Abstract
Receptor-like proteins (RLPs) are cell surface receptors that typically consist of an extracellular leucine-rich repeat domain, a transmembrane domain, and a short cytoplasmatic tail. In several plant species, RLPs have been found to play a role in disease resistance, such as the tomato (Solanum lycopersicum) Cf and Ve proteins and the apple (Malus domestica) HcrVf2 protein that mediate resistance against the fungal pathogens Cladosporium fulvum, Verticillium spp., and Venturia inaequalis, respectively. In addition, RLPs play a role in plant development; Arabidopsis (Arabidopsis thaliana) TOO MANY MOUTHS (TMM) regulates stomatal distribution, while Arabidopsis CLAVATA2 (CLV2) and its functional maize (Zea mays) ortholog FASCINATED EAR2 regulate meristem maintenance. In total, 57 RLP genes have been identified in the Arabidopsis genome and a genome-wide collection of T-DNA insertion lines was assembled. This collection was functionally analyzed with respect to plant growth and development and sensitivity to various stress responses, including susceptibility toward pathogens. A number of novel developmental phenotypes were revealed for our CLV2 and TMM insertion mutants. In addition, one AtRLP gene was found to mediate abscisic acid sensitivity and another AtRLP gene was found to influence nonhost resistance toward Pseudomonas syringae pv phaseolicola. This genome-wide collection of Arabidopsis RLP gene T-DNA insertion mutants provides a tool for future investigations into the biological roles of RLPs.
For decades, it was thought that the communication between plant cells occurs through the cell wall-spanning cytoplasmic bridges called plasmodesmata. However, since the identification of the first plant cell surface receptor (Walker and Zhang, 1990), it has been known that, similar to other multicellular organisms, plants can perceive extracellular signals at the plasma membrane. Since then, many plant cell surface receptors have been found to play key roles in very diverse processes ranging from growth and development, in which they perceive endogenous self signals, to recognition of other organisms, in which they perceive exogenous nonself signals (Diévart and Clark, 2004).
A common structural element of many plant cell surface receptors is the extracellular Leu-rich repeat (eLRR) domain that is generally thought to mediate ligand perception (Kobe and Kajava, 2001; Kinoshita et al., 2005). These eLRRs are composed of 23 to 25 amino acids with the conserved consensus sequence LxxLxxLxLxxNxLt/sgxIpxxLG (Jones and Jones, 1997). The largest group of eLRR-containing cell surface receptors is formed by the receptor-like kinases (RLKs) that are composed of an eLRR domain, a single-pass transmembrane domain, and a cytoplasmic kinase domain, with over 200 representatives in the Arabidopsis (Arabidopsis thaliana) genome (Shiu and Bleecker, 2003). The second largest group of eLRR-containing cell surface receptors is formed by the receptor-like proteins (RLPs) that differ from RLKs in that they lack the cytoplasmic kinase domain and only have a short cytoplasmic tail that lacks obvious motifs for intracellular signaling except for the putative endocytosis motif found in some members (Joosten and de Wit, 1999; Kruijt et al., 2005). Typically, the amino acid sequence of RLPs has been divided into the conserved domains A through G with a putative signal peptide (A), a Cys-rich domain (B), the LRR domain (C), a spacer (D), an acidic domain (E), the transmembrane domain (F), and a short cytoplasmic region (G). Furthermore, the LRR-containing C domain is subdivided into three domains in which the non-LRR island C2 domain interrupts the C1 and C3 LRR regions (Jones and Jones, 1997).
Recently, considerable advances have been made in our understanding of the role and function of RLKs and how they relay extracellular signals to initiate an intracellular response (Nürnberger and Kemmerling, 2006; Li and Jin, 2007). By contrast, very little is known about RLP signaling (Fritz-Laylin et al., 2005; Kruijt et al., 2005). The first RLP gene identified was tomato (Solanum lycopersicum) Cf-9 that mediates resistance against strains of the leaf mold fungus Cladosporium fulvum that carry the avirulence gene Avr9 (Jones et al., 1994). C. fulvum is a biotrophic pathogen that is characterized by strictly apoplastic growth (Thomma et al., 2005). To date, several Cf resistance genes have been cloned from tomato that all to belong to the RLP gene family (Dixon et al., 1996, 1998; Thomas et al., 1997; Takken et al., 1999). In addition to Cf genes, the RLP gene family in tomato comprises two Ve genes that have been reported to provide resistance against vascular wilt pathogens of the genus Verticillium (Kawchuk et al., 2001) that, like C. fulvum, grow extracellularly without penetrating plant cells (Fradin and Thomma, 2006). Finally, the tomato RLP family comprises two LeEIX genes that encode receptors for the ethylene-inducible xylanase produced by extracellularly growing Trichoderma biocontrol fungi (Ron and Avni, 2004).
In addition to tomato, RLPs have been implicated in disease resistance in other plant species (Kruijt et al., 2005). Apple (Malus domestica) HcrVf-2 confers resistance to the apple scab fungus Venturia inaequalis (Belfanti et al., 2004). Furthermore, an Arabidopsis chitin-inducible RLP gene has been implicated in resistance against the powdery mildew pathogen Erysiphe cichoracearum (Ramonell et al., 2005).
RLPs also play significant roles in plant development. For example, Arabidopsis CLAVATA2 (CLV2) was found to be crucial for maintaining a balanced meristematic stem cell population and is required for the accumulation and stability of CLV1, which is an RLK (Jeong et al., 1999). It has been proposed that CLV1 and CLV2 undergo a physical interaction to form a heterodimer to act as receptor for the predicted extracellular peptide ligand CLV3 (Trotochaud et al., 1999; Rojo et al., 2002; Ogawa et al., 2008). Upon ligand perception by the ectodomain (Ogawa et al., 2008), the kinase domain of CLV1 is thought to be activated to initiate the downstream signaling that is required to maintain the stem cell population (Rojo et al., 2002; Diévart and Clark, 2004). In maize (Zea mays), an ortholog of the CLV2 gene has been identified as FASCINATED EAR2 (Taguchi-Shiobara et al., 2001). Furthermore, the RLK thick tassel dwarf1 has been identified as a CLV1 ortholog, suggesting that the CLAVATA signaling pathway is conserved between monocots and dicots (Bommert et al., 2005). Another RLP gene, TOO MANY MOUTHS (TMM), is involved in plant development in Arabidopsis and regulates stomatal distribution across the epidermis (Nadeau and Sack, 2002). Although a physical interaction between TMM and any other RLP or RLK has not been established, TMM was found to negatively regulate three RLKs of the ERECTA family (Shpak et al., 2005).
Previously, in the Arabidopsis genome, 56 putative RLP genes (AtRLPs) were identified that are assembled at 33 loci (Fritz-Laylin et al., 2005). So far, a function has been assigned only to the three AtRLP genes described above (Jeong et al., 1999; Nadeau and Sack, 2002; Ramonell et al., 2005), implicating that the other RLPs are orphan proteins. In the complete genome sequence of the monocot plant rice (Oryza sativa), 90 RLP genes have been identified (Fritz-Laylin et al., 2005). Genes involved in plant development are presumably under evolutionary pressure to maintain a specific function that reduces sequence drift across orthologs, while disease resistance genes are under strong diversifying selection to produce highly divergent sequences with distinct recognition capacities (Fritz-Laylin et al., 2005). Based on the sequence comparison between Arabidopsis and rice RLP genes, and building on the hypothesis that developmental genes are less likely to be duplicated and undergo diversifying selection than are disease resistance genes (Leister, 2004), nine AtRLP genes were proposed as putative developmental orthologous genes, while the remaining AtRLP genes were proposed to be candidate disease resistance genes (Fritz-Laylin et al., 2005). In this article, we report on the assembly and functional analysis of a genome-wide collection of AtRLP family T-DNA knockout lines. This collection has been screened for altered phenotypes in growth and development but also alterations in response to pathogen challenge. Our analysis has revealed novel phenotypes linked with mutations in the well-studied AtRLPs TMM and CLV2. Furthermore, one AtRLP gene is found to play a role in abscisic acid (ABA) signaling, a process in which RLP activity has not been implicated previously. Remarkably, despite an extensive list of pathogens tested, including adapted and nonadapted pathogens of Arabidopsis, we have been able to identify only one AtRLP gene with a role in basal nonhost resistance against the nonadapted bacterial pathogen Pseudomonas syringae pv phaseolicola (Psp). The described AtRLP T-DNA collection is a valuable source for future investigations into the biological roles of RLPs.
RESULTS
AtRLP Gene Structure and AtRLP Protein Analysis
At the onset of this project, a bioinformatic analysis to investigate the structure of all the AtRLP genes was undertaken. To this end, BLAST searches were performed on the Arabidopsis genome sequence using the predicted protein sequences of the previously characterized RLPs CLV2, TMM, and Cf-9 as queries. The set of Arabidopsis genes obtained in this way was further analyzed for the presence of a signal peptide, eLRRs, a transmembrane domain, and a short cytoplasmic tail lacking kinase motifs in the predicted protein. Although a previously published study has identified in total 56 AtRLP genes (Fritz-Laylin et al., 2005), our analysis revealed a set of 57 putative AtRLP genes (Table I). All 57 AtRLP genes were assigned AtRLP numbers in consecutive order according to their gene numbers along the Arabidopsis genome (Table I). The additional AtRLP gene identified here, denoted as AtRLP5, corresponds to At1g34290, and, although it carries only two eLRRs, the predicted protein complies with the canonical RLP domain composition.
Table I.
List of the AtRLP genes and corresponding T-DNA insertion lines used in this study
| Gene Namea | AGI Code | T-DNA Line Ordered | Mutant Name |
|---|---|---|---|
| AtRLP1 | at1g07390 | SALK_059920b | Atrlp1-1 |
| SALK_116923 | Atrlp1-2 | ||
| AtRLP2 | at1g17240 | SALK_049366c | Atrlp2-1 |
| AtRLP3 | at1g17250 | SALK_051677 | Atrlp3-1 |
| SAIL_204_D01d | Atrlp3-2 | ||
| AtRLP4 | at1g28340 | SALK_039264e | Atrlp4-1 |
| AtRLP5 | at1g34290 | SALK_112291 | Atrlp5-1 |
| AtRLP6 | at1g45616 | SALK_080898 | Atrlp6-1 |
| SAIL_84_E01d | Atrlp6-2 | ||
| SALK_020071f | |||
| AtRLP7 | at1g47890 | SALK_030269 | Atrlp7-1 |
| AtRLP8 | at1g54480 | SM_3_38632 | Atrlp8-1 |
| SM_3_20200 | Atrlp8-2 | ||
| AtRLP9 | at1g58190 | SALK_061979 | Atrlp9-1 |
| SALK_023419 | Atrlp9-2 | ||
| AtRLP10 | at1g65380 | GABI_686A09 | Atrlp10-1 |
| (CLV2) | clv2-3 (EMS)g | clv2-3 | |
| AtRLP11 | at1g71390 | SALK_013218 | Atrlp11-1 |
| AtRLP12 | at1g71400 | SALK_151456 | Atrlp12-1 |
| AtRLP13 | at1g74170 | SALK_020984 | Atrlp13-1 |
| AtRLP14 | at1g74180 | SAIL_513_A08d | Atrlp14-1 |
| AtRLP15 | at1g74190 | SALK_041143 | Atrlp15-1 |
| GABI_077G01f | |||
| AtRLP16 | at1g74200 | SALK_032150 | Atrlp16-1 |
| AtRLP17 | at1g80080 | FLAG_014F03d | Atrlp17-1 |
| (TMM) | tmm-1 (EMS)h | tmm-1 | |
| SAIL_165_F02df | |||
| AtRLP18 | at2g15040 | SAIL_400_H02d | Atrlp18-1 |
| AtRLP19 | at2g15080 | FLAG_524A03dij | Atrlp19-1 |
| AtRLP20 | at2g25440 | SALK_130147c | Atrlp20-1 |
| AtRLP21 | at2g25470 | SAIL_693_F05 | Atrlp21-1 |
| SALK_133403f | |||
| AtRLP22 | at2g32660 | SALK_125231 | Atrlp22-1 |
| AtRLP23 | at2g32680 | SALK_034225 | Atrlp23-1 |
| AtRLP24 | at2g33020 | SALK_046236 | Atrlp24-1 |
| AtRLP25 | at2g33030 | SALK_048434i | Atrlp25-1 |
| AtRLP26 | at2g33050 | SALK_104127c | Atrlp26-1 |
| SALK_026997f | |||
| AtRLP27 | at2g33060 | SALK_029443 | Atrlp27-1 |
| AtRLP28 | at2g33080 | SM_3_1740 | Atrlp28-1 |
| AtRLP29 | at2g42800 | SALK_022220 | Atrlp29-1 |
| AtRLP30 | at3g05360 | SALK_122528 | Atrlp30-1 |
| SALK_008911 | Atrlp30-2 | ||
| SALK_122536 | Atrlp30-3 | ||
| SALK_145342 | Atrlp30-4 | ||
| AtRLP31 | at3g05370 | SALK_058586 | Atrlp31-1 |
| SALK_094160 | Atrlp31-2 | ||
| AtRLP32 | at3g05650 | FLAG_588C11d | Atrlp32-1 |
| AtRLP33 | at3g05660 | FLAG_048F06d | Atrlp33-1 |
| SALK_087631 | Atrlp33-2 | ||
| SALK_085252 | Atrlp33-3 | ||
| AtRLP34 | at3g11010 | SALK_067155 | Atrlp34-1 |
| SALK_085506f | |||
| AtRLP35 | at3g11080 | SALK_096171 | Atrlp35-1 |
| SALK_016143 | Atrlp35-2 | ||
| AtRLP36 | at3g23010 | SALK_086147 | Atrlp36-1 |
| AtRLP37 | at3g23110 | SALK_041785 | Atrlp37-1 |
| SALK_012745j | Atrlp37-2 | ||
| AtRLP38 | at3g23120 | SALK_017819 | Atrlp38-1 |
| GT_5_105490df | |||
| AtRLP39 | at3g24900 | SALK_126505 | Atrlp39-1 |
| SALK_126504f | |||
| AtRLP40 | at3g24982 | GABI_564D03 | Atrlp40-1 |
| AtRLP41 | at3g25010 | SALK_024020 | Atrlp41-1 |
| SM_3_20242 | Atrlp41-2 | ||
| SM_3_38956 | Atrlp41-3 | ||
| AtRLP42 | at3g25020 | SALK_080324b | Atrlp42-1 |
| SALK_094190b | Atrlp42-2 | ||
| AtRLP43 | at3g28890 | SALK_041685 | Atrlp43-1 |
| AtRLP44 | at3g49750 | SALK_097350i | Atrlp44-1 |
| SALK_045246c | ATrlp44-2 | ||
| AtRLP45 | at3g53240 | GABI_620G05 | Atrlp45-1 |
| FLAG_339H12cd | Atrlp45-2 | ||
| AtRLP46 | at4g04220 | SALK_048207i | Atrlp46-1 |
| SAIL_15_A02df | |||
| AtRLP47 | at4g13810 | SALK_105921 | Atrlp47-1 |
| AtRLP48 | at4g13880 | SALK_036842 | Atrlp48-1 |
| AtRLP49 | at4g13900 | SALK_067372 | Atrlp49-1 |
| SALK_116910 | Atrlp49-2 | ||
| AtRLP50 | at4g13920 | SALK_070876i | Atrlp50-1 |
| AtRLP51 | at4g18760 | SALK_143038 | Atrlp51-1 |
| SAIL_740_C06i | Atrlp51-2 | ||
| AtRLP52 | at5g25910 | SALK_107922 | Atrlp52-1 |
| SALK_054976f | |||
| AtRLP53 | at5g27060 | SALK_124008 | Atrlp53-1 |
| AtRLP54 | at5g40170 | SAIL_306_E09d | Atrlp54-1 |
| AtRLP55 | at5g45770 | SALK_139161b | Atrlp55-1 |
| SALK_076590 | Atrlp55-2 | ||
| AtRLP56 | at5g49290 | SALK_129306 | Atrlp56-1 |
| SALK_010565 | Atrlp56-2 | ||
| AtRLP57 | at5g65830 | SALK_077716 | Atrlp57-1 |
In chronological order along the five Arabidopsis chromosomes.
T-DNA insertion site within 300 nucleotides downstream of the open reading frame.
T-DNA insertion site between 300 and 1,000 nucleotides upstream of the open reading frame.
SAIL-lines are in CS8846, FLAG-lines in WS-2 and GT-line is in Ler background.
T-DNA insertion site within an intron.
T-DNA insertion site could not be confirmed by PCR; no homozygous T-DNA insertion line was obtained.
EMS mutant clv2-3 (Jeong et al., 1999).
EMS mutant tmm-1 (Nadeau and Sack, 2002).
T-DNA insertion site within 300 nucleotides upstream of the open reading frame.
No homozygous line for the T-DNA insertion was be obtained.
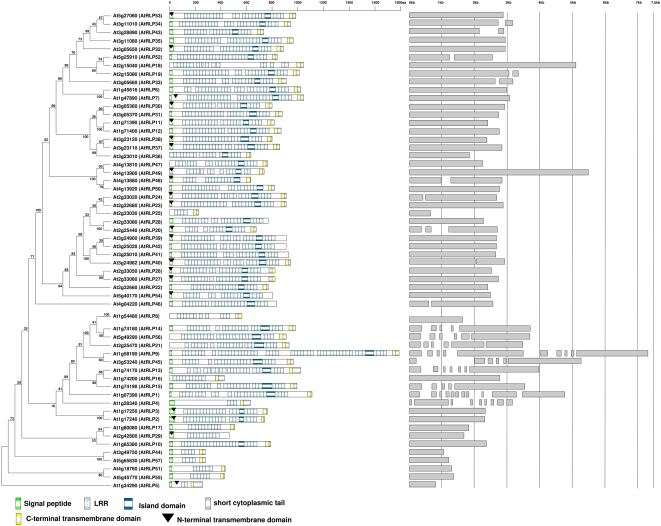
Pairwise amino acid sequence comparison revealed that AtRLPs display low overall sequence identity, with only 10 pairwise combinations that share over 70% identity (Supplemental Table S1). Of these, the proteins encoded by the neighboring genes AtRLP41 and AtRLP42 share the highest level of identity (86%). Furthermore, both proteins are highly similar to AtRLP39 (85% and 82% identity, respectively), and the corresponding genes reside in close proximity to each other, suggesting recent gene multiplication. Two other AtRLP proteins, AtRLP44 and AtRLP57, are found to be similar in length and domain composition, sharing 80% identity (Fig. 1; Supplemental Table S1), although the genes that encode these proteins are located on different chromosomes. To further assess the structures of AtRLP genes, the exon boundaries and corresponding flanking intron sequences were determined. While only 21% of the genes in the Arabidopsis genome are composed of a single exon (Arabidopsis Genome Initiative, 2000), 37 of the 57 (65%) AtRLP-encoding genes were found to contain a single exon (Fig. 1). Interestingly, within the group of genes that contain multiple exons, AtRLP9, AtRLP14, AtRLP15, AtRLP21, and AtRLP56 have introns at similar positions in the genes (Fig. 1). Similarly, the introns of AtRLP19, AtRLP33, and AtRLP34 are also localized at comparable positions (Fig. 1). Furthermore, all the AtRLP genes that contain multiple exons cluster in a phylogenetic tree (Fig. 1).
Figure 1.
A phylogenetic view of AtRLP protein domain configurations and the corresponding RLP gene structures as shown by exon/intron boundaries. Left, Phylogenetic tree of the AtRLP family that also includes CLV2 (AtRLP10) and TMM (AtRLP17). The tree was generated from the alignment of C3-F domains of all AtRLPs with 100 bootstrap replicates as indicated on the branch of the tree. The AGI code and AtRLP gene number are indicated on the left. Genes are organized according to the order along the chromosomes. Middle, Domain organizations as predicted by SMART/Pfam. Each colored box represents a domain as indicated. The arrowhead shows the putative N-terminal transmembrane domain. The open box indicates an amino acid fragment not showing any significant motif or domain. Right, RLP gene structure presented by gray boxes for exons and spaces for the introns.
Next, the domain composition was analyzed for all predicted AtRLP proteins. As has been noted previously (Fritz-Laylin et al., 2005), the AtRLPs exhibit great variation at the sequence level and also in the numbers of eLRRs (Fig. 1). The predicted sizes of the AtRLPs range from 218 amino acids (AtRLP25) to 1,784 amino acids (AtRLP9), whereas the eLRRs vary in number from two (AtRLP5) to 49 (AtRLP9; Fig. 1). Of the 57 AtRLPs, 18 are predicted to contain two transmembrane domains, one at the N terminus and one at the C terminus, although it is presently unclear whether the N terminal transmembrane domain indeed functions as such. Furthermore, it has previously been noted that not all RLPs contain an island domain (C2) within the eLRR region, with TMM as an example (Nadeau and Sack, 2002). Of the 57 AtRLPs, 45 are predicted to contain a C2 island domain nested in between two eLRR blocks (C1 and C3). Remarkably, in 42 of those RLPs, the island domain is followed by a C3 domain that contains exactly four eLRRs (Fig. 1). This distinct domain organization has not only been observed for some functionally characterized RLPs but also for some RLKs (Jones et al., 1994; Song et al., 1995; Clark et al., 1997; Li and Chory, 1997; Jeong et al., 1999; Gómez-Gómez and Boller, 2000; Taguchi-Shiobara et al., 2001). For all AtRLP genes, corresponding cDNA sequences, EST sequences, Massively Parallel Signature Sequencing data, and/or microarray data are deposited in public databases, demonstrating that all 57 AtRLP genes are actively transcribed (Supplemental Figs. S1 and S2).
Assembly of a Genome-Wide Collection of AtRLP Gene T-DNA Insertion Mutants
To identify putative T-DNA insertion lines for all the AtRLP genes, we queried the T-DNA Express database of the SALK Institute Genome Analysis Laboratory (SIGnAL; http://signal.salk.edu). Because often several different insertion lines could be identified for each AtRLP gene, insertion lines were selected based on the position of the T-DNA insertion within the coding sequence to enhance the likelihood of successful disruption of gene function. Preferably, T-DNA insertion lines of the Columbia (Col-0) ecotype were selected with exon insertions (Table I). However, if not available, lines with predicted intron (one line), promoter (11 lines), or terminator (four lines) insertions were chosen. For the 57 AtRLP genes, 89 T-DNA insertion lines were selected (Table I) that were evaluated for presence of the predicted T-DNA insertion using PCR (Supplemental Table S2). Ten lines did not have the predicted insertion, whereas 79 were confirmed to carry a T-DNA insertion in the gene of interest and for which homozygosity of the T-DNA insert was pursued. For two T-DNA insertion lines, FLAG_524A03 and SALK_012745 with an insertion in AtRLP19 and AtRLP37, respectively, only heterozygous insertion lines were obtained, suggesting that homozygosity of these T-DNA mutations caused embryonic lethality. However, subsequent segregation and complementation analysis could not confirm embryo lethality caused by T-DNA homozygosity in these lines, and they were not used for further analysis. Although we were able to identify another T-DNA insertion line for AtRLP37 that was carried to homozygosity (Table I), unfortunately, no alternative T-DNA insertion line was available for AtRLP19. Overall, in the complete collection of 77 homozygous AtRLP T-DNA insertion lines, at least one line was obtained for 56 of the 57 AtRLP genes, while for 19 AtRLP genes multiple mutants were identified (Table I).
Phenotypic Alterations in Growth and Development of AtRLP Gene T-DNA Insertion Mutants
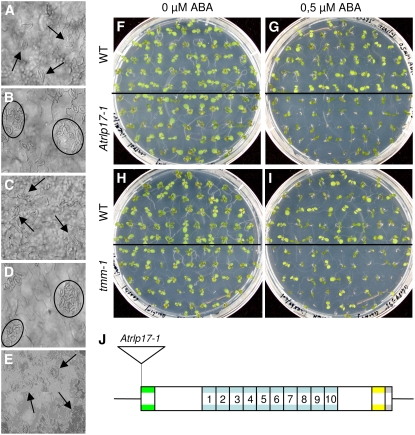
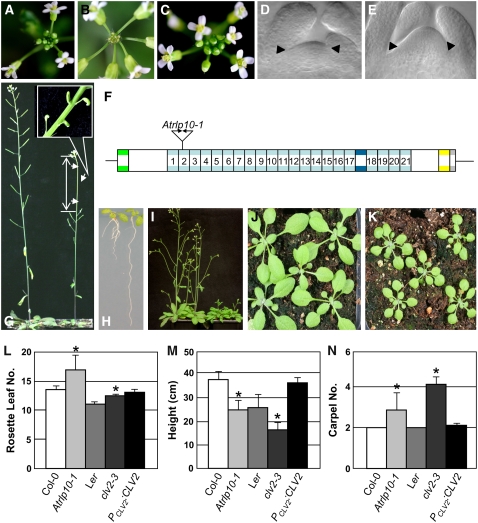
We examined the phenotypes of the complete collection of homozygous T-DNA insertion lines with respect to various characteristics related to plant growth and development. The T-DNA lines were examined for root development, rosette growth, inflorescence emergence, and the development and appearance of flowers and seed. In addition, stomatal patterning across the cotyledons and leaves, formation of the leaf cuticle, and the leaf vascular patterns were analyzed. Two AtRLP genes, CLV2 (AtRLP10) and TMM (AtRLP17), have previously been implicated in plant development (Jeong et al., 1999; Nadeau and Sack, 2002). Our analysis showed that the T-DNA insertion lines Atrlp10-1 and Atrlp17-1 for the CLV2 and the TMM gene, respectively, displayed phenotypes that have previously been reported for a number of mutants in these genes (Yang and Sack, 1995; Kayes and Clark, 1998; Jeong et al., 1999; Nadeau and Sack, 2002). Similar to the ethyl methanesulfonate (EMS) mutant tmm-1, the stomata of the knockout allele Atrlp17-1 that carries a T-DNA in the ATG start codon of the coding sequence were found to cluster across the leaf epidermis (Fig. 2, A–D and J). Complementation of Atrlp17-1 with the wild-type TMM allele resulted in disappearance of the stomatal clustering phenotype (Fig. 2E), showing that Atrlp17-1 is a true TMM mutant allele. In addition, as expected, the Atrlp10-1 mutant with a knockout in the CLV2 gene displayed enlarged shoot meristem (Fig. 3, D and E) and alterations in the development of the gynoecia, flowers, carpels, pedicels, and stamens (data not shown). Like other CLV2 mutants, the Atrlp10-1 mutant fails to respond to in vitro treatment with a synthetic peptide that corresponds to the conserved CLE motif that is present in the CLV3-like peptide ligands (Fig. 3H; Fiers et al., 2005). However, while the previously described CLV2 mutants (clv2-1 to clv2-5) generally have four carpels (Kayes and Clark, 1998), Atrlp10-1 shows only a mild carpel phenotype with 2.6 carpels on average (Fig. 3N).
Figure 2.
Characterization of the Atrlp17-1 mutant allele. A to E, Comparison of stomata distribution of wild-type (A and C) with the Atrlp17-1 mutant (B), tmm-1 mutant (D), and the Atrlp17-1 mutant after complementation with a wild-type TMM allele (E). The arrows indicate single stomata, while the circles indicate stomatal clusters. F to I, Comparison of ABA response of wild-type (top half of the plate) with Atrlp17-1 (F and G; bottom half of the plate) or tmm-1 (H and I; bottom half of the plate) in the absence (F and H) and presence (G and I) of ABA. J, Location of the T-DNA insertion in Atrlp17-1.
Figure 3.
Characterization of the Atrlp10-1 mutant allele. A, Wild-type inflorescence meristem. B, Atrlp10-1 inflorescence meristem. C, Atrlp10-1 inflorescence meristem upon complementation with a wild-type CLV2 allele. D and E, Cleared shoot meristem of wild type (D) and Atrlp10-1 (E). Arrowheads indicate meristem borders. F, Location of T-DNA insertion in Atrlp10 (CLV2). G, Comparison of inflorescence development of wild type (left) with Atrlp10-1 mutant (right). The zoom-in picture indicated no siliques were developed because of the temporary termination of inflorescence meristem of Atrlp10-1 mutant. H, The 8-d, wild-type seedling (left) showed a short root phenotype, while Atrlp10-1 (right) shows no effect with 10 μm CLV3p treatment. I, Comparison of 4-week-old plants of wild type (left) with Atrlp10-1 mutant (right). J and K, Comparison of 2-week-old plants of wild type (J) with Atrlp10-1 mutant (K). L to N, The mean rosette leaf number (L), height of the primary stem (M), and carpel number (N) of wild types, clv2-3, Atrlp10-1, and Atrlp10-1 upon complementation with a wild-type CLV2 allele. Asterisks indicate significant differences (P < 0.01) compared to the respective wild types.
Interestingly, despite the relatively weak carpel phenotype, Atrlp10-1 exhibits a number of phenotypes that have not previously been reported for any of the CLV2 mutants (Fig. 3). Plants from the Atrlp10-1 T-DNA insertion line grow slower, develop more rosette leaves and shorter stems, and flower at a later stage than wild-type plants and the clv2-3 mutant (Fig. 3, I–M). During flowering, the meristem of the main inflorescence stops producing flowers for a short period, upon which flowering is resumed (Fig. 3, A, B, and G). However, side stems do not show this temporary termination of the flower meristem. Linkage analysis in a segregating population has demonstrated that the temporary termination of flowering phenotype is linked to a homozygous T-DNA knockout in Atrlp10-1. Moreover, complementation of Atrlp10-1 with the wild-type CLV2 allele restored all clv2 mutant phenotypes (Fig. 3, C and L–N).
Conditional Phenotypic Alterations of AtRLP Gene T-DNA Insertion Mutants
We tested the collection of T-DNA lines for altered conditional developmental phenotypes, including gravitropism, response to darkness or treatment with different hormones, and a CLV3-like peptide ligand (Supplemental Table S3). For most of the treatments, no consistent differential responsiveness within the collection of AtRLP gene knockout lines was observed (data not shown). The only treatment that resulted in a reliable phenotype was a treatment with the plant hormone ABA. In addition to the previously described stomatal clustering phenotype, tmm-1 and Atrlp17-1 that both carry a mutation in the AtRLP gene TMM displayed decreased sensitivity to ABA. Although seedlings of nontreated Atrlp17-1 and tmm-1 mutants were phenotypically indistinguishable from control plants (Fig. 2, F and H), exogenous application of ABA induced chlorosis in control plants but not in mutants and reduced the growth of Atrlp17-1 and tmm-1 mutants (Fig. 2, G and I) in comparison to the respective control plants. These results indicate that TMM plays a role in ABA-induced chlorosis and growth reduction in Arabidopsis.
Assessment of the Roles of AtRLP Genes in Plant Defense
To determine whether AtRLP genes play a role in the perception and signaling of abiotic stress signals, we have tested the sensitivity of the collection of T-DNA insertion lines for several abiotic stress inducers. These included inducers of salt stress, osmotic stress, drought stress, reactive oxygen stress, and heavy metal stress (Supplemental Table S3). No consistent phenotypic alterations were observed for any of these abiotic stress stimuli within the collection of T-DNA mutant lines in comparison to wild-type plants.
We have also investigated the possible roles of AtRLP genes in the recognition of plant pathogens. The collection of T-DNA insertion lines was assessed for altered phenotypic responses upon pathogen challenge with a diverse range of host-adapted and nonadapted necrotrophic or biotrophic pathogens (Thomma et al., 2001). Nonadapted pathogens are pathogenic on other hosts but normally unable to colonize Arabidopsis. The bacterial pathogens Pectobacterium atrosepticum, P. syringae pv tomato (Pst) DC3000, and Xanthomonas campestris pv campestris; the fungal pathogens Alternaria brassicicola, Botrytis cinerea, Cladosporium cucumerinum, C. fulvum, Colletotrichum destructivum, Oidium neolycopersici, Plectosphaerella cucumerina, Sclerotinia sclerotiorum, and Verticillium dahliae; and the oomycetes Phytophthora infestans and Hyaloperonospora parasitica were among the pathogens tested (Supplemental Tables S3 and S4). Remarkably, none of the T-DNA insertion lines showed a significant phenotypic alteration in their sensitivity toward these pathogens.
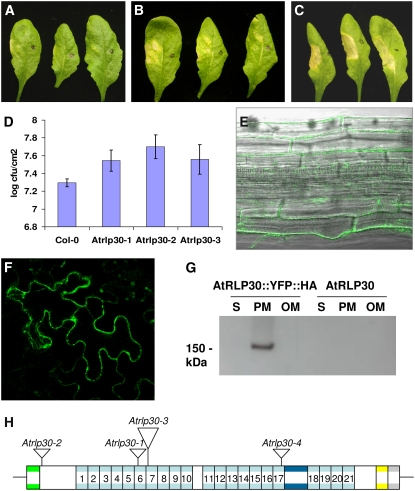
Examination of nonhost interactions was extended using the nonpathogenic bean pathogen Psp strain 1448A that is unable to colonize wild-type Col-0 due to changes to the challenged plant cell wall rather than a hypersensitive response (Soylu et al., 2005; de Torres et al., 2006). Colonization by Psp 1448A is known to be enhanced in Col-0 fls2 mutants that lack the ability to perceive flagellin, irrespective of whether inocula are applied to the leaf surface or infiltrated directly into the mesophyll (Zipfel et al., 2004; de Torres et al., 2006). The response of AtRLP T-DNA insertion lines to infiltration with bacterial suspensions was examined, and symptom development was compared with both wild-type and fls2 mutant plants in each set of experiments. We initially recorded the development of yellowing and patchy collapse of infiltrated tissues using an incremental seven-point scoring system. Lines revealing differences in reaction compared with the wild-type Col-0 in the first experiment were further assessed by repeated tests, including measurement of bacterial multiplication. The mutant Atrlp30-1 recorded consistently enhanced symptom development and more bacterial multiplication with Psp 1448A (Fig. 4, A–C). Subsequently, additional insertion mutants in AtRLP30 recovered from SALK stocks were likewise examined for their reaction to Psp 1448A (Fig. 4, D and H; Table II). In all cases, enhanced symptom development was recorded (Table II) that was associated with the recovery of higher mean numbers of bacteria from infiltrated tissue (Fig. 4D). Student's t tests indicated that all of the mutants allowed significantly higher multiplication than Col-0 (P = 0.05, 0.01, and 0.08 for Atrlp30-1, Atrlp30-2, and Atrlp30-3, respectively). In all cases, the enhanced symptom development in AtRLP30 T-DNA mutant lines was lower than observed in the Col-0 fls2 mutant (Fig. 4, A–C; Table II). Similar as for Atrlp30 mutants, enhanced susceptibility toward Psp 1448A was recorded for Atrlp18-1 mutants. However, we were unable to further confirm the phenotype due to absence of additional lines with T-DNA insertions in At2g15040.
Figure 4.
AtRLP30 is involved in bacterial resistance and localized at the plasma membrane. A to C, Symptom development in Arabidopsis leaves 4 d after inoculation with Psp. Areas in half leaves of Col-0 (A), RLP30-1 (B), and Col-0 fls2 (C) were syringe inoculated after wounding. Full details of symptom scores are recorded in Table II. D, Comparative analysis of the multiplication of Psp 1448A in Col-0 and Atrlp30 mutant plants. Infiltrated leaves were examined 3 d after inoculation; results are means from four replicates with ses. Statistical analysis using Student's t tests showed significantly higher numbers of bacteria in the mutants (P = 0.047, 0.014, and 0.088 for Atrlp30-1, -2, and -3, respectively). E to G, Localization of GFP-tagged AtRLP30 in leaf epidermis and petiole tissue as determined using confocal microscopy (E and F) and western blotting with an antibody directed against the HA tag (G). H, Locations of the T-DNA insertions in AtRLP30.
Table II.
Symptom development in leaves of Col-0 and mutant lines after syringe inoculation with Psp 1448A
DPI, Days postinoculation.
| Plant | DPI | Frequency of Lesion Typea | Mean Score (sd)b | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | |||
| Col-0 | 4 | 4 | 5 | 15 | 1.46 (0.8) | |||||
| 6 | 3 | 2 | 9 | 8 | 2 | 2.29 (1.0) | ||||
| Atrlp30-1 | 4 | 1 | 2 | 9 | 2 | 10 | 2.75 (1.2)* | |||
| 6 | 5 | 5 | 6 | 7 | 1 | 3.75 (1.2)* | ||||
| Atrlp30-3 | 4 | 1 | 3 | 11 | 3 | 6 | 2.42 (1.1)* | |||
| 6 | 2 | 2 | 7 | 7 | 6 | 3.54 (1.2)* | ||||
| Atrlp30-4 | 4 | 2 | 2 | 7 | 6 | 7 | 2.58 (1.2)* | |||
| 6 | 1 | 3 | 4 | 7 | 9 | 3.83 (1.2)* | ||||
| Col-0 fls2 | 4 | 2 | 4 | 9 | 7 | 2 | 4.13 (1.0)* | |||
| 6 | 3 | 3 | 8 | 6 | 4 | 5.21 (1.3)* | ||||
Three half leaves on eight plants were infiltrated with bacteria at OD600 0.25 (approximately 2 × 108 cells mL−1). Symptom development was scored after 4 and 6 d and sites assigned to each progressive category: 0, no symptoms; 1, very pale yellowing; 2, pale yellowing; 3, yellowing over most of the area infiltrated; 4, pale yellowing with patchy collapse; 5, yellow with patchy collapse; 6, collapse of more than 50% of infiltration site; and 7, collapse of all the infiltrated area. Lack of a number means no sites in the category.
Asterisks indicate significant differences (P < 0.1) compared to Col-0 at the respective time points.
Examination of the enhanced susceptibility phenotype of Atrlp30 mutants was extended by examining Pst strains that carry the avirulence genes AvrRpm1, AvrRpt2, AvrRps4, AvrPto, and AvrPtoB, and also hrpA and hrcC mutants of Pst, a coronatine-deficient Pst mutant, and the nonadapted strain P. syringae pv tabaci (Supplemental Table S3). However, Atrlp30 mutants did not display enhanced susceptibility to any of these bacterial strains.
Because of its potential role in basal defense, we examined the subcellular localization of the AtRLP30 protein in Arabidopsis. Transgenic plants expressing C-terminal GFP-tagged AtRLP30 were generated and examined by confocal microscopy. A clear localization of GFP-tagged AtRLP30 to the plasma membrane was, as predicted, observed in the leaf epidermis (Fig. 4F) and petiole tissue (Fig. 4E), which could also be confirmed by western analysis using an antibody directed against the hemagglutinin (HA) tag (Fig. 4G).
The enhanced susceptibility of the Atrlp30 and Atrlp18-1 T-DNA insertion mutants to Psp 1448A could be explained by an altered responsiveness to the pathogen-associated molecular pattern (PAMP) flagellin. Examination of expression data showed that AtRLP30 is induced by various PAMPs, including flg22 (Supplemental Fig. S3). We therefore compared the effect of the flg22 flagellin peptide derived from Psp 1448A on the seedling growth of Col-0 and the Atrlp30-1 T-DNA insertion mutant, but no differences were observed (Supplemental Fig. S3). The reduced basal defense observed in the AtRLP30 mutant was therefore through a route other than flagellin perception. The analysis of response to flg22 was extended to the whole collection of AtRLP T-DNA insertion mutants. In no case was any significant alteration in the inhibition of seedling growth observed (Supplemental Table S5). Similarly, none of the Atrlp mutant lines had a significant alteration in its response to the necrosis-inducing elicitor protein from B. cinerea, BcNEP1 (Schouten et al., 2008), compared to the controls.
Mining of AtRLP Expression Data to Uncover Additional AtRLP-Regulated Biological Processes
In our unbiased screenings, few novel biological roles have been uncovered for AtRLP genes. To gain additional insight into the possible biological processes in which AtRLP genes are involved, the Genevestigator online search tool Meta-Analyzer (Zimmermann et al., 2004) was used (Supplemental Fig. S2). This analysis revealed that the expression of the AtRLP genes in the context of different organs, growth stages, and stress responses is very diverse. Most AtRLP genes are expressed in many organs and developmental stages. AtRLP4, which was predicted as a putative developmental ortholog (Fritz-Laylin et al., 2005), is ubiquitously and highly expressed across almost all the developmental stages and organs, confirming a potential basic function in plant development (Supplemental Figs. S1 and S2). However, the development of the Atrlp4-1 mutant is indistinguishable from that of wild-type plants. Some AtRLP genes are specifically expressed in only one or a few organs, such as AtRLP5, AtRLP8, AtRLP11, AtRLP45, and AtRLP48 that are mainly expressed in pollen (Supplemental Fig. S2), suggesting they may play a role at the reproductive stage. However, no defective pollen phenotypes were observed for mutants in those respective genes. The stress response expression data upon challenge with pests and pathogens, hormones, and abiotic stress factors (Supplemental Fig. S2) show differential expression patterns for all AtRLP genes. Strikingly, AtRLP48 is highly induced only upon hormone treatment, and for two hormone treatments (ABA and zeatin), AtRLP48 is the only AtRLP gene induced. Nevertheless, Atrlp48-1 showed no phenotype upon treatment with these hormones (data not shown).
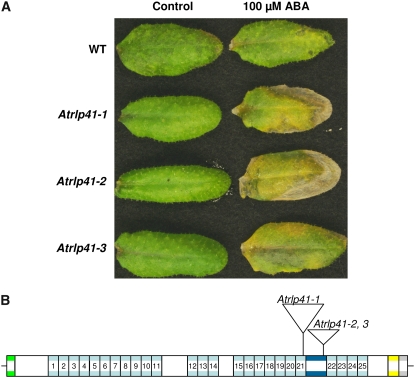
As many as 25 AtRLP genes (AtRLP2–4, 7, 13, 19, 20, 22, 23, 26, 28, 34–38, 40–43, 46, 47, 50, 52, and 54) are predominantly expressed in senescent leaves (Supplemental Fig. S2). Of these, five AtRLP-encoding genes (AtRLP7, 20, 28, 36, and 42) are almost exclusively induced in senescent leaves (Supplemental Fig. S2), suggesting a possible function in senescence-related processes. Therefore, we tested whether the 25 AtRLP genes are involved in senescence-related processes by subjecting leaves of the corresponding mutants to submergence in ABA. Most of the mutants did not show any altered phenotypes. However, three independent T-DNA insertion lines (Salk_024020, SM_3_20242, and SM_3_38956) of AtRLP41 displayed enhanced sensitivity upon exogenous application of 100 μm ABA, because the mutant leaves were bleached while wild-type leaves remained green (Fig. 5A). Therefore, our results indicate that AtRLP41 plays a role in ABA responses.
Figure 5.
Characterization of the Atrlp41 mutant alleles. A, Comparison of the leaf phenotype of wild type with mutants Atrlp41-1, Atrlp41-2, and Atrlp41-3 after exogenous application of ABA (right). B, The location of T-DNA in Atrlp41-1, Atrlp41-2, and Atrlp41-3.
Previously, AtRLP51 was reported to be locally induced in roots by the nonpathogenic, root-colonizing rhizobacterium Pseudomonas fluorescens WCS417r (Verhagen et al., 2004). This bacterium activates induced systemic resistance (ISR) against a broad range of pathogens (Pieterse et al., 1996). To investigate the role of AtRLP51 in activation of ISR, we tested the two T-DNA insertion mutants Atrlp51-1 and Atrlp51-2 for their ability to express ISR upon treatment with P. fluorescens WCS417r. After treatment, plants were inoculated with Pst DC3000 or with B. cinerea. While wild-type and mutant plants grown in noninfested control soil showed full susceptibility, both wild type and the mutants developed similar levels of ISR toward these pathogens in soil infested with P. fluorescens, indicating that AtRLP51 is not involved in ISR (data not shown).
DISCUSSION
We have undertaken a reverse genetic approach to genome-wide study the role of RLP genes in Arabidopsis. Previously, a total of 56 AtRLP genes have been identified (Fritz-Laylin et al., 2005). In this study, we identified one additional putative AtRLP gene, AtRLP5, which corresponds to At1g34290. Although this gene carries only two eLRRs, it complies with the canonical RLP domain composition. Moreover, it has been noted that the number of LRR units of resistance genes and resistance gene analogs can be highly variable, ranging from one to over two dozen, which is likely to be caused by illegitimate recombination (Wicker et al., 2007). We assembled a genome-wide collection of T-DNA knockout mutants that comprises at least one insertion mutant for 56 of the 57 AtRLP genes. We could not obtain any insertion line for just one of the RLP genes, AtRLP19, which may indicate that insertions in this specific AtRLP gene cause lethality. In total, 77 homozygous insertion lines in AtRLP genes have been collected that have all been assessed for phenotypic alterations in plant growth and development and for altered responsiveness to various external stimuli, including abiotic stress triggers and microbial pathogens. Previously, biological roles have been assigned to only two AtRLP genes, CLV2 and TMM (Jeong et al., 1999; Nadeau and Sack, 2002), while the biological functions of the remaining 55 AtRLP genes have remained elusive so far.
In this study, a number of additional novel phenotypes were found for insertion mutants in the CLV2 and TMM genes. Previous studies have demonstrated that mutations in any of the three CLV genes result in enlargement of meristems and increased floral organ numbers (Clark et al., 1993, 1995). Our CLV2 T-DNA insertion allele (Atrlp10-1) was found to grow slower, develop more rosette leaves and shorter stems, and develop flowers at a later stage than wild-type plants or clv2-3 mutants. Furthermore, the meristem of the main inflorescence was found to terminate flowering for a short period, upon which flowering resumed, resulting in an irregular distribution of siliques over the main stem. These novel phenotypes were found to be linked to the T-DNA insertion in CLV2 and may be attributed to the genetic background of the mutation, as the T-DNA insertion is a mutant of the Col-0 ecotype, while all other previously described clv2 mutants are backcrossed into the Landsberg erecta (Ler) ecotype (Kayes and Clark, 1998). The progeny of crosses between Atrlp10-1 and Ler wild-type plants developed a strong carpel phenotype that is comparable to clv2 alleles in the Ler ecotype: more rosette leaves and reduced height without transient termination of the main inflorescence (G. Wang, unpublished data). This suggests that the transient termination of the main inflorescence in Atrlp10-1 is most likely due to interplay within the genetic background of Col-0.
Previously, TMM has been shown to control the initiation of stomatal precursor cells and determine the orientation of the asymmetric divisions that pattern stomata (Geisler et al., 2000; Nadeau and Sack, 2002). In our TMM T-DNA insertion mutant (Atrlp17-1), we also observed the typical stomatal clustering phenotype. In addition, we found that mutations in TMM also displayed altered sensitivity to ABA. Growth of the TMM mutants was reduced upon exogenous application of ABA, while the induced chlorosis that is observed in control plants after ABA treatment was not observed. It has long been known that during early stages of drought, plant roots produce ABA that is transported with the transpiration stream and acts as a physiological signal to close stomata. The actual closure is established by an increase of the Ca2+ concentration in the guard cell cytoplasm (Schroeder and Hagiwara, 1989). At present, it is not known how TMM regulates stomatal distribution, but ABA sensitivity might be a crucial factor in this process. Apart from TMM, a visible altered phenotype upon ABA treatment could be identified for AtRLP41, because the corresponding mutants Atrlp41-1 to Atrlp41-3 showed enhanced sensitivity to exogenous application of ABA. Nevertheless, for these mutants no abnormalities in stomatal patterning could be observed. AtRLP41 appeared to be highly induced during plant senescence, and, because ABA is known to be able to act as an inducer of senescence, it is tempting to speculate that AtRLP41 is involved in ABA-induced senescence responses, although Atrlp41 mutants did not show any phenotypic alterations at this stage. However, ABA also plays important roles in other processes, including seed development and dormancy (Christmann et al., 2006), which might explain why expression at senescence stages has been reported. Although ABA receptors have not been identified yet, it has been demonstrated that an RLK called RPK1 is involved in early ABA perception in Arabidopsis (Osakabe et al., 2005). Reminiscent to the situation as occurs with the RLK CLV1 that interacts with the RLP CLV2, RPK1 may interact with TMM1 or AtRLP41 to constitute an ABA receptor complex.
Interestingly, it was recently shown that TMM negatively regulates three RLKs during the process of stomatal differentiation, one of which is ERECTA that also controls organ size and shape (Torii et al., 1996; Shpak et al., 2005). In addition, it was recently found that ERECTA also regulates plant transpiration efficiency, as ERECTA was found to modulate stomatal density through a role in epidermal pavement cell expansion (Masle et al., 2005). Possibly, TMM functions as an anchor protein for multiple RLKs in different signaling processes. A similar situation has recently been demonstrated for the RLK protein BAK1/SERK3 that not only interacts with the RLK BRI1 to modulate brassinosteroid signaling and thus regulate brassinosteroid-dependent growth (Li et al., 2002; Russinova et al., 2004) but also interacts with the RLK FLS2 that acts as a PAMP receptor for bacterial flagellin and functions in innate immunity in a brassinosteroid-independent manner (Chinchilla et al., 2007; Heese et al., 2007). It is anticipated that BAK1 interacts with additional innate immune receptors, because it also regulates full responses to PAMPs that are not related to flagellin, the containment of microbial infection-induced cell death, and restriction of various bacterial, fungal, and oomycete infections (Chinchilla et al., 2007; Heese et al., 2007; Kemmerling et al., 2007). The participation of specific receptor proteins in different receptor complexes may explain why some of these receptors play roles in processes as diverse as plant development and pathogen defense. This is not only the case for BAK1 but also for ERECTA that, in addition to development (Torii et al., 1996; Masle et al., 2005; Shpak et al., 2005), also plays a role in defense (Godiard et al., 2003; Llorente et al., 2005).
Remarkably, among the genome-wide collection of AtRLP T-DNA insertion mutants, visibly altered phenotypes were observed for only the four genes CLV2, TMM, AtRLP41, and AtRLP30, even though a wide range of developmental stages and treatments were tested. In other plant species, by far most RLP genes have been implicated in mediating microbial perception, mostly as pathogen resistance genes (Kruijt et al., 2005). In Arabidopsis, AtRLP52 has been implicated in resistance against the powdery mildew pathogen E. cichoracearum (Ramonell et al., 2005). Interestingly, it was observed that this specific AtRLP is also required for full resistance against the barley (Hordeum vulgare) pathogen Blumeria graminis f. sp. hordei (J. Mansfield, unpublished data). However, in this study, it is rather surprising that only two of the T-DNA insertion lines in the AtRLP genes, AtRLP18 and AtRLP30, displayed altered susceptibility upon pathogen challenge. Four independent mutations in AtRLP30 were found to affect Arabidopsis nonhost defense against the nonadapted bean (Phaseolus vulgaris) pathogen Psp, although the mutants were not as susceptible as fls2 mutants defective in the perception of bacterial flagellin. This suggests that, rather than acting as a true resistance gene like all other RLPs that have been characterized in plant defense, both AtRLP18 and AtRLP30 act as components of basal defense. Interestingly, defense against another nonadapted P. syringae strain (pv tabaci) was not compromised, while defense against weakly pathogenic Pst strains (hrpA, hcrC, and coronatine mutants) also appeared to be intact. In tomato, the RLP genes Ve1 and Ve2 have been implicated in resistance against race 1 strains of the vascular pathogen V. dahliae (Kawchuk et al., 2001), which also is a pathogen of Arabidopsis (Fradin and Thomma., 2006). Nevertheless, none of the AtRLP insertion lines was found to display altered V. dahliae susceptibility. Based on sequence comparison and bioinformatic analysis, it has been suggested that the vast majority of the AtRLP genes were likely to act as disease resistance genes. Despite screening a broad spectrum of pathogens with different colonization and feeding styles, we have so far not been able to support this hypothesis. Possibly, this is the consequence of not having used the correct pathogen strains against which these genes are active. Alternatively, the AtRLP genes may not act as race-specific disease resistance genes but rather play a role in nonhost resistance or basal host defense. In such cases, the array of potential microbial targets may be dramatically increased and the response to more microbes or even insects and nematodes should be tested (Stout et al., 2006).
The lack of identification of biological functions for AtRLP genes may also be explained by functional redundancy, a phenomenon that typically obscures studies employing reverse genetics strategies, as has been described for MADS-box transcription factors (Pařenicová et al., 2003) and RLK gene family members (Albrecht et al., 2005; DeYoung et al., 2006; Hord et al., 2006). It has been suggested that CLV1 and CLV2 heterodimerize to form a receptor complex for the secreted CLV3 signaling peptide (Jeong et al., 1999; Ogawa et al., 2008). However, when compared to clv1 and clv3 alleles, clv2 mutants display relatively weak phenotypes, because fasciation in clv2 mutants is rarely observed and only under short-day growth conditions (Kayes and Clark, 1998). This may suggest that the role of CLV2 is indeed redundant, although the finding that CLV2, but not CLV1, can perceive the conserved CLE motif of CLV3-like peptides argues against this hypothesis (Fiers et al., 2005). Current strategies employ RNA interference experiments to interfere with the expression of multiple AtRLP genes at the same time and thus possibly overcome functional redundancy among AtRLP genes. The RNA interference lines that are silenced for multiple AtRLP genes can be screened with the various abiotic and biotic stress factors to find biological roles for these AtRLP genes.
MATERIALS AND METHODS
Bioinformatic Analysis
To investigate the structure of AtRLP genes, BLAST queries were performed using Arabidopsis (Arabidopsis thaliana) CLV2 and TMM and tomato (Solanum lycopersicum) Cf-9 predicted protein sequences to search translated sequences from the Arabidopsis genome. SMART (http://smart.embl-heidelberg.de), PFAM (http://pfam.janelia.org), SignalP (http://www.cbs.dtu.dk/services/SignalP), and TMHMM (http://www.cbs.dtu.dk/services/TMHMM) were used for domain predictions. The exon/intron boundaries were investigated using GenScan (http://genes.mit.edu/GENSCAN.html), refined using SeqViewer at The Arabidopsis Information Resource (www.arabidopsis.org), and visualized using Jellyfish software (Riethof and Balakrishnan, 2001).
Identification and Analysis of T-DNA Insertion Mutants
The database at SIGnAL (Alonso et al., 2003; http://signal.salk.edu) was searched to identify putative T-DNA insertion mutants, the available lines of interest of which were obtained from the Nottingham Arabidopsis Stock Center (NASC; http://www.arabidopsis.info), GABI-Kat (Rosso et al., 2003; http://www.gabi-kat.de/), or Genoplante FLAGdb/FST (Balzergue et al., 2001; http://urgi.infobiogen.fr). Correct insertion of the T-DNA in these lines was determined with PCR. Genomic DNA was isolated from individual plants that belong to the respective T-DNA insertion lines and used in two separate PCR reactions with different primer sets (Supplemental Table S2). One contained a gene-specific primer and a T-DNA-specific primer to check for the presence of the insertion, and the second PCR contained two gene-specific primers spanning the proposed insertion site to check for nondisrupted alleles. Plants for which the PCR with a gene-specific primer and a T-DNA-specific primer yielded a product, while the PCR with the two gene-specific primers did not yield a product, were considered homozygous insertion lines, which was confirmed in plants from the subsequent generation.
Plant Growth Conditions
Arabidopsis plants of the ecotypes Col-0, Wassilewskija, and Ler were used. Soil-grown plants were cultured either in a growth chamber at 22°C, 72% relative humidity, and usually a 16-h photoperiod, or in a greenhouse at 21°C during the 16-h day period and 19°C during the night period at 72% relative humidity. In the greenhouse, supplemental light (100 Wm−2) was used when the sunlight influx intensity was below 150 Wm−2.
For in vitro growth of Arabidopsis, seeds were surface sterilized and sown on Murashige and Skoog (MS) medium (Duchefa) solidified with 1.5% plant agar (Duchefa). After sowing, the plates were incubated at 4°C in the dark for 3 d and subsequently transferred to the growth chamber.
Phenotypic Evaluations of Plant Growth and Development
For phenotypic evaluations of plant growth and development, Arabidopsis plants were grown on half-strength MS medium supplemented with 1% Suc and 0.5 g/L MES, pH 5.8. After 2 weeks, plants were transferred to soil for further observations. To assess seed morphology, siliques from the primary inflorescences were screened for seed abortion using a dissection microscope (Tzafrir et al., 2004). Seeds at different developmental stages were mounted in clearing solution (Sabatini et al., 1999), and cleared samples were observed using a Nikon optiphot microscope equipped with Normarski optics. To score vascular patterning and stomatal distribution, cotyledon and rosette leaves were cleared by immersion in ethanol:acetic acid (3:1), subsequently rinsed in 70% ethanol, and incubated in 100% ethanol at 4°C overnight (Jun et al., 2002). The leaves were observed using a dissecting microscope for vascular patterning and Normaski optics for the stomatal distribution. Finally, root geotropism was studied by growing seedlings on vertically oriented half-strength MS plates that were rotated 90° after 6 d of growth. After 10 h, the bending angle of the root was measured (Sedbrook et al., 2002).
Conditional Phenotype Assays
To assess susceptibility toward abiotic stress, seeds were sown on MS plates amended with NaCl (100 or 150 mm), LiCl (20 or 30 mm), mannitol (150 or 200 mm), or hydrogen peroxide (3.3 or 6.7 mm) and evaluated for aberrant growth. To assay heavy metal resistance, plants were grown vertically on half-strength MS medium amended with 2% (w/v) Suc and 85 μm CdCl2 (Lee et al., 2003).
To test whether AtRLP genes are involved in responsiveness to hormones, the sterilized seeds were grown on vertically oriented half-strength MS plates containing different hormones at different concentrations (Supplemental Table S3).
To screen whether AtRLP genes are involved into leaf senescence, detached leaves were floated on 3 mm MES [2-(N-morpholino)ethanesulfonic acid monohydrate] buffer, pH 5.8, in the presence of 50 μm or 100 μm ABA, 50 μm methyl jasmonate, 5 μm ethylene, or 1 μm epibrassinolide (He et al., 2001).
Pathogen Cultivation
Alternaria brassicicola (strain MUCL20297; Mycotheque Université Catholique de Louvain, Louvain-la-Neuve, Belgium), Cladosporium cucumerinum, Cladosporium fulvum, Plectospaerella cucumerina (Thomma et al., 2000), Sclerotinia sclerotium strain ND30, and Verticillium dahliae strain ST37.01 were maintained on potato dextrose agar (Oxoid). Botrytis cinerea (Brouwer et al., 2003) was grown on half-strength potato dextrose agar amended with 5 g/L agar and 150 g/L blended tomato leaves. Colletotrichum destructivum (strain IMI349061; CABI Bioscience) was grown on Mathur's agar (Mathur et al., 1950). All fungal in vitro cultures were grown at 22°C. Oidium neolycopersisi (Bai et al., 2005) was maintained on Moneymaker tomato plants in the greenhouse. Two GFP transformants of the oomycete Phytophthora infestans strains 14.3 (Dr. Govers, Wageningen University, The Netherlands) and 208M2 (Dr. S. Kamoun, Ohio State University) were maintained on rye-agar at 18°C in the dark. Isolates of Hyaloperonospora parasitica were maintained as described (Tör et al., 2002). Pseudomonas syringae pv tomato DC3000 with or without avrRpt2, avrRpm1, or avrRps4 were grown on King's B agar (King et al., 1954) supplemented with the appropriate antibiotics (25 μg/mL rifampicin and 100 μg/mL kanamycin). Pectobacterium atrosepticum strain LMG 6669 (Coordinated Collections of Micro-organisms, Ghent, Belgium) was maintained on nutrient agar (Oxoid). Xanthomonas campestris pv campestris (strain 568) was grown on Kado's medium agar (Kado and Heskett, 1970). All bacterial strains were grown overnight at 28°C.
Pathogen Inoculations
All pathogen (except V. dahliae and H. parasitica) inoculations were performed using soil-grown plants with fully expanded rosette leaves. Inoculum of all in vitro-cultured fungi (except S. sclerotiorum) was prepared as previously described (Broekaert et al., 1990) and used as a suspension of 106 conidia/mL in water. Inoculations with A. brassicicola, B. cinerea, C. destructivum, and P. cucumerina were performed by placing a 6-μL drop of the conidial suspensions on each expanded leaf (Thomma et al., 1998, 2000; O'Connell et al., 2004; Brouwer et al., 2003). C. fulvum and C. cucumerinum suspensions were sprayed as a mist on the adaxial sides of the leaves. For V. dahliae inoculations, 2-week-old Arabidopsis plants were up-rooted, root tips were cut off, and incubated in the conidial suspension for 1 min. Subsequently, the plants were replanted into fresh soil. For S. sclerotiorum, three mycelium plugs from a culture plate were placed in a 300-mL flask containing 100 mL of potato dextrose broth (Difco) and grown for 3 d at 22°C with 150 rpm. Afterward, the mycelium was homogenized in a blender. Leaves were inoculated by placing a 10-μL drop of mycelium fragments (OD600 = 3.5) on each of the fully expanded leaves. For P. infestans, a rye-agar plate with 10-d-old mycelium was incubated with sterile water at 4°C for 2 h to release zoospores from zoosporangia. One 5-μL drop of a suspension of 105 zoospores/mL in water was placed on each fully expanded leaf. To avoid background fluorescence from superficial growing P. infestans, the drops were removed by drying with tissue paper after 36 h. For Oidium neolycopersici, 105 conidia/mL was used. The inoculation was performed as described by Bai et al. (2005). Inoculations of Arabidopsis seedlings with H. parasitica were performed as described (Tör et al., 2002).
For all bacterial inoculations, bacteria were grown overnight at 28°C in the appropriate medium supplemented with the appropriate antibiotics. Strains of P. syringae (except P. syringae pv phaseolicola [Psp]) and P. atrosepticum were spray inoculated with a bacterial suspension of OD600 0.3 supplemented with 0.05% [v/v] Silwet L-77 (van Meeuwen Chemicals). For X. campestris, two different inoculation methods were carried out (Meyer et al., 2005): infiltration of a concentrated bacterial suspension or wound inoculation.
For Psp 1448A, three half leaves on eight plants were infiltrated with bacteria at OD600 0.25 (approximately 2 × 108 cells/mL). Symptom development was scored after 4 and 6 d and sites assigned to each progressive category: 0, no symptoms; 1, very pale yellowing; 2, pale yellowing; 3, yellowing over most of the area infiltrated; 4, pale yellowing with patchy collapse; 5, yellow with patchy collapse; 6, collapse of more than 50% of infiltration site; and 7, collapse of all the infiltrated area. Bacterial numbers were recorded as described by de Torres et al. (2006).
For all inoculations, except those with O. neolycopersici and V. dahliae, plants were kept in boxes with transparent lids at high relative humidity for the remainder of the experiment. As a positive control for the inoculations with A. brassicicola, B. cinerea, and P. cucumerina, pad3-1 mutant plants were used (Thomma et al., 1999, 2000; Kliebenstein et al., 2005). For P. infestans, the pen2-1 mutant was used (Lipka et al., 2005), while for the Pseudomonas strains the genotypes NahG and npr1-1 were used (Thomma et al., 1998). Finally, for X. campestris the ecotype Kas was used as positive control (Xu et al., 2008).
To test whether AtRLP51 is involved in ISR expression, the ISR bioassay was performed as described by Pieterse et al. (1996) except for the challenge inoculation. For P. syringae and for B. cinerea, the inoculations were performed as mentioned previously. Except for P. syringae, a lower concentration of a bacterial suspension of OD600 0.3 five times diluted was used.
Response to Pathogen Elicitors
Flg22-induced seedling growth inhibition assays (Gomez-Gomez et al., 1999) were performed essentially as described (Pfund et al., 2004). After germination of Arabidopsis seeds for 5 d at 22°C, two seedlings were transferred to 750 mL of liquid MS medium in a 25-well plate either with or without 2 mg/L flg22 peptide (sequence, TRLSSGKINSAKDDAAGL). Each treatment was replicated five times. After 2 weeks further growth, the weights of the seedlings were recorded. Wassilewskija-0, Col-0 fls2 (insensitive to flg22), and Col-0 (susceptible to flg22 growth inhibition) were used as controls in each experiment.
Leaves of Arabidopsis plants were pressure infiltrated with the B. cinerea elicitor protein BcNEP1 that was isolated from a Pichia pastoris culture heterologously expressing BcNEP1. A raw protein extract from culture filtrate containing the BcNEP1 protein was isolated as described (Schouten et al., 2008) and was 10 times diluted in MMA (5 g/L MS salts [Duchefa], 1.9 g/L MES).
Localization of AtRLP30
AtRLP30 is predicted to contain a single exon, which was confirmed by sequencing full-length cDNA from Col-0 amplified using reverse transcription-PCR. The resulting cDNA was cloned into the gateway entry vector pDONR/Zeo using BP clonase (Invitrogen) and subsequently transferred to the gateway-compatible binary vector pEarleyGate101 (Earley et al., 2006) using LR clonase (Invitrogen). This resulted in a plasmid with AtRLP30 fused to the coding sequence of YFP∷HA and expression was driven by the cauliflower mosaic virus 35S promoter. The T-DNA insertion line Salk_122528, homozygous for the insertion in AtRLP30, was transformed with this plasmid using the floral dip method (Clough and Bent, 1998). Transgenic plants were selected on soil soaked with 150 mg/L Basta herbicide (glufosinate-ammonium, Bayer CropScience) and confirmed by PCR. Plants were checked for fluorescence using an Olympus IX70 microscope equipped with a Fluroview 300 confocal laser scanning unit. AtRLP30∷YFP∷HA fluorescence was excited with a 488-nm argon laser and fluorescence was detected between 510 nm and 530 nm.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. cDNA, EST, and Massively Parallel Signature Sequencing expression data for AtRLP genes.
Supplemental Figure S2. Expression profile of AtRLP genes in various organs and growth stages and upon stress responses.
Supplemental Figure S3. Expression of AtRLP30 after PAMP treatment.
Supplemental Table S1. Pairwise alignment of AtRLP amino acid sequences.
Supplemental Table S2. Primers used to check for the presence of the predicted T-DNA insertions.
Supplemental Table S3. Conditional phenotype assays for AtRLP mutants.
Supplemental Table S4. Interaction phenotypes of AtRLP mutants with isolates of H. parasitica.
Supplemental Table S5. Screening of RLP mutants with flg22 using seedling assays.
Supplementary Material
Acknowledgments
We thank Drs. Shiu, Fritz-Laylin, and Yang for valuable discussion. We are grateful to NASC, GABI-Kat, and Genoplante FLAGdb/FST for providing plant materials. We further acknowledge Drs. Rao Uppalapati, Yuling Bai, Francine Govers, Thomas Kroj, Bart Lievens, and Berlin Nelson for providing pathogen strains, and Blaise Alako, Bert Essenstam, Terry Amatulli, Ann Baker, Nina Grabov, and Zhao Zhang for technical assistance.
This work was supported by the Dutch Graduate School of Experimental Plant Sciences, by the Research Council for Earth and Life Sciences of the Netherlands Organization for Scientific Research (VIDI grant to B.P.H.J.T.), by the UK Biotechnology and Biological Sciences Research Council (to J.W.M., B.K., A.W.-T., and M.T.), by the Gatsby Charitable Foundation (to C.Z. and J.D.G.J.), and by a postdoctoral long-term fellowship from the European Molecular Biology Organization (to C.Z.).
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) are: Bart P.H.J. Thomma (bart.thomma@wur.nl) and Mahmut Tör (mahmut.tor@warwick.ac.uk).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Albrecht C, Russinova E, Hecht V, Baaijens E, De Vries S (2005) The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES1 and 2 control male sporogenesis. Plant Cell 17 3337–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815 [DOI] [PubMed] [Google Scholar]
- Bai YL, van der Hulst R, Bonnema G, Marcel BC, Meijer-Dekens F, Niks RE, Lindhout P (2005) Tomato defense to Oidium neolycopersici: dominant Ol genes confer isolate-dependent resistance via a different mechanism than recessive ol-2. Mol Plant Microbe Interact 18 354–362 [DOI] [PubMed] [Google Scholar]
- Balzergue S, Dubreucq B, Chauvin S, Le-Clainche I, Le Boulaire F, De Rose R, Samson F, Biaudet V, Lecharny A, Cruaud C, et al (2001) Improved PCR-walking for large-scale isolation of plant T-DNA borders. Biotechniques 30 496–504 [DOI] [PubMed] [Google Scholar]
- Belfanti E, Silfverberg-Dilworth E, Tartarini S, Patocchi A, Barbieri M, Zhu J, Vinatzer BA, Gianfranceschi L, Gessler C, Sansavini S (2004) The HcrVf2 gene from a wild apple confers scab resistance to a transgenic cultivated variety. Proc Natl Acad Sci USA 101 886–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommert P, Satoh-Nagasawa N, Jackson D, Hirano HY (2005) Genetics and evolution of inflorescence and flower development in grasses. Plant Cell Physiol 46 69–78 [DOI] [PubMed] [Google Scholar]
- Broekaert WF, Terras FRG, Cammue BPA, Vanderleyden J (1990) An automated quantitative assay for fungal growth inhibition. FEMS Microbiol Lett 69 55–59 [Google Scholar]
- Brouwer M, Lievens B, Van Hemelrijck W, Van Den Ackerveken G, Cammue BPA, Thomma BPHJ (2003) Quantification of disease progression of several microbial pathogens on Arabidopsis thaliana using real-time fluorescence PCR. FEMS Microbiol Lett 228 241–248 [DOI] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JDG, Felix G, Boller T (2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448 497–500 [DOI] [PubMed] [Google Scholar]
- Christmann A, Moes D, Himmelbach A, Yang Y, Tang Y, Grill E (2006) Integration of abscisic acid signalling into plant responses. Plant Biol 8 314–325 [DOI] [PubMed] [Google Scholar]
- Clark SE, Running MP, Meyerowitz EM (1993) CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119 397–418 [DOI] [PubMed] [Google Scholar]
- Clark SE, Running MP, Meyerowitz EM (1995) CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121 2057–2067 [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM (1997) The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89 575–585 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- de Torres M, Mansfield JW, Grabov N, Brown IR, Ammouneh H, Tsiamis G, Forsyth A, Robatzek S, Grant M, Boch J (2006) Pseudomonas syringae effector AvrPtoB suppresses basal defence in Arabidopsis. Plant J 47 368–382 [DOI] [PubMed] [Google Scholar]
- DeYoung BJ, Bickle KL, Schrage KJ, Muskett P, Patel K, Clark SE (2006) The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J 45 1–16 [DOI] [PubMed] [Google Scholar]
- Diévart A, Clark SE (2004) LRR-containing receptors regulating plant development and defense. Development 131 251–261 [DOI] [PubMed] [Google Scholar]
- Dixon MS, Hatzixanthis K, Jones DA, Harrison K, Jones JDG (1998) The tomato Cf-5 disease resistance gene and six homologs show pronounced allelic variation in leucine-rich repeat copy number. Plant Cell 10 1915–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon MS, Jones DA, Keddie JS, Thomas CM, Harrison K, Jones JDG (1996) The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell 84 451–459 [DOI] [PubMed] [Google Scholar]
- Earley K, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45 616–629 [DOI] [PubMed] [Google Scholar]
- Fiers M, Golemiec E, Xu J, Van Der Geest L, Heidstra R, Stiekema W, Liu CM (2005) The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell 17 2542–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin EF, Thomma BPHJ (2006) Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol Plant Pathol 7 71–86 [DOI] [PubMed] [Google Scholar]
- Fritz-Laylin LK, Krishnamurthy N, Tör M, Sjölander KV, Jones JDG (2005) Phylogenomic analysis of the receptor-like proteins of rice and Arabidopsis. Plant Physiol 138 611–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler M, Nadeau J, Sack FD (2000) Oriented asymmetric divisions that generate the stomatal spacing pattern in Arabidopsis are disrupted by the too many mouths mutation. Plant Cell 12 2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godiard L, Sauviac L, Torii KU, Grenon O, Mangin B, Grimsley NH, Marco Y (2003) ERECTA, an LRR receptor-like kinase protein controlling development pleiotropically affects resistance to bacterial wilt. Plant J 36 353–365 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, Boller T (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5 1003–1011 [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez L, Felix G, Boller T (1999) A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J 18 277–284 [DOI] [PubMed] [Google Scholar]
- He Y, Tang W, Swain J, Green A, Jack T, Gan S (2001) Networking senescence-regulating pathways by using Arabidopsis enhancer trap lines. Plant Physiol 126 707–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese A, Hann DR, Gimenez-Ibanez S, Jones AM, He K, Li J, Schroeder JI, Peck SC, Rathjen JP (2007) The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA 104 12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hord CL, Chen C, Deyoung BJ, Clark SE, Ma H (2006) The BAM1/BAM2 receptor-like kinases are important regulators of Arabidopsis early anther development. Plant Cell 18 1667–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S, Trotochaud AE, Clark SE (1999) The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11 1925–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DA, Jones JDG (1997) The role of leucine-rich repeat proteins in plant defences. Adv Bot Res 24 89–167 [Google Scholar]
- Jones DA, Thomas CM, Hammond-Kosack KE, Balint-Kurti PJ, Jones JDG (1994) Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266 789–793 [DOI] [PubMed] [Google Scholar]
- Joosten MHAJ, de Wit PJGM (1999) The tomato-Cladosporium fulvum interaction: a versatile experimental system to study plant-pathogen interactions. Annu Rev Phytopathol 37 335–367 [DOI] [PubMed] [Google Scholar]
- Jun JH, Ha CM, Nam HG (2002) Involvement of the VEP1 gene in vascular strand development in Arabidopsis thaliana. Plant Cell Physiol 43 323–330 [DOI] [PubMed] [Google Scholar]
- Kado CI, Heskett MG (1970) Selective media for isolation of Agrobacterium, Corynebacterium, Erwinia, Pseudomonas, and Xanthomonas. Phytopathology 60 969–976 [DOI] [PubMed] [Google Scholar]
- Kawchuk LM, Hachey J, Lynch DR, Kulcsar F, Van Rooijen G, Waterer DR, Robertson A, Kokko E, Byers R, Howard RJ, et al (2001) Tomato Ve disease resistance genes encode cell surface-like receptors. Proc Natl Acad Sci USA 98 6511–6515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayes JM, Clark SE (1998) CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development 125 3843–3851 [DOI] [PubMed] [Google Scholar]
- Kemmerling B, Schwedt A, Rodriguez P, Mazzotta S, Frank M, Qamar SA, Mengiste T, Betsuyaku S, Parker JE, Mussig C, et al (2007) The BRI1-associated kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Curr Biol 17 1116–1122 [DOI] [PubMed] [Google Scholar]
- King EO, Ward MK, Raney DE (1954) Two simple media for the demonstration of phycocyanin and fluorescin. J Lab Clin Med 44 301–307 [PubMed] [Google Scholar]
- Kinoshita T, Caño-Delgado A, Seto H, Hiranuma S, Fujioka S, Yoshida S, Chory J (2005) Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433 167–171 [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, Rowe HC, Denby KJ (2005) Secondary metabolites influence Arabidopsis/Botrytis interactions: variation in host production and pathogen sensitivity. Plant J 44 25–36 [DOI] [PubMed] [Google Scholar]
- Kobe B, Kajava AV (2001) The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol 11 725–732 [DOI] [PubMed] [Google Scholar]
- Kruijt M, De Kock MJD, de Wit PJGM (2005) Receptor-like proteins involved in plant disease resistance. Mol Plant Pathol 6 85–97 [DOI] [PubMed] [Google Scholar]
- Lee S, Moon JS, Ko TS, Petros D, Goldsbrough PB, Korban SS (2003) Overexpression of Arabidopsis phytochelatin synthase paradoxically leads to hypersensitivity to cadmium stress. Plant Physiol 131 656–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister D (2004) Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance genes. Trends Genet 20 116–122 [DOI] [PubMed] [Google Scholar]
- Li CM, Brown I, Mansfield J, Stevens C, Boureau T, Romantschuk M, Taira S (2002) The Hrp pilus of Pseudomonas syringae elongates from its tip and acts as a conduit for translocation of the effector protein HrpZ. EMBO J 21 1909–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chory J (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90 929–938 [DOI] [PubMed] [Google Scholar]
- Li J, Jin H (2007) Regulation of brassinosteroid signaling. Trends Plant Sci 12 37–41 [DOI] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110 213–222 [DOI] [PubMed] [Google Scholar]
- Lipka V, Dittgen J, Bednarek P, Bhat R, Wiermer M, Stein M, Landtag J, Brandt W, Rosahl S, Scheel D, et al (2005) Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310 1180–1183 [DOI] [PubMed] [Google Scholar]
- Llorente F, Alonso-Blanco C, Sanchez-Rodriguez C, Jorda L, Molina A (2005) ERECTA receptor-like kinase and heterotrimeric G protein from Arabidopsis are required for resistance to the necrotrophic fungus Plectosphaerella cucumerina. Plant J 43 165–180 [DOI] [PubMed] [Google Scholar]
- Masle J, Gilmore SR, Farquhar GD (2005) The ERECTA gene regulates plant transpiration efficiency in Arabidopsis. Nature 436 866–870 [DOI] [PubMed] [Google Scholar]
- Mathur RS, Barnett HL, Lilly VG (1950) Sporulation of Colletotrichum lindemuthianum in culture. Phytopathology 40 104–114 [Google Scholar]
- Meyer D, Lauber E, Roby D, Arlat M, Kroj T (2005) Optimization of pathogenicity assays to study the Arabidopsis thaliana-Xanthomonas campestris pv. campestris pathosystem. Mol Plant Pathol 6 327–333 [DOI] [PubMed] [Google Scholar]
- Nadeau JA, Sack FD (2002) Control of stomatal distribution on the Arabidopsis leaf surface. Science 296 1697–1700 [DOI] [PubMed] [Google Scholar]
- Nürnberger T, Kemmerling B (2006) Receptor protein kinases: pattern recognition receptors in plant immunity. Trends Plant Sci 11 519–522 [DOI] [PubMed] [Google Scholar]
- O'Connell R, Herbert C, Sreenivasaprasad S, Khatib M, Esquerre-Tugaye MT, Dumas B (2004) A novel Arabidopsis-Colletotrichum pathosystem for the molecular dissection of plant-fungal interactions. Mol Plant Microbe Interact 17 272–282 [DOI] [PubMed] [Google Scholar]
- Ogawa M, Shinohara H, Sakagami Y, Matsubayashi Y (2008) Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319 294. [DOI] [PubMed] [Google Scholar]
- Osakabe Y, Maruyama K, Seki M, Satou M, Shinozaki K, Yamaguchi-Shinozaki K (2005) Leucine-rich repeat receptor-like kinase1 is a key membrane-bound regulator of abscisic acid early signaling in Arabidopsis. Plant Cell 17 1105–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pařenicová L, de Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, Cook HE, Ingram RM, Kater MM, Davies B, et al (2003) Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis. Plant Cell 15 1538–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfund C, Tans-Kersten J, Dunning FM, Alonso JM, Ecker JR, Allen C, Bent AF (2004) Flagellin is not a major defense elicitor in Ralstonia solanacearum cells or extracts applied to Arabidopsis thaliana. Mol Plant Microbe Interact 17 696–706 [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, Van Wees SCM, Hoffland E, Van Pelt JA, Van Loon LC (1996) Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis-related gene expression. Plant Cell 8 1225–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramonell K, Berrocal-Lobo M, Koh S, Wan JR, Edwards H, Stacey G, Somerville S (2005) Loss-of-function mutations in chitin responsive genes show increased susceptibility to the powdery mildew pathogen Erysiphe cichoracearum. Plant Physiol 138 1027–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riethof DA, Balakrishnan R (2001) LabVelocity: online tools for life science products, protocols, technical information, MEDLINE searches, and laboratory calculations. Biotechniques 30 1310–1315 [DOI] [PubMed] [Google Scholar]
- Rojo E, Sharma VK, Kovaleva V, Raikhel NV, Fletcher JC (2002) CLV3 is localized to the extracellular space, where it activates the Arabidopsis CLAVATA stem cell signaling pathway. Plant Cell 14 969–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron M, Avni A (2004) The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 16 1604–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53 247–259 [DOI] [PubMed] [Google Scholar]
- Russinova E, Borst JW, Kwaaitaal M, Cano-Delgado A, Yin Y, Chory J, de Vries SC (2004) Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16 3216–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, et al (1999) An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99 463–472 [DOI] [PubMed] [Google Scholar]
- Schouten A, van Baarlen P, van Kan JA (2008) Phytotoxic Nep1-like proteins from the necrotrophic fungus Botrytis cinerea associate with membranes and the nucleus of plant cells. New Phytol 177 493–505 [DOI] [PubMed] [Google Scholar]
- Schroeder JL, Hagiwara S (1989) Cytosolic calcium regulates ion channels in the plasma membrane of Vicia faba guard cells. Nature 338 427–430 [Google Scholar]
- Sedbrook JC, Carroll KL, Hung KF, Masson PH, Somerville CR (2002) The Arabidopsis SKU5 gene encodes an extracellular glycosyl phosphatidylinositol-anchored glycoprotein involved in directional root growth. Plant Cell 14 1635–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB (2003) Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol 132 530–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak ED, McAbee JM, Pillitteri LJ, Torii KU (2005) Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309 290–293 [DOI] [PubMed] [Google Scholar]
- Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, Gardner J, Wang B, Zhai WX, Zhu LH, et al (1995) A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270 1804–1806 [DOI] [PubMed] [Google Scholar]
- Soylu S, Brown IR, Mansfield JW (2005) Cellular reactions in Arabidopsis following challenge by strains of Pseudomonas syringae: from basal resistance to compatibility. Physiol Mol Plant Pathol 66 232–243 [Google Scholar]
- Stout MJ, Thaler JS, Thomma BPHJ (2006) Plant-mediated interactions between pathogenic microorganisms and herbivorous arthropods. Annu Rev Entomol 51 663–689 [DOI] [PubMed] [Google Scholar]
- Taguchi-Shiobara F, Yuan Z, Hake S, Jackson D (2001) The fasciated ear2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize. Genes Dev 15 2755–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takken FLW, Thomas CM, Joosten MHAJ, Golstein C, Westerink N, Hille J, Nijkamp HJJ, De Wit PJGM, Jones JDG (1999) A second gene at the tomato Cf-4 locus confers resistance to Cladosporium fulvum through recognition of a novel avirulence determinant. Plant J 20 279–288 [DOI] [PubMed] [Google Scholar]
- Thomas CM, Jones DA, Parniske M, Harrison K, Balint-Kurti PJ, Hatzixanthis K, Jones JDG (1997) Characterization of the tomato Cf-4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognitional specificity in Cf-4 and Cf-9. Plant Cell 9 2209–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BPHJ, Eggermont K, Broekaert WF, Cammue BPA (2000) Disease development of several fungi on Arabidopsis can be reduced by treatment with methyl jasmonate. Plant Physiol Biochem 38 421–427 [Google Scholar]
- Thomma BPHJ, Eggermont K, Penninckx IAMA, Mauch-Mani B, Vogelsang R, Cammue BPA, Broekaert WF (1998) Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci USA 95 15107–15111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BPHJ, Nelissen I, Eggermont K, Broekaert WF (1999) Deficiency in phytoalexin production causes enhanced susceptibility of Arabidopsis thaliana to the fungus Alternaria brassicicola. Plant J 19 163–171 [DOI] [PubMed] [Google Scholar]
- Thomma BPHJ, Penninckx IAMA, Broekaert WF, Cammue BPA (2001) The complexity of disease signaling in Arabidopsis. Curr Opin Immunol 13 63–68 [DOI] [PubMed] [Google Scholar]
- Thomma BPHJ, Van Esse HP, Crous PW, de Wit PJGM (2005) Cladosporium fulvum (syn. Passalora fulva), a highly specialized plant pathogen as a model for functional studies on plant pathogenic Mycosphaerellaceae. Mol Plant Pathol 6 379–393 [DOI] [PubMed] [Google Scholar]
- Tör M, Gordon P, Cuzick A, Eulgem T, Sinapidou E, Mert F, Can C, Dangl JL, Holub EB (2002) Arabidopsis SGT1b is required for defence signaling conferred by several downy mildew (Peronospora parasitica) resistance genes. Plant Cell 14 993–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y (1996) The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotochaud AE, Hao T, Wu G, Yang Z, Clark SE (1999) The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and a Rho-related protein. Plant Cell 11 393–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I, Pena-Muralla R, Dicherman A, Berg M, Rogers R, Hutchens S, Sweeney TC, McElver J, Aux G, Patton D, et al (2004) Identification of genes required for embryo development in Arabidopsis. Plant Physiol 135 1206–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhagen BWM, Glazebrook J, Zhu T, Chang HS, Van Loon LC, Pieterse CMJ (2004) The transcriptome of rhizobacteria-induced systemic resistance in Arabidopsis. Mol Plant Microbe Interact 17 895–908 [DOI] [PubMed] [Google Scholar]
- Walker JC, Zhang R (1990) Relationship of a putative receptor protein-kinase from maize to the S-locus glycoproteins of Brassica. Nature 345 743–746 [DOI] [PubMed] [Google Scholar]
- Wicker T, Yahiaoui N, Keller B (2007) Illegitimate recombination is a major evolutionary mechanism for initiating size variation in plant resistance genes. Plant J 51 631–641 [DOI] [PubMed] [Google Scholar]
- Xu RQ, Blanvillain S, Feng JX, Jiang BL, Li XZ, Wei HY, Kroj T, Lauber E, Roby D, Chen B, et al (2008) AvrAC(Xcc8004), a type III effector with a leucine-rich repeat domain from Xanthomonas campestris pathovar campestris confers avirulence in vascular tissues of Arabidopsis thaliana ecotype Col-0. J Bacteriol 190 343–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Sack FD (1995) The too many mouths and four lips mutations affect stomatal production in Arabidopsis. Plant Cell 7 2227–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JDG, Felix G, Boller T (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428 764–767 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.