). Recently, the accumulation of a new class of sRNA, 30 to 40 nucleotides in length, termed long-siRNAs (lsiRNAs), was associated with P. syringae infection. One of these lsiRNAs, AtlsiRNA-1, contributes to plant bacterial resistance by silencing AtRAP, a negative regulator of plant defense (Katiyar-Agarwal et al., 2007). A Pseudomonas bacterial flagellin-derived peptide is found to induce the accumulation of miR393 in Arabidopsis. miR393 negatively regulates mRNAs of F-box auxin receptors, resulting in increased resistance to the bacterium (P. syringae), and the overexpression of miR393 was shown to reduce the plant's bacterial titer by 5-fold (Navarro et al., 2006). A recent report showed that specific miRNAs are associated with disease development in pine (Pinus spp.) following fusiform rust infection, suggesting that the miRNA pathway may also be involved in plant interactions with fungal pathogens (Lu et al., 2007). Evidence for the involvement of miRNAs in plant defense against nonviral pathogens is further provided by recent deep sequencing of sRNAs in plants, resulting in the identification of at least two miRNAs that target NBS-LRR disease resistance genes in Arabidopsis (Rajagopalan et al., 2006; Howell et al., 2007). Besides bacteria and fungi, a recent report shows that silencing of the RDR protein RDR1 in Nicotiana attenuata, either with a virus-induced silencing vector or with an IR transgene, significantly increases the susceptibility of the plant to attack by herbivorous insects such as Manduca sexta, mirids, beetles, and grasshoppers (Pandey and Baldwin, 2007). As RDRs are important components in siRNA biogenesis, this indicates that siRNAs may also play a role in plant defense mechanisms mounted against insect pests.
While more research is required to establish specific roles of RNA silencing pathways in plant defense against nonviral pathogens and insects, it can be anticipated that gene silencing-based technologies could potentially be developed to control bacterial infection, fungal diseases, and insect infestation of agronomically important crop species. Possible approaches include the overexpression or down-regulation of host-encoded gene silencing factors known to be involved in disease resistance pathways. Alternatively, the overexpression or knockdown of sRNA species already shown to be involved in plant defense pathways could also give rise to resistance against nonviral pathogens in plants.
CONCLUSION
Plant biologists pioneering in homology-dependent transgene silencing and pathogen-derived virus resistance research in the early 1990s could not have realized at the time that they had stumbled on one of the most fundamental and conserved gene control mechanisms in eukaryotic organisms. What they saw, but could not fully understand at the time, including cosuppression, RNA-mediated virus resistance, and RdDM, represent the core aspects of what we know today about the mechanisms and functions of RNA silencing. The revelation of the dsRNA-induced mechanism in 1998 was a watershed event, leading to a vast expansion of interest in researching the molecular details and biological functions of RNA silencing in all eukaryotes. This and subsequent discoveries of the various related sRNA pathways revolutionized the way we study gene regulation and developmental control in plants and animals.
Although there is still much to learn about the molecular processes and biological roles of RNA silencing in plants, our current understanding of this RNA-mediated gene control mechanism has already provided new platforms for developing molecular tools for gene function studies and crop improvements. For instance, the hpRNA and artificial miRNA systems, developments based on our knowledge of two basic sRNA pathways in plants, have already proven to be effective tools for reverse genetic analysis of gene function and for genetic engineering of virus resistance and the manipulation of metabolic pathways to improve agronomic traits and to produce products of pharmaceutical value in plants. The continued efforts to solve the remaining puzzles in the RNA silencing pathways, such as virus-induced gene silencing and dsRNA-induced TGS, are likely to generate even further technologies. And from the recent discovery that RNA silencing pathways play a role in both biotic and abiotic stress responses in plants, we can hope that RNA silencing-based technologies will help humankind to face the challenges of productive agriculture in the increasingly unfavorable environmental conditions associated with climate change.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Andrew Eamens (andrew.eamens@csiro.au).
References
- Allen RS, Millgate AG, Chitty JA, Thisleton J, Miller JA, Fist AJ, Gerlach WL, Larkin PJ (2004) RNAi-mediated replacement of morphine with the nonnarcotic alkaloid reticuline in opium poppy. Nat Biotechnol 22 1559–1566 [DOI] [PubMed] [Google Scholar]
- Angell SM, Baulcombe DC (1997) Consistent gene silencing in transgenic plants expressing a replicating potato virus X RNA. EMBO J 16 3675–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Synder JA, Bartel DP (2007) Common functions for diverse small RNAs of land plants. Plant Cell 19 1750–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum JA, Bogaert T, Clinton W, Heck GR, Feldmann P, Ilagan O, Johnson S, Plaetinck G, Munyikwa T, Pleau M, et al (2007) Control of coleopteran insect pests through RNA interference. Nat Biotechnol 25 1322–1326 [DOI] [PubMed] [Google Scholar]
- Baumberger N, Baulcombe DC (2005) Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA 102 11928–11933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409 363–366 [DOI] [PubMed] [Google Scholar]
- Bohmert K, Camus I, Bellini C, Bouchez D, Caboche M, Benning C (1998) AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J 17 170–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu J-K (2005) Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosnan CA, Mitter N, Christie M, Smith NA, Waterhouse PM, Carroll BJ (2007) Nuclear gene silencing directs reception of long-distance mRNA silencing in Arabidopsis. Proc Natl Acad Sci USA 104 14741–14746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummell DA, Balint-Kurti PJ, Harpster MH, Palys JM, Oeller PW, Gutterson N (2003) Inverted repeat of a heterologous 3′-untranslated region for high-efficiency, high-throughput gene silencing. Plant J 33 793–800 [DOI] [PubMed] [Google Scholar]
- Cao X, Aufsatz W, Zilberman D, Mette MF, Huang MS, Matzke M, Jacobsen SE (2003) Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr Biol 13 2212–2217 [DOI] [PubMed] [Google Scholar]
- Chan SW, Henderson IR, Jacobsen SE (2005) Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet 6 351–360 [DOI] [PubMed] [Google Scholar]
- Chen X, Liu J, Cheng Y, Jia D (2002) HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development 129 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood DJ (2003) Research on plant-parasitic nematode biology conducted by the United States Department of Agriculture-Agricultural Research Service. Pest Manag Sci 59 748–753 [DOI] [PubMed] [Google Scholar]
- Cigan AM, Unger-Wallace E, Haug-Collet K (2005) Transcriptional gene silencing as a tool for uncovering gene function in maize. Plant J 43 929–940 [DOI] [PubMed] [Google Scholar]
- Cox KM, Sterling JD, Regan JT, Gasdaska JR, Frantz KK, Peele CG, Black A, Passmore D, Moldovan-Loomis C, Srinivasan M, et al (2006) Glycan optimization of a human monoclonal antibody in the aquatic plant Lemna minor. Nat Biotechnol 24 1591–1597 [DOI] [PubMed] [Google Scholar]
- Dorlhac de Borne F, Vincentz M, Chupeau Y, Vaucheret H (1994) Co-suppression of nitrate reductase host genes and transgenes in transgenic tobacco plants. Mol Gen Genet 243 613–621 [DOI] [PubMed] [Google Scholar]
- Dunoyer P, Himber C, Voinnet O (2005) DICER-LIKE4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat Genet 37 1356–1360 [DOI] [PubMed] [Google Scholar]
- Escobar MA, Civerolo EL, Summerfelt KR, Dandekar AM (2001) RNAi-mediated oncogene silencing confers resistance to crown gall tumorigenesis. Proc Natl Acad Sci USA 98 13437–13442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbairn DJ, Cavalloro AS, Bernard M, Mahalinga-Iyer J, Graham MW, Botella JR (2007) Host-delivered RNAi: an effective strategy to silence genes in plant parasite nematodes. Planta 226 1525–1533 [DOI] [PubMed] [Google Scholar]
- Finnegan EJ, Margis R, Waterhouse PM (2003) Posttranscriptional gene silencing is not compromised in the Arabidopsis CARPEL FACTORY (DICER-LIKE1) mutant, a homolog of Dicer-1 from Drosophila. Curr Biol 13 236–240 [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391 806–811 [DOI] [PubMed] [Google Scholar]
- Fischer U, Kuhlmann M, Pecinka A, Schmidt R, Mette MF (2007) Local DNA features affect RNA-directed transcriptional gene silencing and DNA methylation. Plant J 53 1–10 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39 1033–1037 [DOI] [PubMed] [Google Scholar]
- Fuchs M, Gonsalves D (2007) Safety of virus-resistant transgenic plants two decades after their introduction: lessons from realistic field risk assessment studies. Annu Rev Phytopathol 45 173–202 [DOI] [PubMed] [Google Scholar]
- Fusaro AF, Matthew L, Smith NA, Curtin SJ, Dedic-Hagan J, Ellacott GA, Watson JM, Wang MB, Brosnan C, Carroll BJ, et al (2006) RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep 7 1168–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Perez RD, Houdt HV, Depicker A (2004) Spreading of post-transcriptional gene silencing along the target gene promotes systemic silencing. Plant J 38 594–602 [DOI] [PubMed] [Google Scholar]
- Golden TA, Schauer SE, Lang JD, Pien S, Mushegian AR, Grossniklaus U, Meinke DW, Ray A (2002) SHORT INTEGUMENTS1/SUSPENSOR1/CARPEL FACTORY, a Dicer homolog, is a maternal effect gene required for embryo development in Arabidopsis. Plant Physiol 130 808–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286 950–952 [DOI] [PubMed] [Google Scholar]
- Han Y, Grierson D (2002) The influence of inverted repeats on the production of small antisense RNAs involved in gene silencing. Mol Genet Genomics 267 629–635 [DOI] [PubMed] [Google Scholar]
- Heilersig BH, Loonen AE, Janssen EM, Wolters AM, Visser RG (2006) Efficiency of transcriptional gene silencing of GBSSI in potato depends on the promoter region that is used in an inverted repeat. Mol Genet Genomics 275 437–449 [DOI] [PubMed] [Google Scholar]
- Hoffman Y, Aflalo C, Zarka A, Gutman J, James TY, Boussiba S (2008) Isolation and characterization of a novel chytrid species (phylum Blastocladiomycota), parasitic on the green alga Haematococcus. Mycol Res 112 70–81 [DOI] [PubMed]
- Howell MD, Fahlgren N, Chapman EJ, Cumbie JS, Sullivan CM, Givan SA, Kasschau KD, Carrington JC (2007) Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell 19 926–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Running MP, Meyerowitz EM (1999) Disruption of an RNA helicase/RNase III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 126 5231–5243 [DOI] [PubMed] [Google Scholar]
- Jones AL, Thomas CL, Maule AJ (1998) De novo methylation and co-suppression induced by a cytoplasmically replicating plant RNA virus. EMBO J 17 6385–6393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L, Ratcliff F, Baulcombe DC (2001) RNA-directed transcriptional gene silencing in plants can be inherited independently of the RNA trigger and requires Met1 for maintenance. Curr Biol 11 747–757 [DOI] [PubMed] [Google Scholar]
- Jorgensen RA, Cluster PD, English J, Que Q, Napoli CA (1996) Chalcone synthase cosuppression phenotypes in petunia flowers: comparison of sense vs. antisense constructs and single-copy vs. complex T-DNA sequences. Plant Mol Biol 31 957–973 [DOI] [PubMed] [Google Scholar]
- Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, Daxinger L, Kriel DP, Matzke M, Matzke AJM (2005) Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet 37 761–765 [DOI] [PubMed] [Google Scholar]
- Kanno T, Mette MF, Kreil DP, Aufsatz W, Matzke M, Matzke AJM (2004) Involvement of a putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr Biol 14 801–805 [DOI] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Gao S, Vivian-Smith A, Jin H (2007) A novel class of bacteria-induced small RNAs in Arabidopsis. Genes Dev 21 3123–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Morgan R, Dahlbeck D, Borsani O, Villegas A Jr, Zhu J-K, Staskawicz BJ, Jin H (2006) A pathogen-inducible endogenous siRNA in plant immunity. Proc Natl Acad Sci USA 103 18002–18007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumoto Y, Fukuchi-Mizutani M, Fukui Y, Brugliera F, Holton TA, Karan M, Nakamura N, Yonekura-Sakakibara K, Togami J, Pigeaire A, et al (2007) Engineering of the rose flavonoid biosynthetic pathway successfully generated blue-hued flowers accumulating delphinidin. Plant Cell Physiol 48 1589–600 [DOI] [PubMed] [Google Scholar]
- Kennerdell JR, Carthew RW (1998) Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell 95 1017–1026 [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-4. Cell 75 843–854 [DOI] [PubMed] [Google Scholar]
- Lindbo JA, Silva-Rosales L, Proebsting WM, Dougherty WG (1993) Induction of highly specific antiviral state in transgenic plants: implications for regulation of gene expression and virus resistance. Plant Cell 5 1749–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Singh SP, Green AG (2002) High-stearic and high-oleic cottonseed oils produced by hairpin RNA-mediated post-transcriptional gene silencing. Plant Physiol 129 1732–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Fedoroff N (2000) A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 12 2351–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SF, Sun YH, Amerson H, Chiang VL (2007) MicroRNAs in loblolly pine (Pinus taeda L.) and their association with fusiform rust gall development. Plant J 51 1077–1098 [DOI] [PubMed] [Google Scholar]
- Mallory AC, Vaucheret H (2006) Functions of microRNAs and related small RNAs in plants. Nat Genet 38 S31–S36 [DOI] [PubMed] [Google Scholar]
- Margis R, Fusaro AF, Smith NA, Curtin SJ, Watson JM, Finnegan EJ, Waterhouse PM (2006) The evolution and diversification of Dicers in plants. FEBS Lett 580 2442–2450 [DOI] [PubMed] [Google Scholar]
- Matzke MA, Priming M, Trnovsky J, Matzke AJM (1989) Reversible methylation and inactivation of marker genes in sequentially transformed tobacco plants. EMBO J 8 643–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mette MF, Aufsatz W, van der Winden J, Matzke MA, Matzke AJ (2000) Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J 19 5194–5201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzlaff M, O'Dell M, Cluster PD, Flavell RB (1997) RNA-mediated RNA degradation and chalcone synthase A silencing in petunia. Cell 88 845–854 [DOI] [PubMed] [Google Scholar]
- Millar AA, Waterhouse PM (2005) Plant and animal microRNAs: similarities and differences. Funct Integr Genomics 5 129–135 [DOI] [PubMed] [Google Scholar]
- Mussgnug JH, Thomas-Hall S, Rupprecht J, Foo A, Klassen V, McDowall A, Schenk PM, Kruse O, Hankamer B (2007) Engineering photosynthetic light capture: impacts on improved solar energy to biomass conversion. Plant Biotechnol J 5 802–814 [DOI] [PubMed] [Google Scholar]
- Napoli C, Lemieux C, Jorgensen R (1990) Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasini N, Estelle M, Vionnet O, Jones JD (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signalling. Science 312 436–439 [DOI] [PubMed] [Google Scholar]
- Ngo H, Tschudi C, Gull K, Ullu E (1998) Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc Natl Acad Sci USA 95 14687–14692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu QW, Lin SS, Reyes JL, Chen KC, Wu HW, Yeh SD, Chua NH (2006) Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat Biotechnol 24 1420–1428 [DOI] [PubMed] [Google Scholar]
- Okano Y, Miki D, Shimamoto K (2008) Small interfering RNA (siRNA) targeting of endogenous promoters induces DNA methylation, but not necessarily gene silencing, in rice. Plant J 53 65–77 [DOI] [PubMed] [Google Scholar]
- Palauqui JC, Elmayan T, Pollien JM, Vaucheret H (1997) Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J 16 4738–4745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palauqui JC, Vaucheret H (1995) Field trial analysis of nitrate reductase co-suppression: a comparative study of 38 combinations of transgene loci. Plant Mol Biol 29 149–159 [DOI] [PubMed] [Google Scholar]
- Pandey SP, Baldwin IT (2007) RNA-directed RNA polymerase 1 (RdR1) mediates the resistance of Nicotiana attenuata to herbivore attack in nature. Plant J 50 40–53 [DOI] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X (2002) CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12 1484–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu J, Ye J, Fang RX (2007) Artificial microRNA-mediated virus resistance in plants. J Virol 81 6690–6699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP (2006) A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev 20 3407–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff F, Harrison BD, Baulcombe DC (1997) A similarity between viral defence and gene silencing in plants. Science 276 1558–1560 [DOI] [PubMed] [Google Scholar]
- Regina A, Bird A, Topping D, Bowden S, Freeman J, Barsby T, Kosar-Hashemi B, Li Z, Rahman S, Morell M (2006) High-amylose wheat generated by RNA interference improves indices of large-bowel health in rats. Proc Natl Acad Sci USA 103 3546–3551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Vijn I, Rebocho A, van Blokland R, Roelofs D, Mol JN, Kooter JM (2001) Transcriptional and posttranscriptional gene silencing are mechanistically related. Curr Biol 11 436–440 [DOI] [PubMed] [Google Scholar]
- Smith NA, Singh SP, Wang MB, Stoutjesdijk PA, Green AG, Waterhouse PM (2000) Total silencing by intron-spliced hairpin RNAs. Nature 407 319–320 [DOI] [PubMed] [Google Scholar]
- Stam M, de Bruin R, Kenter S, van der Hoorn RAL, van Blokland R, Mol JNM, Kooter JM (1997) Post-transcriptional silencing of chalcone synthase in Petunia by inverted transgene repeats. Plant J 12 63–82 [Google Scholar]
- Swaney S, Powers H, Goodwin J, Rosales LS, Dougherty WG (1995) RNA-mediated resistance with nonstructural genes from the tobacco etch virus genome. Mol Plant Microbe Interact 8 1004–1011 [DOI] [PubMed] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC (1999) The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99 123–132 [DOI] [PubMed] [Google Scholar]
- Thierry D, Vaucheret H (1996) Sequence homology requirements for transcriptional silencing of 35S transgenes and post-transcriptional silencing of nitrite reductase (trans)genes by the tobacco 271 locus. Plant Mol Biol 32 1075–1083 [DOI] [PubMed] [Google Scholar]
- van der Krol AR, Mur LA, Beld M, Mol JNM, Stuitje AR (1990) Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell 2 291–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H (2005) RNA polymerase IV and transcriptional silencing. Nat Genet 37 659–60 [DOI] [PubMed] [Google Scholar]
- Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert J-L, Bartel DP, Crete P (2004) Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell 16 69–79 [DOI] [PubMed] [Google Scholar]
- Voinnet O (2005) Non-cell autonomous RNA silencing. FEBS Lett 579 5858–5871 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Lederer C, Baulcombe DC (2000) A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103 157–67 [DOI] [PubMed] [Google Scholar]
- Wang MB, Abbott DC, Waterhouse PM (2000) A single copy of a virus-derived transgene encoding hairpin RNA gives immunity to barley yellow dwarf virus. Mol Plant Pathol 1 347–356 [DOI] [PubMed] [Google Scholar]
- Wang MB, Metzlaff M (2005) RNA silencing and antiviral defense in plants. Curr Opin Plant Biol 8 216–222 [DOI] [PubMed] [Google Scholar]
- Wang MB, Waterhouse PM (2000) High-efficiency of silencing of a β-glucuronidase gene in rice is correlated with repetitive transgene structure but is independent of DNA methylation. Plant Mol Biol 43 67–82 [DOI] [PubMed] [Google Scholar]
- Wassenegger M (2005) The role of the RNAi machinery in heterochromatin formation. Cell 122 13–16 [DOI] [PubMed] [Google Scholar]
- Wassenegger M, Hiemes S, Riedel L, Sänger HL (1994) RNA-directed de novo methylation of genomic sequences in plants. Cell 76 567–576 [DOI] [PubMed] [Google Scholar]
- Waterhouse PM, Graham MW, Wang MB (1998) Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc Natl Acad Sci USA 95 13959–13964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27 581–590 [DOI] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC (2004) Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2 E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Liu Z, Lu F, Dang A, Huang H (2006) SERRATE is a novel nuclear regulator in primary miRNA processing in Arabidopsis. Plant J 47 841–850 [DOI] [PubMed] [Google Scholar]
- Yu L, Yu X, Shen R, He Y (2005) HYL1 gene maintains venation and polarity of leaves. Planta 221 231–242 [DOI] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Jacobsen SE (2003) ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299 716–719 [DOI] [PubMed] [Google Scholar]