Abstract
The cuticle, covering the surface of all primary plant organs, plays important roles in plant development and protection against the biotic and abiotic environment. In contrast to vegetative organs, very little molecular information has been obtained regarding the surfaces of reproductive organs such as fleshy fruit. To broaden our knowledge related to fruit surface, comparative transcriptome and metabolome analyses were carried out on peel and flesh tissues during tomato (Solanum lycopersicum) fruit development. Out of 574 peel-associated transcripts, 17% were classified as putatively belonging to metabolic pathways generating cuticular components, such as wax, cutin, and phenylpropanoids. Orthologs of the Arabidopsis (Arabidopsis thaliana) SHINE2 and MIXTA-LIKE regulatory factors, activating cutin and wax biosynthesis and fruit epidermal cell differentiation, respectively, were also predominantly expressed in the peel. Ultra-performance liquid chromatography coupled to a quadrupole time-of-flight mass spectrometer and gas chromatography-mass spectrometry using a flame ionization detector identified 100 metabolites that are enriched in the peel tissue during development. These included flavonoids, glycoalkaloids, and amyrin-type pentacyclic triterpenoids as well as polar metabolites associated with cuticle and cell wall metabolism and protection against photooxidative stress. Combined results at both transcript and metabolite levels revealed that the formation of cuticular lipids precedes phenylpropanoid and flavonoid biosynthesis. Expression patterns of reporter genes driven by the upstream region of the wax-associated SlCER6 gene indicated progressive activity of this wax biosynthetic gene in both fruit exocarp and endocarp. Peel-associated genes identified in our study, together with comparative analysis of genes enriched in surface tissues of various other plant species, establish a springboard for future investigations of plant surface biology.
The anatomical structure of tomato (Solanum lycopersicum) fruit is composed of several different tissue types (Montgomery et al., 1993). It consists of a pericarp, formed from the ovary walls surrounding the placental tissue, the locular tissue, and the seeds. The pericarp is further subdivided into several cell types and layers. A single interior cell layer, termed the endocarp, limits the pericarp and is adjacent to the locular region, while the thick part of the pericarp, the mesocarp, encompasses layers of large, highly vacuolated parenchymatous cells and contains vascular bundles. The exocarp is composed of several layers of collenchymatous cells and a single layer of epidermal cells, which in turn is covered, and in some cases encased, in a waxy cuticle that thickens as the fruit ages (Lemaire-Chamley et al., 2005). The term “peel,” while not a botanical term, is also sometimes used to denote the outer layers; the peel is typically composed of multiple cell types, including epidermis, collenchyma, and even parenchyma, depending on how the peels are removed. Similarly, the term “flesh,” as used here, refers to the pericarp material from which the peel has been removed and that therefore is predominantly composed of parenchyma and collenchyma.
The cuticle plays a key role in the survival of plants, serving as the interface between plants and their biotic and abiotic environment. The primary physiological function of the plant cuticle is to seal the tissue against a relatively dry atmosphere, preventing desiccation by minimizing nonstomatal water loss (Kerstiens, 1996a; Riederer and Schreiber, 2001; Riederer and Burghardt, 2006). It has been shown that the transpiration barrier is largely provided by the cuticular waxes (Schonherr, 1976) and to a much lesser degree by cutin. It is yet unclear whether other cuticle components, such as terpenoids and flavonoids, may also contribute to the transpiration barrier. In addition, plant surfaces are of importance in ecological interactions and pest resistance, as they necessarily represent the first line of contact with other organisms and thus may have a crucial role in protection against herbivores (Eigenbrode and Espelie, 1995). Another major function of the cuticle is protection against mechanical damage (Kerstiens, 1996a, 1996b; Heredia, 2003), invoked from outside by biting insects or growing fungal hyphae as well as from inside by increasing turgor pressure during organ growth. It is generally accepted that the cutin matrix, a polymer formed by three-dimensional cross-links of covalent bonds, contributes to the mechanical strength of the cuticle (Kolattukudy, 1980). In a recent study, López-Casado et al. (2007) demonstrated that polysaccharides incorporated into the cutin matrix are responsible for the elastic modulus, stiffness, and linear elastic behavior of the whole cuticle, while the viscoelastic behavior of the cuticular membrane (low elastic modulus and high strain values) can be assigned to the cutin. Mechanical failure of fruit cuticles leads to substantial losses of production through fruit splitting or cuticle cracking. The fruit surface influences the outward appearance of the fruit (color, glossiness, texture, and uniformity), efficacy of postharvest treatments, storage, transport, and shelf life. Hence, the knowledge regarding fruit surface properties is fundamental for the improvement of fruit quality traits.
The metabolites composing the cuticle are synthesized by the epidermis layer(s) and secreted to the extracellular matrix of all primary aboveground plant organs (Samuels et al., 2005). Acyl lipids, isoprenoids, and phenylpropanoids are the three major groups of metabolites involved in the construction of the cuticle. The C16 and C18 fatty acyl-acyl carrier proteins (ACPs) formed by the fatty acid synthesis in the plastids are subsequently cleaved from ACP by thioesterases and exported to the endoplasmic reticulum as fatty acyl-CoAs (Schnurr et al., 2004). At this point, the pool of acyl-CoAs is split to various metabolic pathways, including the synthesis of membrane lipids, storage lipids (in seeds), cutin, and cuticular waxes.
A major component of the cuticle, the polyester cutin is insoluble in organic solvents and consists of oxygenated fatty acids with a chain length of 16 or 18 carbons (Kolattukudy, 2001; Nawrath, 2006). As mentioned above, the precursors for cutin synthesis are derived from plastidic de novo fatty acid synthesis, which generates palmitic (16:0), stearic (18:0), and oleic (18:1) acids attached to ACP. Cutin monomers are synthesized from fatty acyl-CoAs by multiple hydroxylation and epoxidation reactions (Kolattukudy, 1981) in which cytochrome P450-dependent enzymes are primary candidates to be involved. It was suggested that the biosynthesis of cutin monomers would also involve lipoxygenases, peroxygenases, and epoxidases (Blee and Schuber, 1993) and that the cutin polymer is formed by linking CoA-bound monomers to free hydroxyl groups in the polymer, a reaction catalyzed by acyl-CoA:cutin transferase (Reina and Heredia, 2001). Recently, Li et al. (2007) reported the identification of a pair of glycerol-3-P acyltransferases (GPAT4 and GPAT8) that are essential for cutin biosynthesis, and their overexpression in Arabidopsis (Arabidopsis thaliana) resulted in an 80% increase in C16 and C18 cutin monomer production.
Embedded in the cutin matrix, but also deposited on the outer surface of the cuticle (i.e. epicuticular), are the waxes, which are complex mixtures of very long chain fatty acid (VLCFA) derivatives. C16 and C18 acyl-CoAs are used to generate VLCFAs through sequential additions of two-carbon units in a reaction catalyzed by membrane-bound, multienzyme acyl elongase systems. The formed VLCFA CoA esters can either be hydrolyzed to free fatty acids, reduced to aldehydes that are further derivatized to alkanes, secondary alcohols, and ketones (in the decarbonylation pathway), or used for the generation of wax esters by condensing primary alcohols (derived from acyl-CoA precursors by reduction) with acyl-CoAs (in the acyl reduction pathway; Millar et al., 1999; Jetter et al., 2006; Rowland et al., 2006).
Cuticular waxes often contain nonacyl lipids, including, for example, pentacyclic triterpenoids derived from the cytosolic isoprenoid pathway (i.e. the mevalonate pathway). Triterpenoids have been identified in cuticles of many plant species and in several cases accumulate to high concentrations. Triterpenoid biosynthesis is independent of acyl lipid biosynthesis, starting with the transformation of acetyl-CoA into mevalonate and then into farnesyl pyrophosphate. With the ensuing formation of squalene and then of 2,3-oxidosqualene, catalyzed by squalene synthase and squalene epoxidase, respectively, the pathway diverts from other terpenoid pathways. Finally, the cyclization of oxidosqualene, catalyzed by oxidosqualene cyclases, is the branch point for the biosynthesis of phytosterols and triterpenoids (Xu et al., 2004).
In some cases, additional secondary metabolites, mostly phenolics such as flavonoids, also constitute plant cuticles. Flavonoids are synthesized by the phenylpropanoid pathway in which the amino acid Phe is used to produce 4-coumaroyl-CoA. This can be combined with malonyl-CoA to yield chalcones (i.e. flavonoid precursors with two phenyl rings). Conjugate ring closure of chalcones results in a three-ring structure, the typical form of flavonoids. The metabolic pathway continues through a series of enzymatic modifications to yield several flavonoid classes, including the flavonols, dihydroflavonols, and anthocyanins. Many other products can be formed along this pathway, including the flavan-3-ols, proanthocyanidins (tannins), and additional polyphenolics. Some other classes of chemicals, including sterols and alkaloids, were also identified as cuticular components in plants (Jetter et al., 2006).
The composition of tomato surface and intracuticular waxes is dominated by very long chain alkanes (predominantly n-hentriacontane, C31H64), fatty acids (C16, C18, and C24), C32 n-aldehyde, and triterpenoids (Baker, 1982; Bauer et al., 2004a; Vogg et al., 2004; Leide et al., 2007). The latter components were exclusively detected in the intracuticular wax, including high quantities of three major pentacyclic triterpenoids α-, β-, and δ-amyrin (Vogg et al., 2004). The main cutin monomers in tomato fruit cuticles include 16-hydroxyhexadecanoic acid, 10,16-dihydroxyhexadecanoic acid, and 18-hydroxyoctadecanoic acid.
As mentioned above, phenolics are also present in the tomato cuticle (Hunt and Baker, 1980). Various flavonoids accumulate in the peel during tomato fruit ripening (Verhoeyen et al., 2002). The main flavonoid in tomato peel is naringenin chalcone (a flavonone), accumulating to approximately 1% of the peel dry weight during the orange stage of fruit development (Muir et al., 2001). The flavonols quercetin-rutinoside (rutin) and kaempferol-rutinoside were found to accumulate to a lesser degree exclusively in ripening tomato peel (together approximately 0.1% dry weight of overripe peel tissue; Muir et al., 2001; Bovy et al., 2002; Schijlen et al., 2007). Other phenolics detected in the cuticle of tomato fruit peel were m- and p-coumaric acids, reaching maximum levels in early fruit development (the green stage; Hunt and Baker, 1980).
Tomato has long served as a model system for examining the fruit ripening process (Giovannoni et al., 1995; Giovannoni, 2007). In recent years, an array of tools were developed and used for the study of tomato fruit, allowing the examination of the cell at the transcript as well as at the protein and metabolite levels (Alba et al., 2004, 2005; Fei et al., 2004; Fernie et al., 2004; Rose et al., 2004; Moore et al., 2005). Vogg et al. (2004) studied the SlCER6 gene (previously referred to by Vogg et al. [2004] as LeCER6), a tomato homolog of the Arabidopsis CER6 involved in VLCFA elongation. It was shown that mutation of the SlCER6 gene leads to an alteration of the cuticular wax composition and water permeability. Lemaire-Chamley et al. (2005) used global analysis of gene expression to identify genes linked with the differentiation of specialized tissues in the early development of tomato fruit. Genes that were found to be preferentially expressed in the exocarp (defined as the epidermis and additional layers of outer pericarp cells) were categorized into two groups based on their putative biological functions. Among these were genes putatively involved in protection of the fruit against pathogens and stress tolerance, the formation of the cuticle (lipid transfer proteins and cell wall-related proteins), and enzymes required for flavonoid synthesis and ascorbic acid. A second category of genes associated with the exocarp at early fruit development were related to fruit growth and included proteins involved in polysaccharide synthesis, cell wall structure, cell adhesion, and cell wall relaxation. Recently, Saladie et al. (2007) described the Delayed Fruit Deterioration (DFD) cultivar, which exhibits reduced softening and normal ripening at the same time. While DFD did not show any clear cell wall-related phenotype compared with a normally softening cultivar, it showed minimal transpirational water loss, elevated cellular turgor, and altered chemical features of its cuticle. The authors suggested that the cuticle affects the softening of tomato fruit directly by providing a physical support and also indirectly by regulating the water status.
Despite the availability of molecular tools and the importance of the peel tissue, both in terms of fruit biology and in relation to fruit quality traits, only a limited number of studies have investigated the fruit peel biology at the molecular level to date. In this study, we performed extensive profiling and comparison of tomato fruit peel versus flesh tissues at both the metabolite and transcript levels. By doing so, we revealed unique sets of genes and metabolic pathways that are active in the peel at various stages of fruit development. A large portion of the peel-associated transcripts showed homology with genes involved in the assembly of the cuticle and with genes prevalent in epidermal layers of other plant species, including in vegetative organs (e.g. in Arabidopsis stems). Hence, our results lay the basis for a comparative analysis between reproductive and vegetative organs and for elucidating the molecular events implicated in cuticle formation and function.
RESULTS
Microscopic Examination of Tomato Fruit Surface
Tomato fruit development can be divided into four main phases: cell differentiation, cell division, cell expansion, and ripening (Gillaspy et al., 1993). In this study, we focused on profiling the latter two phases, starting from cell expansion to the late ripening phase. We selected five representative stages of tomato fruit development and ripening: immature green (IG), mature green (MG), breaker (Br), orange (Or), and red (Re). In the first phase of this study, we carried out structural characterization of the respective surface structures by the use of scanning electron microscopy. At the IG stage, the tomato fruit surface is covered by a relatively dense mixture of type VI and type I trichomes (Fig. 1, A–D). These develop on top of conical epidermal pavement cells (Fig. 1, E and F) resembling those typically formed on the surface of petals (e.g. in Antirrhinum majus; Noda et al., 1994). As the fruit enlarges and matures, trichomes could only be detected sporadically on the fruit surface, probably because the total number of trichomes is determined early on during fruit development, while the pavement cells in between them continue to expand. Young and mature fruit did not exhibit major differences in the shape of pavement cells. Compared with the cuticle of the model plant Arabidopsis, the cuticle of tomato fruit is a relatively thick structure encasing the epidermis cells. Occasionally, the cuticular layer surrounds more than a single cell layer below the outermost epidermis (Fig. 1, G and H).
Figure 1.
The surface and peel of tomato during consecutive stages of fruit development. A to H, Electron micrographs of Alisa Craig tomato fruit surfaces. At the IG developmental stage, the tomato fruit surface is covered by a relatively dense mixture of type VI and type I trichomes (A–D). Young (E) and mature (F) fruit do not exhibit major differences in the shape of their conical epidermal pavement cells. At the Re stage, the tomato fruit cuticle is relatively thick and surrounds the entire epidermal cells (G and H). I, Light microscopy images of fruit peel cross sections at the five developmental stages examined in the study revealed the presence of several collenchyma and parenchyma layers beneath the epidermis. Cuticle lipids were stained by Sudan IV. ep, Epidermis; tsc, trichome support cells; t-I, type I trichome; t-VI, type VI trichome.
Large-Scale Analyses of Transcripts and Metabolites in Tomato Fruit Peel
The main aim of this study was to conduct large-scale transcriptome and metabolome analyses of fleshy fruit outer tissues. Therefore, we manually dissected the fruit into peel and flesh tissues (without the seeds and gel) at all five selected fruit developmental stages (see above). Light microscopy study of peel sample sections revealed that these peel samples contained two to three collenchymatous cell layers as well as three to five layers of parenchyma cells beneath the epidermis (Fig. 1I). Overall, no significant difference was observed in cell layer proportion and composition between the peels isolated from fruit at different developmental stages. Thus, although they contain multiple cell types, the investigated peel samples were essentially enriched with epidermis and cuticular material. Transcriptome analysis was conducted using the Tomato Genome Array representing approximately 8,000 nonredundant tomato transcripts. A total of 30 arrays were used to monitor gene expression at five stages of fruit development in either peel or flesh tissue, with three biological replicates.
Metabolite analysis was conducted on the same set of samples as described above for transcriptome analysis. Three different analytical methods were employed in order to cover a wide range of compound classes present in tomato fruit peel. In the first method, ultra-performance liquid chromatography coupled to a quadrupole time-of-flight mass spectrometer (UPLC-QTOF-MS) was used to detect mainly semipolar components (in both electrospray ionization [ESI]-positive and ESI-negative mode). The high mass resolution and accuracy of the UPLC-QTOF-MS system and MS/MS analysis allow structural elucidation of unknown peaks, although in a large number of cases the identification might be ambiguous (e.g. in the case of isomers). In order to profile polar compounds, in particular primary metabolites, we used the previously established gas chromatography-mass spectrometry (GC-MS) analysis of derivatized fruit extracts (Fernie et al., 2004). In tomato fruit, this technology allowed us to monitor the levels of 56 metabolites, including amino acids, organic acids, sugar alcohols, tricarboxylic acid cycle intermediates, soluble sugars, sugar phosphates, and a few secondary metabolites. Profiling of wax, cutin, and triterpenoid levels in isolated fruit cuticles at different developmental stages was performed by a third analytical method using GC-FID (flame ionization detection) and GC-MS.
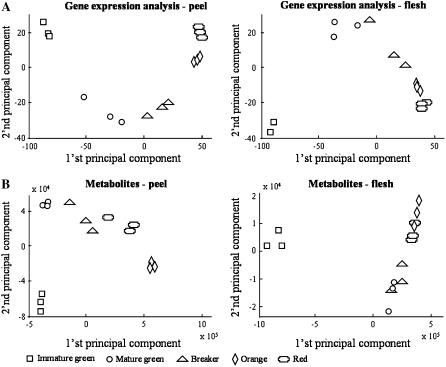
To obtain a broad view of the differences in the transcript and metabolite profiles of fruit peel and flesh tissues, we conducted principal component analysis on the data sets derived from the metabolite profiling using UPLC-QTOF-MS (operated in the ESI-negative mode) and gene expression analysis (Fig. 2). Gene and metabolite expression profiles could be clearly distinguished in either one of the tested fruit tissues. In terms of fruit developmental stages, gene expression and metabolite level profiles of IG fruit were largely dissimilar to those derived from fruit of other developmental stages, and the Or and Re stages appeared very close in both transcript and metabolite profiles.
Figure 2.
Gene expression and metabolic profiles in tomato peel and flesh tissues during fruit development. A, Principal component analysis of transcriptome assays carried out using the tomato GeneChip, with samples from peel (left) and flesh (right) tissues. B, Principal component analysis of metabolic profiles obtained by UPLC-QTOF-MS analysis, with samples from peel (left) and flesh (right) tissues. Transcriptome and metabolome analyses were carried out on the same set of samples (n = 3 for every sample type [stage × tissue]).
The Tomato Fruit Peel Transcriptome
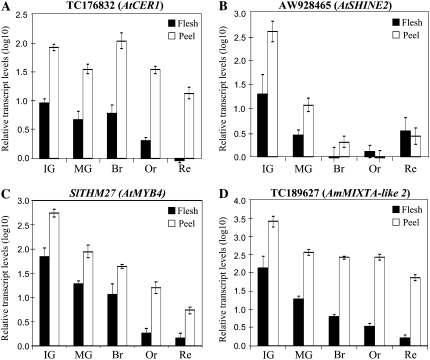
To generate a detailed picture of the tomato fruit peel transcriptome, we discerned transcripts that were highly expressed in the peel from those abundant in the flesh tissue. Following replicate reproducibility and variance filtering procedures (see “Materials and Methods”), 4,582 gene probes were retained for further analysis. After applying a 2-fold threshold for differential expression between samples, 574 nonredundant transcripts showed a 2-fold or higher increased expression in the peel versus the flesh in at least one stage of fruit development, and the up-regulation of 284 of these was found to be significant (P < 0.05 compared with its corresponding sample in the flesh). Of these, 184 were up-regulated in the peel in a single stage of fruit development, 41 in two stages, 32 in three stages, 14 in four stages, and 13 were up-regulated in the peel at all five tested stages of fruit development (a full list of the peel up-regulated genes is presented in Supplemental Table S1). Relative expression of 19 of these transcripts examined by means of quantitative real-time PCR (RT-PCR) analyses confirmed the gene expression results obtained by array analysis. Six of the 19 (putative tomato CER1, SHINE2 [SHN2], MIXTA-LIKE2, and CHS1, as well as SlTHM27 and SlCER6) are presented in Figure 3 and in Figure 6 below.
Figure 3.
RT-PCR expression analyses of selected transcripts confirmed their up-regulation in tomato fruit peel as detected by the microarray analysis. A, Tomato TC176832, a putative AtCER1 ortholog. B, Tomato AW928465, a putative AtSHN2 ortholog. C, SlTHM27 (TC174616), a putative AtMYB4 ortholog. D, Tomato TC189627, a putative snapdragon (A. majus) MIXTA-LIKE2 ortholog. Besides SlSHN2 expression at the Br, Or, and Re stages, significantly higher expression levels were detected in the peel for all tested transcripts at all other stages (Student's t test; n = 3; P < 0.05).
Figure 6.
Expression of SlCER6 and SlCHS1 as detected by a reporter gene assay and RT-PCR. A, Reporter GUS expression driven by the upstream region of peel-associated SlCER6 leads to staining of the exocarp (Ex) and endocarp (En) tissues in fruit slice at the Or stage. B, Reporter GUS exocarp expression driven by the upstream region of the peel-associated SlCHS1 gene in fruit slice at the Br stage. A light microscopy section image, magnifying the region of the stained exocarp, reveals discernible GUS reporter staining in the epidermis cells and one or two additional cell layers beneath them (image in the open rectangle). C, RT-PCR expression analysis of tomato SlCER6 (TC172551), a putative AtCER6 (CUT1) ortholog, and TC170658, a putative AtCHS1 ortholog, during the five tested fruit developmental stages. Significantly higher expression levels of both transcripts were detected in the peel compared with the flesh at all tested developmental stages (Student's t test; n = 3; P < 0.05).
Genes Associated with the Biosynthesis of Cuticular Components Are Predominantly Expressed in the Peel Tissue
To examine the relative abundance of transcripts preferentially expressed in the peel tissue and putatively involved in plant surface formation, we sorted the peel enriched transcripts according to their corresponding putative functional categories (Fig. 4). The results indicated high levels of peel-associated transcripts in pathways leading to the biosynthesis of cuticle constituents. Genes associated with one of the three functional categories, namely, wax/cutin, phenylpropanoid/flavonoid, and fatty acid metabolism, represented 3%, 6%, and 8%, respectively, of the 574 nonredundant transcripts that showed a 2-fold or higher peel increased expression in at least one stage of fruit development (Fig. 4).
Figure 4.
Functional categories representation of the peel-associated transcripts detected by the array analysis. The 574 transcripts that showed a 2-fold or higher expression in the peel versus flesh (in at least one tested stage of tomato fruit development) are represented. In most cases, the putative functional categories were assigned according to sequence homologies with previously studied genes from other species. The distribution of categories is given in percentage of the total 574 transcripts.
Genes associated with cutin, wax, and fatty acid metabolism represented 15% of the total 284 transcripts that were significantly peel up-regulated (P < 0.05 compared with its corresponding sample in the flesh) in at least one out of the five tested fruit developmental stages (Table I). The putative functions of most of these gene products were previously determined or suggested in studies of Arabidopsis cuticular mutants. For example, HOTHEAD (HTH), LONG CHAIN ACYL-COA SYNTHASE (LACS), GDSL-MOTIF LIPASE/HYDROLASE, and EXTRACELLULAR LIPASE (EXL) genes were predicted to encode proteins involved in cutin biosynthesis (Schnurr et al., 2004; Kurdyukov et al., 2006; Kannangara et al., 2007).
Table I.
Selected peel up-regulated (2-fold or higher; P < 0.05) transcripts identified in this study as putatively associated with cuticle metabolism in tomato fruit peel
| Identifier (TCa) | Annotation | Clusterb | |
|---|---|---|---|
| Phenylpropanoid/flavonoid metabolism | |||
| TC174247 | Cinnamyl alcohol dehydrogenase (CAD) | 6 | |
| TC174616 | MYB transcription factor (THM27) | 6 | |
| TC172671 | Acyltransferase | 6 | |
| BT014421 | Isoflavone reductase-like | 9 | |
| BI929841 | UDP-Xyl phenolic glycosyltransferase | 13 | |
| TC170008 | Phospho-2-dehydro-3-deoxyheptonate aldolase (DAHP synthase) | 14 | |
| TC172766 | Prephenate dehydratase (PDH) | 14 | |
| TC170658 | Putative chalcone synthase 1B | 16 | |
| TC172191 | Chalcone synthase 2 (CHS2) | 16 | |
| TC171118 | 5-Enolpyruvylshikimate-3-P synthase (EPSPS) | 16 | |
| BF096444 | Chorismate mutase 1 (CM1) | 16 | |
| CK715539 | Chorismate mutase 1 (CM1) | 16 | |
| TC172772 | Phe ammonia lyase (PAL) | 16 | |
| BG631837 | 4-Coumarate-CoA ligase 2 (4CL2) | 16 | |
| TC178705 | Putative chalcone isomerase 4 | 16 | |
| TC182590 | Putative chalcone-flavanone isomerase | 16 | |
| BT014352 | Flavanone 3-hydroxylase (F3H) | 16 | |
| TC176277 | Flavonoid 3-glucosyl transferase (3GT) | 16 | |
| TC176549 | Glycosyl transferase (GT) | 16 | |
| TC172800 | Flavonol synthase/flavanone 3-hydroxylase (FLS) | 16 | |
| TC183733 | 4-Coumarate 3-hydroxylase (C3H) | 16 | |
| BT014024 | Anthocyanidin-3-glucoside rhamnosyltransferase | 16 | |
| TC180112 | Cinnamoyl-CoA reductase (CCR) | 16 | |
| TC181932 | Isoflavone reductase-like | 16 | |
| CK715844 | Cinnamoyl-CoA reductase (CCR) | 16 | |
| Wax/cutin pathway | |||
| AW928465 | AP2 transcription factor (SHN2/WIN1-like) | 5 | |
| TC190363 | Putative HTH | 5 | |
| TC174873 | Putative HTH | 5 | |
| TC175044 | Putative HTH | 5 | |
| TC173809 | Enoyl-CoA reductase (AtCER10) | 6 | |
| BT013894 | Acyl-CoA synthetase (ACS) | 13 | |
| TC187142 | Long chain acyl-CoA synthase (LACS) | 16 | |
| TC173257 | 3-Ketoacyl-CoA synthase (SlCER6/AtKCS6/AtCUT1) | 16 | |
| TC172551 | 3-Ketoacyl-CoA synthase (SlCER6/AtKCS6/AtCUT1) | 16 | |
| TC171885 | Fatty acid elongase-like protein (CER2-like) | 16 | |
| TC182022 | CER1-like | 16 | |
| BG130630 | Fiddlehead (FDH) | 16 | |
| TC182303 | Glycerol-3-P dehydrogenase | 6 | |
| Fatty acid metabolism (possibly wax/cutin)c | |||
| TC175280 | Acyl-CoA thioesterase | 2 | |
| TC169929 | ω-3 fatty acid desaturase | 3 | |
| TC172718 | Putative pyruvate dehydrogenase E1 β-subunit | 4 | |
| BG631546 | Extracellular lipase 1 (EXL1) | 5 | |
| TC170157 | Esterase | 5 | |
| TC185612 | Esterase | 6 | |
| TC170192 | Lipid transfer protein | 6 | |
| BT014187 | Lipid transfer protein | 6 | |
| TC176438 | Cytochrome b5 | 6 | |
| BG735394 | Stearoyl-ACP desaturase | 9 | |
| TC170046 | Lipoxygenase | 9 | |
| TC180384 | Esterase/lipase/thioesterase | 13 | |
| TC188586 | Acyl-ACP thioesterase | 13 | |
| AW039066 | Extracellular lipase (EXL1) | 13 | |
| BT012874 | GDSL motif lipase | 13 | |
| TC172499 | GDSL motif lipase | 14 | |
| TC179926 | Acyl-CoA oxidase | 14 | |
| TC172218 | Nonspecific lipid transfer protein | 14 | |
| TC179774 | Lipid transfer protein | 16 | |
| BG628343 | GDSL motif lipase | 16 | |
| BI209975 | Lipase | 16 | |
| TC174706 | 3-Hydroxyisobutyryl-CoA hydrolase | 16 | |
| TC174104 | GDSL motif lipase | 16 | |
| Others | |||
| TC173181 | Strictosidine synthase | Alkaloid metabolism | 5 |
| TC188260 | Tropinone reductase | Alkaloid metabolism | 13 |
| TC169841 | β-Galactosidase | Cell wall | 9 |
| TC189627 | MYB transcription factor (MIXTA-like) | TF activity | 2 |
| TC173525 | MADS box transcription factor (RIN) | TF activity | 22 |
GenBank accession numbers are given when no TC index (The Institute for Genomic Research identifier) is available.
Cluster of expression profile to which the gene belongs (see Supplemental Fig. S1 for clusters).
Transcripts included in the fatty acid metabolism category are homologs/orthologs of genes that were suggested/predicted but not shown to be involved in wax/cutin metabolism.
Apart from cutin metabolism, the biosynthetic pathways for the formation of VLCFAs and their conversion to aliphatic wax constituents were also represented by transcripts preferentially expressed in the tomato peel tissue (Table I). These included genes putatively associated with the metabolism of fatty acids in the plastids prior to their export to the endoplasmic reticulum (i.e. ACYL-ACP THIOESTERASE) and their esterification to CoASH during transport through the plastid envelope (i.e. LACS; Table I). The next step in wax metabolism is the elongation of fatty acids (produced in the plastids) up to C34 VLCFAs. A set of genes putatively belonging to the elongase complex components and to those performing the committed steps in the biosynthesis of wax components with different chain lengths were also found to be peel associated (Table I). These included a putative ENOYL-COA REDUCTASE, 3-KETOACYL-COA SYNTHASE (KCS6; also known as CUT1 or CER6), and FIDDLEHEAD (FDH). Additional peel-associated genes that have been predicted to be involved in cuticle development include various LIPID TRANSFER PROTEINs (LTPs), ESTERASEs, LIPASEs, CER1-LIKE, and CER2-LIKE. Interestingly, we also identified a peel up-regulated transcript that is a putative ortholog of the Arabidopsis SHN2 transcription factor (SHN2/WIN1-LIKE; Table I), a regulator of genes involved in cutin and wax metabolism (Aharoni et al., 2004; Broun et al., 2004; Kannangara et al., 2007). Expression of the tomato SHN2-LIKE was up-regulated in the peel in at least the first two of the five tested developmental stages (see Fig. 3B for RT-PCR results).
Transcripts putatively corresponding to the flavonoid pathway also showed preferential expression in the peel tissue (Table I), including structural genes in the pathway such as CHALCONE SYNTHASE1 (CHS1) and CHS2, CHALCONE ISOMERASE (CHI), FLAVANONE 3-HYDROXYLASE (F3H), FLAVONOL SYNTHASE (FLS), or genes encoding proteins that modify the flavonoid aglycone (ACYLTRANSFERASEs [ACTs], RHAMNOSYLTRANSFERASEs [RTs], and GLYCOSYLTRANSFERASEs [GTs]). Genes putatively associated with the general phenylpropanoid pathway and its lignin biosynthesis branch (4-COUMARATE:COA LIGASE [4CL], PHENYLALANINE AMMONIA LYASE [PAL], CINNAMYL ALCOHOL DEHYDROGENASE [CAD], 4-COUMARATE 3-HYDROXYLASE [C3H], and CINNAMOYL-COA REDUCTASE [CCR]) were also up-regulated in the peel. The shikimate pathway and its downstream branches are the major precursor-supplying routes for the formation of aromatic amino acids, including Phe. Elevated activity of the phenylpropanoid/flavonoid pathway in the peel tissue requires induction of precursor supply from the shikimate pathway, as observed by the peel up-regulation of putative PHOSPHPO-2-DEHYDRO-3-DEOXYHEPTANOATE ALDOLASE (DAHP SYNTHASE), 5-ENOLPYRUVYLSHIKIMATE-3-PHOSPHATE SYNTHASE (EPSPS), and CHORISMATE MUTASE1 (CM1) genes. With respect to transcriptional regulation, branches of the phenylpropanoid pathway are known to be controlled by transcription factors of the R2-R3 MYB family. The transcript of the tomato THM27, an R2-R3 MYB transcription factor (Lin et al., 1996), was found to be strongly up-regulated in the peel at four of the five tested stages of fruit development (see Fig. 3C for RT-PCR results). This tomato gene shows high homology with AtMYB4 and AmMYB308, which were previously suggested to act as regulators of phenolic acid metabolism in the phenylpropanoid pathway (Tamagnone et al., 1998; Jin et al., 2000).
Clusters of Genes Showing Coordinated Expression in the Fruit Peel Tissue
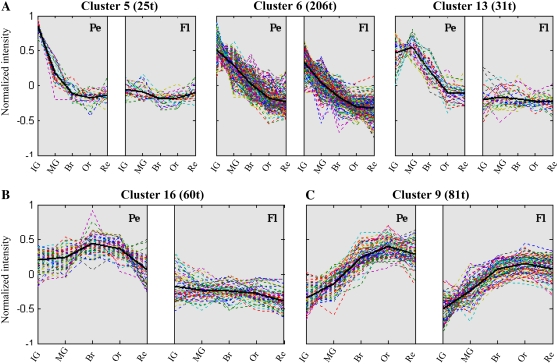
In order to study the expression patterns of genes that are up-regulated in the peel during fruit development, we applied hierarchical cluster analysis to all transcripts having at least one developmental stage with a mean peel-flesh ratio value of 2-fold or more. Thirty clusters containing 574 nonredundant transcripts were created (Supplemental Fig. S1; Supplemental Table S1). Figure 5 displays five selected gene clusters and their expression profiles in the peel and flesh tissues. Clusters 5, 6, and 13 (Fig. 5A) represent transcripts with higher peel expression at early stages of fruit development. While in clusters 5 and 6, gene expression in the peel is down-regulated soon after the IG fruit stage up to the Re stage, in cluster 13 the down-regulation commences later at the MG stage (Fig. 5A). In the flesh samples, genes belonging to cluster 5 show a more constant level of expression during development compared with those in cluster 6, which show a down-regulated expression pattern. Cluster 16 (Fig. 5B) represents transcripts with increased peel expression at the Br stage (middle phase) and declining expression at later stages, and cluster 9 (Fig. 5C) represents transcripts with steadily increasing expression toward late stages of fruit development in both peel and flesh tissues.
Figure 5.
Five selected clusters of gene expression profiles in tomato fruit peel (Pe) and flesh (Fl) tissues along the five tested developmental stages. A, Clusters 5, 6, and 13 represent genes with higher expression at early stages of fruit peel development. B, Cluster 16 represents genes with increased expression at the Br stage of peel development and a lower expression at later stages. C, Cluster 9 includes genes with steadily increasing expression up to the Or and Re developmental stages in both peel and flesh tissues. In parentheses next to each cluster name is the number of transcripts (t) that constitute it. Black lines represent the cluster average profile. The clustering method is described in “Materials and Methods.” [See online article for color version of this figure.]
Cluster 5 (Fig. 5A), including 25 transcripts, is composed of transcripts putatively encoding cell wall-related enzymes (expansin and xyloglucan galactosyltransferase) as well as those associated with cutin metabolism, such as HTH-LIKE, EXL1, and the SHN2 transcription factor ortholog. Cluster 6 (Fig. 5A) includes the highest number of transcripts (206), 32% of them with unknown function. According to the functional categories division, the expression pattern revealed in this cluster is related to transcripts putatively associated with carbohydrate metabolism, cell wall metabolism, and primary metabolism. Furthermore, several transcripts that encode proteins associated with the metabolism of wax/cutin components (both de novo fatty acid biosynthesis and elongation of fatty acids) also take part in this cluster. These include β-ketoacyl-ACP synthase, 3-ketoacyl-CoA thiolase, ATP citrate synthase, enoyl-ACP reductase, β-ketoacyl-CoA synthase, enoyl-CoA reductase, and glycerol-3-P dehydrogenase. The SlTHM27 gene described above as a potential regulator of the phenylpropanoid pathway or one of its branches belongs to cluster 6. The only four peel-associated transcripts putatively related to the cell cycle category are also members of this cluster 6. In addition, transcripts putatively associated with the metabolism of hormones, such as brassinosteroids, cytokinins, and jasmonic acid, showed a similar early expression in the peel during fruit development. A third cluster of this high early expression group is cluster 13 (comprising 31 transcripts), which represents transcripts encoding putative cell wall metabolism enzymes (cellulose synthase and polygalacturonase) and several different types of lipases that might be associated with wax and/or cutin metabolism.
Cluster 16 (Fig. 5B) represents 60 transcripts, 40% of which are putatively related to the biosynthesis of cuticular components (wax, fatty acids, and flavonoid metabolism). Coregulated expression of transcripts putatively belonging to the general phenylpropanoid and flavonoid pathways (PAL, C3H, 4CL, CHS, CHI, FLS, and F3H) was evident in this cluster. The putative CM1 and EPSPS transcripts, both part of the shikimate pathway that generates precursors for phenylpropanoid metabolism, share the same expression pattern. Another set of transcripts putatively associated with fatty acid elongation and wax metabolism (KCS, CER2-LIKE, FDH, LACS, CUT1/CER6/KCS6, and CER1-LIKE genes) is also part of this cluster.
Cluster 9 (Fig. 5C), the fifth selected cluster, includes 81 transcripts, 26 of which could not be assigned to a putative function category. Although this is a relatively large cluster (among the 30 clusters), it does not contain any wax/cutin-related genes, and only two genes that might encode phenylpropanoid/flavonoid-related proteins (MALONYL TRANSFERASE and ISOFLAVONE REDUCTASE-LIKE) are members of this cluster. Transcripts putatively associated with carbohydrate transport and cell wall metabolism (endo-β-mannanase, polygalacturonases, and β-galactosidase) are particularly coexpressed in this “late expression” cluster.
Tomato Genes with Up-Regulated Expression in the Peel Are Also Preferentially Expressed in Tomato Stem Epidermis and in Epidermis-Enriched Tissues of Other Species
An additional array hybridization experiment was conducted in order to identify transcripts that are enriched in tomato stem surface tissue. Three biological repeats of manually dissected stem peel isolated from tomato seedlings at the four-true-leaves developmental stage were used for this analysis. Light microscopy investigation of the isolated stem surface tissue showed that in addition to the epidermis layer, it included several layers of elongated collenchyma cells underneath (Supplemental Fig. S2). Following filtering (see “Materials and Methods”) and the application of a 2-fold threshold for differential expression between samples, 140 nonredundant transcripts were found to be up-regulated in tomato stem epidermis versus the whole stem (Supplemental Table S3). Forty percent (55 of 140) of these were also up-regulated in tomato fruit peel (Supplemental Table S1). Twenty-seven of these 55 transcripts were significantly enriched in both tissues (Table II).
Table II.
List of genes preferentially expressed in both tomato fruit and stem epidermal tissues, classified by putative functional categories
| Identifier | Annotationa | Pathway |
|---|---|---|
| TC172800 | Flavonol synthase/flavanone 3-hydroxylase (FLS) | Phenylpropanoid/flavonoid metabolism |
| TC176277 | Flavonoid 3-glucosyl transferase (3GT) | Phenylpropanoid/flavonoid metabolism |
| BT014024 | Anthocyanidin-3-glucoside rhamnosyltransferase | Phenylpropanoid/flavonoid metabolism |
| BG631837 | 4-Coumarate-CoA ligase 2 (4CL2) | Phenylpropanoid/flavonoid metabolism |
| TC178705 | Putative chalcone isomerase 4 | Phenylpropanoid/flavonoid metabolism |
| TC182590 | Putative chalcone-flavanone isomerase | Phenylpropanoid/flavonoid metabolism |
| TC170658 | Putative chalcone synthase 1B | Phenylpropanoid/flavonoid metabolism |
| TC174873 | Putative HTH | Wax/cutin |
| TC173809 | Enoyl-CoA reductase (AtCER10) | Wax/cutin |
| TC172551 | 3-Ketoacyl-CoA synthase (SlCER6/AtKCS6/AtCUT1) | Wax/cutin |
| TC182303 | Glycerol-3-P dehydrogenase | Wax/cutin |
| BT013894 | Acyl-CoA synthetase (ACS) | Fatty acid metabolism (putative wax/cutin)b |
| BG628343 | GDSL motif lipase | Fatty acid metabolism (putative wax/cutin)b |
| TC170046 | Lipoxygenase | Fatty acid metabolism (putative wax/cutin)b |
| BT012874 | GSDL motif lipase | Fatty acid metabolism (putative wax/cutin)b |
| TC179801 | MATE efflux family protein | Transporter activity |
| TC174622 | α/β-fold family protein | Aromatic compound metabolism |
| TC170338 | Pathogenesis-related protein | Defense response |
| TC177645 | Responsive to desiccation (RD22) | Response to stress/defense |
| TC169946 | α-Galactosidase (SlαGal) | Cell wall |
| TC176590 | Putative 1-deoxy-d-xylulose-5-P synthase 2 | Isoprenoid pathway |
| TC170338 | Pathogenesis-related protein | Response to stress/defense |
| BT013457 | Putative squamosa promoter-binding protein | Transcription factor activity |
| TC175108 | Unknown | – |
| TC172043 | Unknown | – |
| BG630651 | Unknown | – |
| TC175399 | Cytochrome P450 family protein | – |
In most cases, annotation is based on the studied closest homolog/ortholog from other species.
Transcripts included in the fatty acid metabolism category are homologs/orthologs of genes that were suggested/predicted but not shown to be involved in wax/cutin metabolism.
In order to identify common epidermis- and possibly cuticle-related genes, comparisons of the tomato fruit- and stem peel-enriched transcripts were performed against sets of genes that are known to be prevalent in epidermis-related tissues of other plant species, including Arabidopsis stem epidermis (Suh et al., 2005), maize (Zea mays) epidermis (Nakazono et al., 2003), and cotton (Gossypium hirsutum) fibers (Arpat et al., 2004; Samuel Yang et al., 2006), as well as an in silico-generated set of apple (Malus domestica) fruit peel-enriched transcripts. The epidermis-prevalent gene sets were compared by TBLASTX against all tomato consensus contigs present in the J. Craig Venter Institute (JCVI) database (http://www.tigr.org), and their best hits were further analyzed in order to identify homologs/orthologs of tomato peel-enriched genes. Seventy-three (of more than 3,000) genes, whose expression was shown to be increased in Arabidopsis stem epidermis (Suh et al., 2005), had best hits that were also up-regulated in tomato epidermal tissues. Thirty-one of 130 maize epidermis-prevalent genes, and 55 of 925 transcripts enriched in cotton fibers from three different developmental stages (differentiation, expansion/primary cell wall synthesis, and secondary cell wall synthesis), had best tomato hits that were also enriched in tomato epidermal tissues. In silico northern analysis (i.e. electronic northern) performed on apple EST libraries from nine different tissues revealed that homologs/orthologs of 78 tomato epidermis up-regulated genes are expressed in apple fruit peel, 14 of which also seemed to be enriched in this tissue. Overall, 127 of the tomato peel-associated transcripts had orthologs/homologs with up-regulated expression in epidermis-enriched tissues of other species (Supplemental Table S3). Twenty-three of these were up-regulated in three or more epidermis-related tissues (Table III). Genes putatively encoding phenylpropanoid/flavonoid or wax/cutin and fatty acid metabolism dominate the gene groups presented in both Table II and Table III (55% and 47%, respectively).
Table III.
Genes enriched in three or more epidermis-related tissues from several species, classified by putative functional categories
| Tomato | Arabidopsis | Cotton Fibers | Maize | Applea | Annotation | Pathway |
|---|---|---|---|---|---|---|
| TC182590bc | TC32267 | TC316570 | Putative chalcone-flavanone isomerase | Phenylpropanoid/flavonoid metabolism | ||
| TC183733b | TC59138 | TC326272 | 4-Coumarate 3-hydroxylase (C3H) | Phenylpropanoid/flavonoid metabolism | ||
| TC170658bc | TC66153 | Putative chalcone synthase 1B | Phenylpropanoid/flavonoid metabolism | |||
| TC178705bc | TC316570 | Putative chalcone isomerase 4 | Phenylpropanoid/flavonoid metabolism | |||
| TC174873bc | AT1G12570 | TC66521 | Putative HTH | Wax/cutin | ||
| TC173809bc | AT3G55360 | TC66162 | Enoyl-CoA reductase (AtCER10) | Wax/cutin | ||
| TC172551bc | TC27375 | 3-Ketoacyl-CoA synthase (SlCER6/AtKCS6/AtCUT1) | Wax/cutin | |||
| TC172218b | TC27353 | TC351531 | Nonspecific lipid transfer protein | Fatty acid metabolism (putative wax/cutin) | ||
| TC172718b | TC320985 | Enriched | Putative pyruvate dehydrogenase E1 β-subunit | Fatty acid metabolism (putative wax/cutin) | ||
| TC181822c | TC58921 | TC321032 | Putative nonspecific lipid transfer protein | Fatty acid metabolism (putative wax/cutin) | ||
| TC173004c | TC58974 | TC351531 | Nonspecific lipid transfer protein 2 precursor | Fatty acid metabolism (putative wax/cutin) | ||
| TC177222c | AT3G04290 | TC66645 | TC328310 | GDSL motif lipase/hydrolase family protein | Fatty acid metabolism (putative wax/cutin) | |
| TC190023b | AT4G31290 | Enriched | ChaC-like family protein-like | Transporter activity | ||
| TC179801bc | TC67183 | TC324316 | MATE efflux family protein | Transporter activity | ||
| TC190585c | AT1G52830 | TC317300 | Putative auxin-regulated protein | Transcription factor activity | ||
| TC172136c | AT2G06850 | TC59253 | TC325852 | Xyloglucan endotransglycosylase | Cell wall | |
| TC171696c | AT4G26010 | TC59029 | Peroxidase (TPX2) | Phe metabolism | ||
| TC176590bc | Enriched | Putative 1-deoxy-d-xylulose-5-P synthase 2 | Isoprenoid pathway | |||
| TC170313b | TC75875 | TC343146 | Osmotin-like protein | Defense response | ||
| TC172010b | TC73353 | TC331315 | Putative 2-oxoglutarate- dependent dioxygenase | – | ||
| TC174737b | AT2G37540 | TC73501 | Short chain dehydrogenase | – | ||
| TC175399bc | TC59138 | TC326272 | Cytochrome P450 family protein | – | ||
| TC171200c | TC59601 | TC316573 | Putative mannitol dehydrogenase | – |
In silico northern analysis of apple EST libraries from various tissues indicated these transcripts to be enriched in apple fruit peel.
Tomato epidermis-related genes enriched in fruit peel.
Tomato epidermis-related genes enriched in stem epidermis.
Epidermal GUS Expression Driven by Upstream Genomic Regions of Peel-Associated Genes
In order to examine the putative capacity of peel-enriched gene upstream regions to derive epidermal expression, we isolated the upstream genomic fragments of the flavonoid biosynthetic gene SlCHS1 (pSlCHS1; 1,050-bp region) and the wax metabolism gene SlCER6 (pSlCER6; 2,000-bp region). Constructs including these upstream fragments fused to a GUS reporter were transformed into tomato plants (Micro-Tom). In both cases, GUS expression was detected in the peel (exodermis) tissue (Fig. 6, A and B). In fruit of pSlCER6-expressing plants, GUS staining was also observed in the internal cell layer that borders the pericarp and the gel-containing locules (i.e. the endocarp). In the case of pSlCHS1, intense GUS staining was observed in the fruit epidermis (Fig. 6B) of six unrelated examined transgenic plants, albeit in two of these, faint GUS staining was also detected in the vasculature embedded in the pericarp tissue. Light microscopy study revealed discernible GUS staining in the epidermal cells and one or two additional cell layers below. RT-PCR expression analyses corroborated the reporter assays of these two genes, revealing higher expression levels in the peel tissue compared with their expression in flesh (Fig. 6C).
The Tomato Peel Metabolome
As described above, the information obtained from gene expression analysis of tomato peel during fruit development was complemented by metabolic profiling of the same sample set. Several analytical techniques were utilized for the detection of a maximal number of tomato peel metabolites.
UPLC-QTOF-MS Analysis
Nontargeted metabolite analysis performed by UPLC-QTOF-MS resulted in the detection of 7,197 and 3,786 mass signals in positive and negative ionization modes, respectively. To estimate the number of metabolites up-regulated in the peel in at least one of the tested developmental stages, statistical filtering was applied to the mass signals data. Totals of 5,560 and 2,907 mass signals in the positive and negative ionization modes, respectively, were at least 2-fold more abundant in the peel versus the flesh (Supplemental Table S5). This set of differential mass signals was analyzed in order to cluster together masses belonging to the same metabolite (see “Materials and Methods” for details). After clustering of differential masses (combining both ionization modes), 1,551 groups were formed, 740 of which were singletons. The number of groups formed by our clustering analysis (i.e. 1,551) was estimated to represent the amount of total peel up-regulated metabolites.
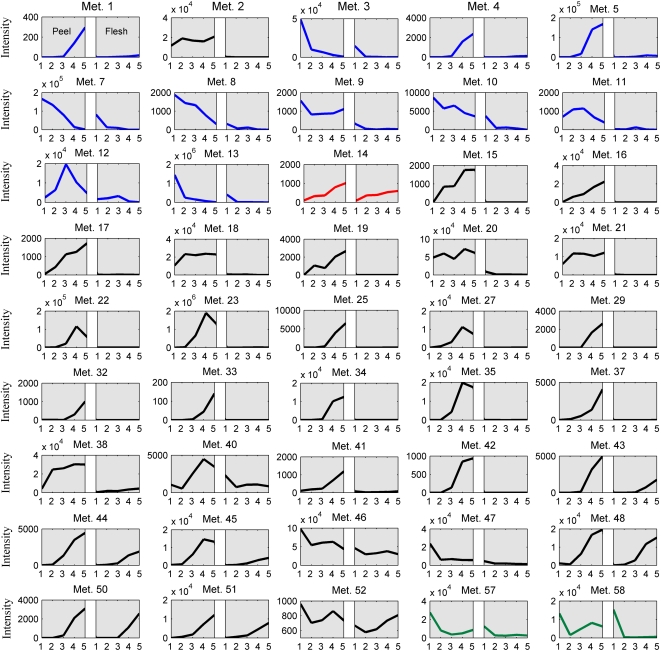
We were able to assign 58 putative tomato peel metabolites based on accurate mass measurements, publicly available information, and MS/MS analyses (see “Materials and Methods”; Table IV; Supplemental Table S6). Following a two-way ANOVA test, 45 of these metabolites were found to be significantly up-regulated in the peel compared with the flesh tissue (Table IV). Most metabolites detected in the peel by the UPLC-QTOF-MS technology were either phenylpropanoids or alkaloids (Fig. 7). Thirty metabolites derived from various branches of the phenylpropanoid pathway were detected, including derivatives either of flavonols (quercetin and kaempferol), naringenin, and naringenin chalcone or of phenolic acids (coumaric acid, ferulic acid, quinic acid, benzoic acid, and caffeic acid). In most cases, the levels of these metabolites increased in the peel during fruit development (Fig. 7).
Table IV.
Putative metabolites identified by UPLC-QTOF-MS in tomato fruit peel and flesh tissues
AAA, Aromatic amino acid; FA, formic acid; Ion. Mode, ionization mode; Neg, negative; Phe/flav, phenylpropanoid/flavonoid; Poly. d., polyamine derivative; Pos, positive.
| Peak No. | Putative Metabolite | Group | Molecular Formula | Retention Time | Molecular Weight (Theoretical Mass) | Mol. Weight (m/z Found) | Ion. Mode |
|---|---|---|---|---|---|---|---|
| min | |||||||
| 1a | Esculeoside B | Alkaloids | C56H93NO28 | 9.62 | 1228.5957 | 1228.5984 | Pos |
| 2a | Kaempferol-Glc-Rha | Phe/flav | C27H30O15 | 11.74 | 595.1658 | 595.1650 | Pos |
| 3a | Dehydrotomatine | Alkaloids | C50H81NO21 | 17.58 | 1032.5374 | 1032.5363 | Pos |
| 4a | Dehydrolycoperoside G, dehydrolycoperoside F, or dehydroesculeoside A | Alkaloids | C58H93NO29 | 13.09 | 1268.5906 | 1268.5931 | Pos |
| 5a | Lycoperoside G, lycoperoside F, or esculeoside A I | Alkaloids | C58H95NO29 | 13.50 | 1270.6063 | 1270.6043 | Pos |
| 6 | Lycoperoside G, lycoperoside F, or esculeoside A II | Alkaloids | C58H95NO29 | 11.04 | 1270.6063 | 1270.6052 | Pos |
| 7a | Lycoperoside A, lycoperoside B, or lycoperoside C I | Alkaloids | C52H85NO23 | 17.94 | 1092.5585 | 1092.5579 | Pos |
| 8a | (Lycoperoside A) FA, (lycoperoside B) FA, or (lycoperoside C) FA II | Alkaloids | (C52H85NO23) HCOOH | 18.48 | 1136.5494 | 1136.5505 | Neg |
| 9a | (Lycoperoside H) FA or (hydroxy-α-tomatine) FA I | Alkaloids | (C50H83NO22) HCOOH | 12.60 | 1094.5389 | 1094.5417 | Neg |
| 10a | Lycoperoside H or hydroxy-α-tomatine II | Alkaloids | C50H83NO22 | 13.71 | 1050.5480 | 1050.5497 | Pos |
| 11a | (Lycoperoside H) FA or (hydroxy-α-tomatine) FA III | Alkaloids | (C50H83NO22) HCOOH | 14.15 | 1094.5389 | 1094.5404 | Neg |
| 12a | Hydroxy-lycoperoside A, hydroxy-lycoperoside B, or hydroxy-lycoperoside C | Alkaloids | C52H85NO24 | 15.08 | 1108.5535 | 1108.5528 | Pos |
| 13a | α-Tomatine | Alkaloids | C50H83NO21 | 18.22 | 1034.5531 | 1034.5519 | Pos |
| 14a | Pantothenic acid hexose | Val metabolite | C15H27NO10 | 3.14 | 380.1562 | 380.1572 | Neg |
| 15a | Quercetin dihexose pentose deoxyhexose | Phe/flav | C38H48O25 | 5.11 | 905.2558 | 905.2582 | Pos |
| 16a | Quercetin dihexose deoxyhexose | Phe/flav | C33H40O21 | 5.70 | 773.2135 | 773.2138 | Pos |
| 17a | Quercetin dihexose deoxyhexose p-coumaric acid | Phe/flav | C42H46O23 | 13.92 | 919.2503 | 919.2526 | Pos |
| 18a | Quercetin hexose deoxyhexose pentose | Phe/flav | C32H38O20 | 9.17 | 743.2029 | 743.2011 | Pos |
| 19a | Quercetin hexose deoxyhexose pentose p-coumaric acid | Phe/flav | C41H44O22 | 13.92 | 889.2397 | 889.2417 | Pos |
| 20a | Rutin | Phe/flav | C27H30O16 | 10.16 | 611.1607 | 611.1611 | Pos |
| 21a | Kaempferol hexose deoxyhexose pentose | Phe/flav | C32H38O19 | 10.42 | 727.2080 | 727.2072 | Pos |
| 22a | Naringenin | Phe/flav | C15H12O5 | 19.17 | 273.0758 | 273.0756 | Pos |
| 23a | Naringenin chalcone | Phe/flav | C15H12O5 | 19.78 | 273.0758 | 273.0752 | Pos |
| 24 | Naringenin chalcone dihexose | Phe/flav | C27H32O15 | 9.97 | 597.1814 | 597.1828 | Pos |
| 25a | Naringenin chalcone hexose I | Phe/flav | C21H22O10 | 13.74 | 433.1140 | 433.1138 | Neg |
| 26 | Naringenin chalcone hexose II | Phe/flav | C21H22O10 | 15.15 | 433.1140 | 433.1140 | Neg |
| 27a | Naringenin chalcone hexose III | Phe/flav | C21H22O10 | 15.39 | 433.1140 | 433.1133 | Neg |
| 28 | Naringenin hexose | Phe/flav | C21H22O10 | 12.75 | 433.1140 | 433.1149 | Neg |
| 29a | Naringenin dihexose I | Phe/flav | C27H32O15 | 6.26 | 597.1814 | 597.1833 | Pos |
| 30 | Naringenin dihexose II | Phe/flav | C27H32O15 | 11.14 | 597.1814 | 597.1822 | Pos |
| 31 | Methyl ether of hydroxylated naringenin or methyl ether of hydroxylated naringenin chalcone I | Phe/flav | C16H14O6 | 19.85 | 303.0863 | 303.0858 | Pos |
| 32a | Methyl ether of hydroxylated naringenin hexose or methyl ether of hydroxylated naringenin chalcone hexose I | Phe/flav | C22H24O11 | 13.48 | 463.1246 | 463.1260 | Neg |
| 33a | Methyl ether of hydroxylated naringenin hexose or methyl ether of hydroxylated naringenin chalcone hexose II | Phe/flav | C22H24O11 | 14.36 | 463.1246 | 463.1256 | Neg |
| 34a | Methyl ether of hydroxylated naringenin or methyl ether of hydroxylated naringenin chalcone II | Phe/flav | C16H14O6 | 20.47 | 303.0863 | 303.0862 | Pos |
| 35a | Hydroxylated naringenin chalcone | Phe/flav | C15H12O6 | 16.95 | 289.0707 | 289.0700 | Pos |
| 36 | Hydroxylated naringenin (eriodictyol) | Phe/flav | C15H12O6 | 16.07 | 289.0707 | 289.0710 | Pos |
| 37a | Hydroxylated naringenin hexose (eriodictyol hexose) | Phe/flav | C21H22O11 | 10.42 | 449.1089 | 449.1080 | Neg |
| 38a | Coumaric acid hexose I | Phe/flav | C15H18O8 | 5.39 | 325.0929 | 325.0921 | Neg |
| 39 | Coumaric acid hexose II | Phe/flav | C15H18O8 | 3.59 | 325.0929 | 325.0923 | Neg |
| 40a | Ferulic acid hexose | Phe/flav | C16H20O9 | 6.17 | 355.1034 | 355.1041 | Neg |
| 41a | Hydroxybenzoic acid hexose | Phe/flav | C13H16O8 | 1.72 | 299.0772 | 299.0784 | Neg |
| 42a | trans-Resveratrol | Phe/flav | C14H12O3 | 13.95 | 227.0713 | 227.0715 | Neg |
| 43a | Dicaffeoylquinic acid I | Phe/flav | C25H24O12 | 12.04 | 515.1195 | 515.1206 | Neg |
| 44a | Dicaffeoylquinic acid II | Phe/flav | C25H24O12 | 12.26 | 515.1195 | 515.1209 | Neg |
| 45a | Dicaffeoylquinic acid III | Phe/flav | C25H24O12 | 13.60 | 515.1195 | 515.1199 | Neg |
| 46a | 3-Caffeoylquinic acid | Phe/flav | C16H18O9 | 4.95 | 353.0878 | 353.0872 | Neg |
| 47a | 4-Caffeoylquinic acid | Phe/flav | C16H18O9 | 6.69 | 353.0878 | 353.0875 | Neg |
| 48a | 5-Caffeoylquinic acid | Phe/flav | C16H18O9 | 5.47 | 353.0878 | 353.0880 | Neg |
| 49 | Caffeic acid hexose I | Phe/flav | C15H18O9 | 3.48 | 341.0878 | 341.0887 | Neg |
| 50a | Caffeic acid hexose II | Phe/flav | C15H18O9 | 4.25 | 341.0878 | 341.0893 | Neg |
| 51a | Caffeic acid hexose III | Phe/flav | C15H18O9 | 4.75 | 341.0878 | 341.0880 | Neg |
| 52a | Benzyl alcohol hexose pentose | Phe/flav | C18H26O10 | 6.36 | 401.1453 | 401.1461 | Neg |
| 53 | Benzyl alcohol dihexose | Phe/flav | C19H28O11 | 5.07 | 431.1559 | 431.1553 | Neg |
| 54 | Phe | Phe/flav | C9H11NO2 | 2.29 | 166.0863 | 166.0860 | Pos |
| 55 | Tricaffeoylquinic acid | Phe/flav | C34H30O15 | 18.31 | 677.1512 | 677.1518 | Neg |
| 56 | Trp | AAA | C11H12N2O2 | 3.98 | 205.0972 | 205.0974 | Pos |
| 57a | N-Feruloylputrescine I | Poly. d. | C14H20N2O3 | 4.13 | 265.1547 | 265.1548 | Pos |
| 58a | N-Feruloylputrescine II | Poly. d. | C14H20N2O3 | 5.07 | 265.1547 | 265.1548 | Pos |
Metabolites that were found to be significantly up-regulated in the peel compared with the flesh tissue.
Figure 7.
Forty-five peel-associated metabolites detected by UPLC-QTOF-MS analysis. On the left side is a plot representing the five developmental stages (IG, MG, Br, Or, and Re numbered as 1–5) in the peel, and on the right side is a plot representing the profile in the flesh. Metabolite numbers (e.g. Met. 1) refer to the metabolite peak numbers in Table IV. Blue, Alkaloids; red, Val metabolism; black, phenylpropanoids/flavonoids; green, amine derivatives.
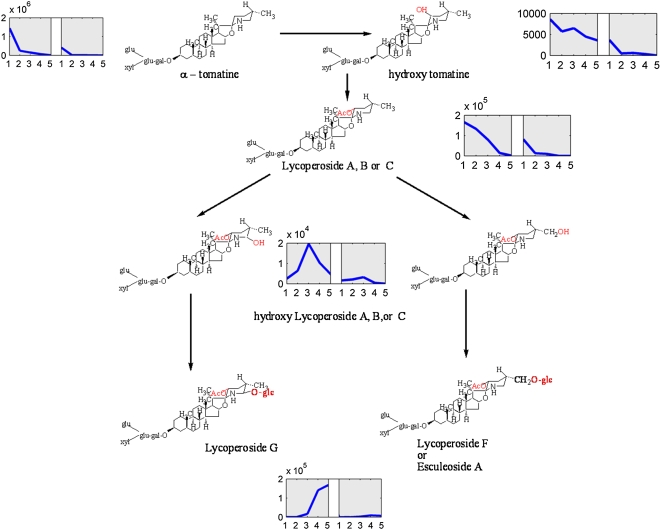
The list of metabolites accumulating in the fruit peel also included 11 glycoalkaloids that exhibited diverse patterns of accumulation during fruit development. Analysis of their structure and accumulation profiles implicated a putative pathway for glycoalkaloid metabolism in tomato peel, in which the glycoalkaloids formed in early fruit development are converted to the glycoalkaloids detected in later stages (i.e. Or and Re) of tomato fruit maturation (Fig. 8). In early development (MG), α-tomatine is present at a very high concentration, and its level dramatically decreases during development and ripening. On the other hand, the acetyl glucosylated forms of α-tomatine (lycoperoside G, lycoperoside F, or esculeoside A) showed an opposite profile, in which metabolite levels increased sharply during ripening. These findings, in combination with additional information regarding three other identified α-tomatine derivatives, namely, hydroxy tomatine and lycoperoside A, B, or C (showing decreasing levels during maturation, similar to α-tomatine), and putative intermediates in the proposed pathway, namely, hydroxy lycoperoside A, B, or C (showing an intermediate pattern of maximum levels at the Br stage followed by lower levels at maturity), indicated that the “late” biosynthesis of lycoperosides might utilize the α-tomatine produced early in fruit development.
Figure 8.
A proposed scheme for glycoalkaloid metabolism in tomato peel during fruit development, starting from α-tomatine. The glycoalkaloids formed in early fruit development are metabolized into the glycoalkaloids detected at later stages of tomato fruit maturation (i.e. Or and Re). The accumulation profile of each metabolite (peel left and flesh right) is presented next to it (in the profile, IG, MG, Br, Or, and Re are numbered as 1–5). [See online article for color version of this figure.]
Apart from phenylpropanoids, flavonoids, and glycoalkaloids, we also identified two amine derivatives (conjugated to ferulic acid; N-feruloylputrescine I and II), the aromatic amino acid Trp and the Val metabolite pantothenic acid hexose, as predominantly produced in the fruit peel compared with the flesh tissue.
GC-MS Analysis of Polar Metabolites
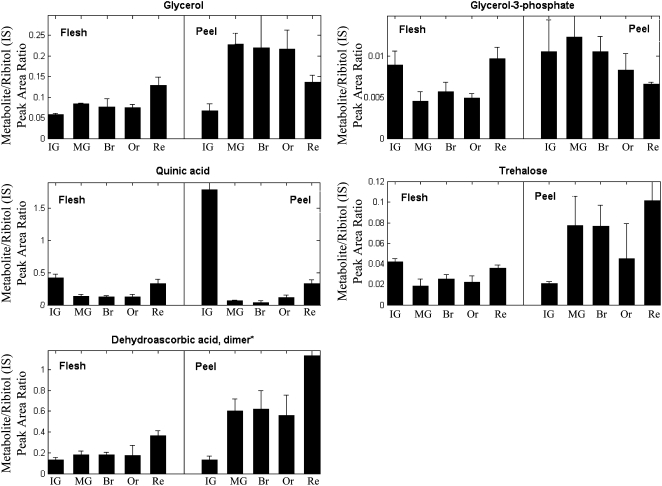
The UPLC-QTOF-MS analysis of mostly secondary metabolites was complemented by GC-MS metabolite profiling of derivatized extracts in which the majority of measured compounds are polar, primary metabolites. A total of 56 identified metabolites were monitored in the same sample sets used for the array and UPLC-QTOF-MS analyses (Supplemental Table S7). Following a two-way ANOVA test, 24 of the 56 detected metabolites, mostly organic acids and sugars, were found to be significantly up-regulated in the peel compared with the flesh tissue (at least in one developmental stage; Table V). Some of these metabolites could be associated with cuticular metabolism in tomato fruit. These included glycerol and glycerol-3-P, both accumulated to their maximum levels in the middle stages of fruit development (MG and Br stages; Fig. 9). Following the Br stage, glycerol showed steady levels in the peel until a sharp decline at the Re stage, while glycerol-3-P levels were decreased continuously from the MG stage up to the Re stage. Quinic acid, serving as a precursor for the formation of phenylpropanoids (Fig. 9), showed a similar accumulation profile in both peel and flesh, with a high level in IG followed by a decline at MG and a rise toward the Re stage. At the IG stage, quinic acid levels in the peel were much higher than those in the flesh. Additional metabolites of interest in the peel tissue included trehalose and dehydroascorbate, which accumulated in the peel during fruit development from the IG to the Re stage (Fig. 9).
Table V.
Peel-enriched metabolites detected by GC-MS analysis in derivatized extracts of tomato fruit peel and flesh during five developmental stages
| Peak No. | Putative Metabolite | Compound Group | Up/Downa |
|---|---|---|---|
| 1 | Glycerol-3-P | Sugar phosphate | Down |
| 2 | myo-Inositol-1-Pb | Sugar phosphate | Down |
| 3 | Glycerolb | Sugar alcohol | No change |
| 4 | myo-Inositolb | Sugar alcohol | Down |
| 5 | Arab | Sugar | Up |
| 6 | Xylb | Sugar | Up |
| 7 | Ribb | Sugar | Up |
| 8 | Trehaloseb | Sugar | Up |
| 9 | Galactopyranoside, methyl | Sugar | – |
| 10 | Galactinolb | Sugar | No change |
| 11 | Raffinoseb | Sugar | – |
| 12 | Threonic acid-1,4-lactone | Organic acid | – |
| 13 | N-Acetyl-Glub | Amino acid | – |
| 14 | Dehydroascorbic acid, dimerc | Organic acid | Up |
| 15 | 2-Keto-gluconic acid | Organic acid | – |
| 16 | Succinic acidb | Organic acid | Down |
| 17 | Quinic acidb | Organic acid | Down |
| 18 | GlcA | Organic acid | – |
| 19 | Galactonic acid | Organic acid | – |
| 20 | Gluconic acidb | Organic acid | Down |
| 21 | GalAb | Organic acid | Up |
| 22 | Putrescineb | Amine | Up |
| 23 | Serotoninb | Amine | – |
| 24 | Naringenin chalconeb | Flavonoid | – |
Changes in metabolite levels during fruit development as detected by GC-MS analysis of derivatized extracts performed by Carrari et al. (2006). Flesh and peel tissues were not separated in this study. –, Metabolites not reported in the Carrari et al. (2006) study.
Compounds were identified on the basis of comparison of their mass spectra and retention indexes with those of authentic standards; other metabolites were putatively identified by the comparison of their retention indexes and mass spectra with those available in spectral databases (see “Materials and Methods”).
Might have been generated from ascorbic acid in the presence of air.
Figure 9.
Accumulation of selected peel-associated polar metabolites detected by GC-MS analysis of tomato flesh and peel-derivatized extracts during tomato fruit development. Samples were similar to the ones used for the array and other types of metabolic analysis (n = 3 for each sample). See Supplemental Table S7 for a full list of GC-MS-detected polar metabolites that were found to be abundant in the peel tissue.
Analysis of Cuticular Wax, Cutin, and Triterpenoids during Fruit Development
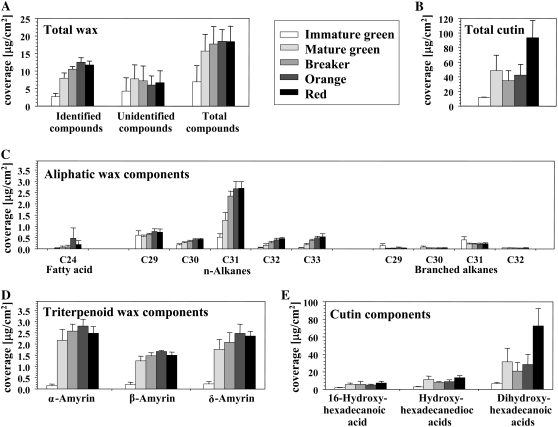
To examine the composition of cuticular components during tomato fruit development, cuticular waxes were extracted from peels derived from various developmental stages and the remaining cutin matrix was then depolymerized by BF3/methanol for cutin analysis. In the wax mixture, a total of 13 compounds were identified, including three triterpenoid alcohols (α-amyrin, β-amyrin, and δ-amyrin), a series of nine branched and unbranched alkanes (C29–C33), and C24 fatty acid. Levels of the three triterpenoids showed the most significant increase between the IG stage and the MG stage and peaked at the Or stage (Fig. 10A). Alkanes with chains longer than C31 showed the most dramatic level changes during fruit development. Among these, the predominant wax compound n-hentriacontane (n-C31) exhibited a more than 5-fold increase from the IG stage (0.5 ± 0.2 μg cm−2) to the Re stage (2.7 ± 0.3 μg cm−2; Fig. 10B). Thus, the total coverage of waxes increased throughout fruit development, most notably between the IG (6.9 ± 4.6 μg cm−2) and the Or (18.5 ± 3.3 μg cm−2; Fig. 10C) stages. Cutin coverage also increased during fruit development, mostly in two steps between immature and mature green and between the Or and Re stages (Fig. 10, A and C). The identified cutin monomers included 16-hydroxyhexadecanoic acid, hydroxyhexadecane-1,16-dioc acid, dihydroxyhexadecanoic acid, and trihydroxyoctadecanoic acid. Isomers of the C16 dihydroxy fatty acid with hydroxyl groups in the 7,16, 8,16, 9,16, and 10,16 positions were also identified, but they could not be separately quantified.
Figure 10.
Composition of compounds identified by GC-MS-FID analysis during the five tested developmental stages of isolated fruit cuticles. A, Total wax amounts. B, Total cutin amounts. C, Amounts of aliphatic wax compounds. D, Amounts of triterpenoid wax amounts. E, Amounts of cutin monomers. Detected cutin constituents include 16-hydroxyhexadecanoic acid, hydroxyhexadecane-1,16-dioc acid, and various dihydroxyhexadecanoic acid isomers. Values are given as averages (n = 3) and sd.
DISCUSSION
To date, most molecular studies in tomato and other fleshy fruit have been performed on the fruit as a whole, without distinguishing its various tissues. Since different fruit tissues play diverse roles in fruit development and ripening, crucial information for our understanding of fruit biology has been overlooked. In this study, we carried out detailed profiling of the tomato fruit outer tissues. The analysis was conducted at five stages of fruit development, starting from the small IG stage (25 DPA) up to the Re stage of fruit development. Transcripts and metabolites that are most likely associated with major activities in the peel during its development have been identified by comparative transcriptome and metabolome analyses between peel and flesh tissues. For gene and metabolite profiling experiments, we used manually dissected fruit flesh and peel. Peel samples contained the epidermis, several cell layers below it, and the cuticle. Thus, a portion of the genes that have been detected as peel associated could be expressed in either the epidermis or the outer mesocarp cells, while metabolites could have originated from either one or more of these layers, including the cuticle. Despite the use of “epidermis-enriched” tissue, many of the genes and metabolites detected here seem to be associated with the formation of the cuticle building blocks. Future research using laser dissection (Nakazono et al., 2003) or cell sorting technologies (Mattanovich and Borth, 2006) might allow exclusive dissection of the epidermal cell layer for more accurate gene and metabolite profiling.
Flavonoids, Glycoalkaloids, and the Amyrin-Type Pentacyclic Triterpenoids Are Dominant Classes of Secondary Metabolites Produced in the Tomato Fruit Peel
The UPLC-QTOF-MS metabolic profiling showed that tomato peel contains an array of secondary metabolites, most of which belong to the flavonoid or alkaloid class. The use of fruit peels rather than isolated cuticles for the UPLC-QTOF-MS analysis enabled gene expression analysis but did not allow the differentiation between metabolites that are embedded in the cuticle (or deposited as epicuticular material) to those accumulating in epidermal cells or other peel cell layers. A previous report described high accumulation levels of the yellow pigment naringenin chalcone in tomato fruit cuticle (Bauer et al., 2004b). However, in order to examine whether the range of phenolic compounds (mainly flavonoid derivatives) and glycoalkaloids detected as peel associated in this study actually accumulate in the cuticle and/or epidermis cells or other peel cell layers, future experiments should involve the metabolic profiling of semipolar metabolites in isolated fruit cuticles. A third class of cuticle-associated secondary metabolites that were found here to have abundantly increased levels in the cuticle following the IG stage of fruit development are the pentacyclic triterpenoids (α-, β-, and δ-amyrin). This finding is in accordance with a previous report by Vogg et al. (2004), who examined the properties of the cuticular barrier in the slcer6 mutant fruit and found a 4-fold increase in water loss (compared with the wild type) as well as a reduction in intracuticular aliphatic waxes and a compensating increase in pentacyclic triterpenoids.
Fruit surface tissues, including the cuticle and the epidermal layer below it, form the interface between the fruit and its environment. Hence, the role of secondary metabolites that accumulate in these tissues is coupled, either directly or indirectly, with this type of interaction. For example, triterpenoids such as the amyrins and the various glycoalkaloids (e.g. α-tomatine and dehydrotomatine) might be potent antifungal agents (Friedman, 2002) that prevent fungi and other pathogens such as bacteria from penetrating the fruit. UPLC-QTOF-MS analysis conducted in our study revealed that the dozen different peel-associated glycoalkaloids exhibit various accumulation profiles during fruit development. The production of α-tomatine in early tomato fruit development and its degradation in the ripening phase have been described before. It was suggested that prior to ripening, it serves as a protecting mechanism against predators and that its decrease in late fruit developmental stages allows the promotion of seed dispersal (Rick et al., 1994). Other alkaloids produced at later stages of fruit development might take part in the defense against pathogens attracted to soft ripening fruit. Our profiling data support the hypothesis of Fujiwara et al. (2004), who suggested that esculeoside A (accumulating at late stages of fruit development) might be produced from tomatine. Moreover, based on chemical structures and patterns of metabolite accumulation during fruit development, we predicted that hydroxy tomatine and lycoperoside A, B, or C (accumulating at early stages of fruit development) are also precursors in this pathway. The metabolite profiling results further indicate that hydroxy lycoperosides A, B, and C are intermediate metabolites for lycoperosides G and F as well as esculeoside A formed at late stages of fruit development.
While the secondary metabolites present in the fruit peel could act as protectants against changing environmental conditions and in deterring pathogens, they could also play a role in attracting seed-dispersing frugivors. For example, the flavonoid naringenin chalcone, accumulating in the fruit peel during the Br stage, could serve in the protection against UV radiation and in the attraction of frugivors by its intense pigmentation (Bovy et al., 2002; Whitney, 2005). Metabolites present in the peel, such as the amyrins and naringenin chalcone, could also serve as structural elements of the cuticle. Their portion in the overall cuticular waxes, their physical characteristics, and their spatial arrangement/assembly within the cuticle may have an effect on properties of the cuticular barrier (e.g. elastic modulus and water loss). Luque et al. (1995) suggested that the flavonoids naringenin and naringenin chalcone play an important role in the control of water transport across the cuticle. More recently, Schijlen et al. (2007) generated transgenic tomato plants in which the expression of CHS1 and CHS2 was silenced by RNA interference. These two chalcone synthases catalyze the formation of naringenin chalcone, whose levels, therefore, were dramatically reduced in the transgenic lines. Upon ripening, the CHS RNA interference tomato fruit were reddish and showed a dull appearance, in contrast to the red and shiny fruit of wild-type plants. Moreover, electron microscopy analysis revealed that CHS-silenced fruit lacked the typical epidermal conical cells detected in wild-type fruit. This suggests that the flavonoid naringenin chalcone influences structural properties of the epidermal cell layer. Additional known tomato varieties and mutants that lack naringenin chalcone in the peel are DFD (Saladie et al., 2007), nor (Bargel and Neinhuis, 2004), y (Duggar, 1913; Lindstrom, 1925), as well as other cultivars and wild tomato species (Z. Dani, personal communication). As suggested previously by Vogg et al. (2004), triterpenoids also affect the tomato fruit cuticular structure and water permeability.
Recent reports provided evidence for an active transport mechanism that governs the secretion of cuticular components from the epidermal cells to the extracellular matrix. Pighin et al. (2004) suggested that the Arabidopsis CER5 protein (WBC12), an ATP-binding cassette-type transporter, is involved in the transport of wax components through the plasma membrane of epidermal cells. Later, a similar Arabidopsis protein (WBC11) was also suggested to act as a transporter of both wax and cutin monomers (Bird et al., 2007; Luo et al., 2007; Panikashvili et al., 2007; Ukitsu et al., 2007). Membrane transport of various secondary metabolites by members of the ATP-binding cassette protein family was previously demonstrated in several cases (Yazaki, 2006). Thus, similar proteins might be involved in cuticular lipid and flavonoid secretion in the tomato fruit surface. The assortment of putative transporter proteins encoded by genes identified here as predominantly expressed in the peel (5% of the total peel-enriched genes; Fig. 4) are good candidates for the identification of such transporters.
A Variety of Genes Encoding Putative Cell Wall-Related Enzymes Are Preferentially Expressed in the Peel during Fruit Development
Epidermal cells exhibit a unique cell wall structure that encompasses several domains between the symplasm and the epicuticular surface. A primary cell wall is the innermost domain that surrounds the epidermis cell. It contains cellulose microfibril layers together with hemicelluloses and is encased by pectins and proteins. A layer of secondary cell wall, present above the primary cell wall, is covered by a pectin-rich layer composed of homogalacturonans and rhamnogalacturonans. Cellulose microfibrils branch through the pectin-rich layer, which is in direct contact with the cuticular lipids. The epidermal cell walls largely contribute to the mechanical properties of the tomato fruit exocarp and the resistance of the turgor-driven fruit growth in tomato (Thompson, 2001; Andrews et al., 2002; Bargel and Neinhuis, 2005). For example, peroxidases and xyloglucan endotransglycosylase have been reported to influence stiffening and weakening of the epidermal cell wall (Thompson et al., 1998; Andrews et al., 2000).
Fifteen different genes putatively encoding cell wall-related enzymes showed up-regulated expression in the tomato peel compared with the flesh. These include polygalacturonase, two expansins, xyloglucan galactosyltransferase, α-galactosidase, xyloglucan endotransglycosylase, pectin methylesterase, β-galactosidase, endo-β-mannanase, UDP-GlcUA decarboxylase, cellulose synthase, arabinosidase, endo-1-4-β-glucanase, and pectin acetyl esterase. These enzymes are likely to be active in the epidermis cell walls. However, some of them could take part in the softening of the outer pericarp tissue, since, as mentioned above, the peel samples contained remainders of the mesocarp cells (collenchyma and parenchyma cells). The link between the activity of cell wall enzymes and the cuticular layer was demonstrated by antisense suppression of the tomato β-GALACTOSIDASE6 gene (Moctezuma et al., 2003). The fruit of these transgenic plants showed extensive cracking, but, more interestingly, it also exhibited doubled cuticle thickness and increased glossiness. The authors suggested that modification to galactan metabolism in the pectin-rich layer of the epidermis cell wall could result in a compensating process by which cuticle deposition is enhanced in order to repair the cracks and/or strengthen a weaker structure.
Accumulation of Polar Metabolites in the Peel Is Associated with Cuticle- and Cell Wall-Related Metabolism and the Protection of Fruit against Photooxidative Stress
GC-MS analysis detected 24 metabolites, mainly organic acids and sugars, that showed increased levels in peel compared with flesh. Two of the peel-enriched metabolites detected in our study were glycerol and glycerol-3-P. Graca et al. (2002) have examined depolymerized cutin from the leaves and fruit of seven plant species and detected glycerol monomers ranging between 1% and 14% of the lysates. They interpreted the abundance of this metabolite as evidence that glycerol molecules take part in the cross-linking of the cutin polymer.
As mentioned above, the cuticle structure is associated with a unique cell wall structure in the epidermal cells. In accordance with the range of genes encoding cell wall enzymes that are preferentially expressed in the peel compared with the flesh, several carbohydrate derivatives, detected in the peel at relatively high levels, could be associated with cell wall metabolism. These included myo-inositol, myo-inositol 1-P, Ara, Xyl, and GlcUA (Fry et al., 2001; Sharples and Fry, 2007). Another interesting observation was the accumulation of GalUA, which is the main component of pectin and might be part of the cuticle-associated pectin-rich layer composed of homogalacturonans and rhamnogalacturonans (see above).
Intense light and elevated temperature conditions cause increased concentrations of reactive oxygen species that result in photooxidative damage in the fruit. In tomato, this was reported to occur mainly in the surface tissues of young green fruit (Andrews et al., 2004). Metabolites produced in either epidermis or the layers below it in the fruit function as antioxidant protectors and should prevent photooxidative damage (Torres and Andrews, 2006; Torres et al., 2006). Several metabolites with putative antioxidant capacity showed preferential accumulation in the peel tissue compared with flesh. One such metabolite that is part of the antioxidant response system and that accumulated in the peel is dehydroascorbic acid (Fig. 9). Supporting evidence for the active metabolism of ascorbic acid in the peel was provided by the peel up-regulated expression of cytochrome b561 (detected in this study) and guanidine diphosphate-Man pyrophosphorylase (GDMP; Lemaire-Chamley et al., 2005). While cytochrome b561 catalyzes ascorbate-driven transmembrane electron transport and contributes to ascorbate-mediated redox reactions in subcellular compartments, the GDMP enzyme is involved in ascorbate biosynthesis. A second antioxidant metabolite that was found to accumulate in the peel tissue is the amine putrescine, which can act as a free radical scavenger either directly or after interacting with other molecules, such as free ferulic and caffeic acids (Bors et al., 1989; Choi et al., 2007). Indeed, among the metabolites detected as peel associated by our UPLC-QTOF-MS analysis were N-feruloylputrescine I and II.
Additional metabolites that were detected as peel associated by UPLC-QTOF-MS analysis could serve as antioxidants in the tomato fruit surface, including the various flavonoids, particularly quercetin and kaempferol derivatives, trans-resveratrol, and chlorogenic acid (i.e. 3-caffeoylquinic acid; Niggeweg et al., 2004) and its related structures (dicaffeoylquinic acids I, II, and III; 3-caffeoylquinic acid, 4-caffeoylquinic acid, and 5-caffeoylquinic acid; and tricaffeoylquinic acid). Quinic acid, which is incorporated into chlorogenic acid and related structures, was also found by the GC-MS analysis to be peel up-regulated. Thus, various peel-enriched metabolites detected in our study by both GC-MS and UPLC-QTOF-MS analyses are likely to be involved in the antioxidative protection mechanism in the fruit surface.
Cuticular Lipid-Related Gene Expression and Metabolism Precede Those Related to Phenylpropanoids and Flavonoids during Fruit Development
Wax and cutin components as well as phenylpropanoid derivatives such as the flavonoids are major constituents of the tomato fruit cuticle. The results obtained in this study suggest that the accumulation of these two main metabolite classes is only partially overlapping during tomato fruit development. As presented in Figure 5, while some transcripts associated with cuticular lipids accumulate already at the early IG stage of fruit development and others during the MG and Br stages, a large cluster of transcripts from the general phenylpropanoid and flavonoid pathways are coordinately expressed and induced at the Br stage. Metabolic analysis corroborates these observations and demonstrates that the activity of pathways leading to both cuticular lipids and phenylpropanoids declines at the late, Re stage of fruit development (Fig. 7). In accordance with our results, Baker (1982) demonstrated a continuous increase in total waxes from immature green to firm red tomato fruit. Recently, Bauer et al. (2004b) and Leide et al. (2007) also showed a continuous increase in wax coverage during fruit ripening stages in two different cultivars. Similar to the data presented by Baker (1982) and Leide et al. (2007), we found that the cutin coverage increased during fruit development and that major components of the cutin monomers are 9,16- and 10,16-dihydroxyhexadecanoic acid (Fig. 10).
Even though glycoalkaloids and pentacyclic triterpenoids accumulate to high levels in tomato fruit, no genes encoding enzymes putatively catalyzing a committed step in the biosynthesis of these compounds have been reported so far. Our set of peel up-regulated transcripts, however, did contain enzymes encoding genes that might be involved in the supply of precursors to the formation of these substances through the mevalonate pathway (Kalinowska et al., 2005). These include a putative 3-hydroxy-3-methylglutaryl-CoA reductase and two successive steps in the same pathway (one step downstream), namely, a putative mevalonate diphosphate decarboxylase and an isopentenyl diphosphate isomerase. Several candidate genes that might be directly involved in these two biosynthetic pathways (based on their homology with characterized alkaloid-related genes), such as strictosidine synthase-like gene (Barleben et al., 2007) and tropinone reductase-like gene (Dräger, 2006), were also included among the peel-associated transcripts detected in this study (Table I).
GUS Expression Driven by the SlCER6 Upstream Region Suggests the Formation of Surface Waxes in the Endocarp of Tomato Fruit
The detection of GUS staining in the internal cell layer that borders the pericarp and the gel-containing locules (i.e. the endocarp) of the pSlCER6 fruit implicates the formation of surface waxes also in this tissue. This is in accordance with the findings of Almeida and Huber (2001), who examined whether liquid from the locular tissue, driven by internal pressure, could infiltrate the pericarp apoplast. Apart from an increase in internal locular pressure beginning at the MG stage, peaking at the pink stage, and strongly declining at the Re stage, the authors also detected alterations in the anatomical features of the endocarp surface. While in early stages of fruit ontogeny the endocarp surface develops apertures in densities ranging from 6.7 to 47.9 apertures mm−2, these apertures were sporadic and entirely lacking in the pink and ripe fruit. Almeida and Huber (2001) further suggested that the apertures are occluded during fruit maturation and ripening, possibly by the progressive deposition of surface waxes, which would prevent or minimize the pressure-driven infiltration of locular fluids into the pericarp apoplast. Thus, the expression pattern of the tomato CER6 gene, encoding a condensing enzyme involved in the synthesis of VLCFA precursors for wax production, strengthens the hypothesis that progressive wax deposition occurs in the endocarp during fruit development. Chemical and anatomical examination of the endocarp tissue of both wild-type and slcer6 fruit (Vogg et al., 2004) may shed more light on this putative endocarp wax accumulation process.
Regulatory Networks in the Tomato Fruit Peel
Genes with putative transcription factor activity are a major portion (10%) of the total peel up-regulated genes. In cluster 5 (represented by high levels of expression at IG followed by a continuous decline until the Re stage), we have identified a putative tomato ortholog of the Arabidopsis SHN2 gene that, together with two other similar genes, was suggested to act as a transcriptional regulator of the cutin and wax biosynthetic pathways. The Arabidopsis SHN1/WIN1, SHN2, and SHN3 genes were shown to induce wax accumulation when overexpressed in Arabidopsis (Aharoni et al., 2004; Broun et al., 2004), and, recently, Kannangara et al. (2007) provided evidence that SHN1/WIN1 is a transcriptional activator acting directly on cutin biosynthetic genes. Several tomato genes identified here as coregulated with SlSHN2 are putative orthologs of genes suggested by Kannangara et al. (2007) to be downstream targets of SHN1/WIN1 in the cutin biosynthetic pathway. These include an HTH-LIKE protein, a GDSL motif extracellular lipase, and a glycerol-3-P dehydrogenase. Thus, SlSHN2 might be the tomato equivalent of SHN/WIN and serve as an activator of the cutin biosynthetic pathway in fruit epidermis. Preliminary results support the latter hypothesis, since SlSHN2 overexpression in Arabidopsis plants (J. Shi and A. Aharoni, unpublished data) displayed a similar phenotype to those obtained by the overexpression of the Arabidopsis SHN1/WIN1, SHN2, and SHN3 factors in Arabidopsis plants (Aharoni et al., 2004; Broun et al., 2004).
As detected in many cuticle mutants, particularly in Arabidopsis, the biosynthesis of wax and cutin is tightly associated with epidermal cell differentiation (Wellesen et al., 2001; Chen et al., 2003; Aharoni et al., 2004; Panikashvili et al., 2007). Here, we have identified a putative tomato ortholog of the AmMIXTA-LIKE2 gene that exhibited preferential expression in the peel compared with the flesh tissue at all five tested stages (Fig. 3D). Expression was particularly high at the IG stage and declined as fruit development proceeded to the late Re stage. Several MIXTA-LIKE MYB transcription factors, defined by a conserved sequence motif downstream of the MYB-binding domain and reported to play a role in epidermal cell differentiation, have been characterized in plants (Kranz et al., 1998). The MIXTA from Antirrhinum majus (AmMYBX) and PhMYB1 from Petunia hybrida were both shown to take part in conical cell formation in the petal epidermis (Avila et al., 1993; Noda et al., 1994; Glover et al., 1998; van Houwelingen et al., 1998; Jaffé et al., 2007). The mixta mutants in both Antirrhinum and Petunia exhibit flat epidermal cells compared with those of wild-type petals. Another MIXTA-LIKE MYB gene from Antirrhinum, AmMYBML1, was shown to play a role in the differentiation of three specialized petal cells, including epidermal conical cells in the ventral petal hinge region, in the elaboration of the mesophyll below the epidermis, and in trichome development in the throat of the flower (Perez-Rodriguez et al., 2005). As described above, the tomato fruit epidermis contains cells similar to the conical cells detected on Antirrhinum and Petunia petals, and the tomato fruit epidermis also develops trichomes. The tomato peel-associated MIXTA-LIKE gene detected here might also be involved in similar differentiation processes of fruit epidermis.
A large number of flavonoid-related regulatory genes, particularly those belonging to the MYB family of transcription factors, were characterized previously (Stracke et al., 2001; Davies and Schwinn, 2003). The high activity level of the flavonoid pathway detected in the peel, both at the level of gene expression and in terms of metabolism, led us to search for candidate genes that might be involved in the regulation of the flavonoid pathway in the peel tissue. One such candidate was the THM27 gene, included among the 14 MYB-type transcription factors previously isolated from various tissues of tomato (Lin et al., 1996). Its closest Arabidopsis homolog is MYB4, which is known to be involved in the UV-B response (Jin et al., 2000; Zhao et al., 2007). The atmyb4 mutant accumulates sunscreens and the phenylpropanoid sinapate esters in its leaves. SlTHM27 exhibits its highest expression levels in the early IG stage, and its expression declines during fruit maturation. Thus, SlTHM27 might regulate genes related to the upstream phenylpropanoid pathway (from PAL down to the CHS step) rather than those related to the committed downstream steps in flavonoid biosynthesis, which are activated only later at the Br stage. This is in accordance with findings for AtMYB4, which was shown to function as a repressor of cinnamate 4-hydroxylase catalyzing a reaction in the general phenylpropanoid pathway (Jin et al., 2000).
As described above, most investigations conducted to date on tomato fruit development and ripening did not distinguish between the various fruit tissues; therefore, very little tissue-specific information was previously acquired. This is also true for fruit ripening regulation, which has been investigated only for pericarp (i.e. flesh) characteristics such as carotenoid accumulation, chlorophyll degradation, flavor and aroma formation, cell wall metabolism, and the softening processes as well as ethylene involvement. Therefore, it is currently not clear to what extent the various processes occurring in the peel, for example, accumulation of the naringenin chalcone pigment, are part of the ripening regulation and whether they are ethylene dependent or independent. The identification of the RIPENING INHIBITOR (RIN) gene, a key regulator of fruit ripening that is preferentially expressed in the peel (Table I), makes this question even more interesting. Future analysis of ripening mutants' transcriptomes and their comparison with the sets of peel-associated genes detected in our study might shed new light on the extent to which peel-associated processes are part of the known ethylene-dependent ripening process considered to date to be flesh associated.
A Common Set of Genes Is Active in Tomato Reproductive and Vegetative Organ Surfaces as Well as in the Surface of Various Other Plant Species
Apart from the specific study of gene expression and metabolic profiles in tomato fruit peel tissue, another important motivation for this work was the comparative analysis between surface tissues of reproductive organs and vegetative organs. Forty percent of the genes up-regulated in tomato stem epidermis were also detected among the genes up-regulated in fruit peel. Assignment of putative functional categories to genes of this set revealed that mostly genes associated with phenylpropanoid/flavonoid, wax, and cutin metabolism are enriched in both surface tissues. Some of the genes up-regulated in both tomato surface tissues (i.e. stem and fruit) were also up-regulated in surface tissues of other plant species. For example, putative homologs of the Arabidopsis epidermis-associated HTH genes, involved in the formation of cutin (Krolikowski et al., 2003; Kurdyukov et al., 2006), were up-regulated in both tomato stem and fruit peel, in Arabidopsis stem peel, and in cotton fibers. We also encountered numerous similarities between epidermis-associated genes of Arabidopsis and tomato and those expressed in developing cotton fibers. Fatty acid biosynthesis and elongation was ranked as the second-most up-regulated biochemical pathway during cotton fiber elongation (Shi et al., 2006), and recent reports by Qin et al. (2007a, 2007b) highlighted the importance of VLCFAs in the extensibility of cotton fibers. These findings might account for the similarity in gene activity between cotton fibers and other epidermis-related tissues. Furthermore, the comparison between genes up-regulated in various surface tissues and cotton fibers enabled the detection of “universal” epidermis-related genes with unknown functions. For example, homologs of a putative cytochrome P450 from the CYP71A family with unknown function were up-regulated in tomato fruit peel and stem epidermis as well as in cotton fibers and in maize epidermis (Table III).
In conclusion, this study made, to our knowledge, the first steps toward understanding the genetic makeup of tomato fruit surface tissues in terms of both gene expression and the activity of metabolic pathways. Genes identified in this work are excellent candidates for more detailed investigations of surface tissue assembly and biological activity in tomato as well as other fleshy fruit species.
MATERIALS AND METHODS
Plant Material
Seeds of tomato (Solanum lycopersicum ‘Alisa Craig’) were obtained from the Tomato Genetics Resource Center (http://tgrc.ucdavis.edu). Flowers of greenhouse-grown plants were marked at anthesis, and fruit were harvested according to appearance and counted as follows: approximately 25 DPA (IG stage), approximately 42 DPA (MG stage), approximately 44 DPA (Br stage), approximately 46 DPA (Or stage), and approximately 48 DPA (Re stage). One biological repeat was a mixture of four to five individual fruit from the same stage of development. Immediately upon harvesting, peel and flesh (without the gel and seeds) were manually dissected and frozen in liquid nitrogen. Transgenic plants (Micro-Tom) were generated as described by Meissner et al. (1997).
Characterization of Upstream Regions and Plasmid Construction
The 2- and 1.05-kb upstream regions of the SlCER6 and SlCHS1 genes, respectively, were isolated from Micro-Tom tomato genomic DNA using the BD GenomeWalker kit (Clontech), the primary PCR oligonucleotides 5′-CAGCTTAACAGAGTTACAGAAATTTGGG-3′ and 5′-GCGTAGATGTTCCAATGGCTAAGATC-3′, and the nested PCR oligonucleotides 5′-GCTTCTGGCATTTTCACAAACAAAC-3′ and 5′-GTTGCGCCTTACGATACTCCTCC-3′ for SlCER6 and SlCHS1 upstream regions, respectively. Amplified genomic fragments were then cloned with a GUS reporter gene into the pBINPLUS vector (Vanengelen et al., 1995).
GeneChip Analysis and Clustering of Expression Profiles
Total RNA was extracted by the hot phenol method (Verwoerd et al., 1989), and the cDNA synthesized by the Invitrogen SuperScript II reverse transcriptase was used as a template to generate biotinylated copy RNA. Biotinylated copy RNA was fragmented and hybridized to the Affymetrix GeneChip Tomato Genome Array as described in the Affymetrix technical manual (available at www.affymetrix.com). Normalization of log2-based expression intensity values was carried out using RMA analysis (Irizarry et al., 2003) implemented in the R microarray analysis package (http://www.R-project.org). Initial filtering of the genes was performed using the absent/present call acquired by MAS5 analysis software (Affymetrix). Only transcripts with at least one stage containing a present call were retained. Next, all expression values below the 10th percentile were set to the 10th percentile value. Transcripts with poor quality spots showing low replicate reproducibility (high root square deviation [RSD]) in at least one-third of the tested stages were eliminated from further analysis. Peel up-regulated genes were defined as those having a greater than or at least 2-fold intensity ratio in peel versus flesh in at least one developmental stage. Two-sample Student's t test with false discovery rate correction was used for the detection of significantly (P < 0.05) differentially expressed genes. Expression profile clustering was performed on the normalized (mean-center) log2-based values of the peel up-regulated transcripts. A Pearson correlation distance measure and average linkage clustering method (implemented in MATLAB version 7.3.0; TheMathWorks) were used. Peel up-regulated genes were defined here as those having a greater than or at least 2-fold ratio value in peel versus flesh in at least one developmental stage.
Comparisons with Epidermis-Enriched Transcripts from Other Species
In order to identify universal epidermis-related genes, tomato fruit and stem peel-enriched transcripts were compared with genes with prevalent expression in the epidermis of other plant species. At the first stage, sets of known epidermis-related genes from other species were compared by TBLASTX against all tomato consensus contigs from the JCVI database (http://www.tigr.org); best hits of these BLAST results were then further analyzed in order to identify orthologs/homologs of the tomato epidermal genes. Alignment cutoffs for the TBLASTX analysis of the Arabidopsis (Arabidopsis thaliana), maize (Zea mays), and cotton (Gossypium hirsutum) sets were set to E = 10−5, E = 10−17, and E = 10−20, respectively. Tomato best hits of these TBLASTX analyses were then compared with the list of tomato peel-enriched transcripts for the identification of relevant homologs/orthologs. We also performed an in silico analysis in order to identify the expression and determine the enrichment of homologs/orthologs of tomato epidermis-associated genes in apple (Malus domestica) fruit peel. ESTs from two apple fruit skin peel libraries (AAFA and AAFB; total of 7,729 ESTs) were compared by TBLASTX against all of the tomato consensus contigs from the JCVI database. Seventy-eight of the tomato best hits, with 10−4 E value alignment cutoff, were identified as tomato epidermis up-regulated genes. These 78 tomato transcripts were then compared by TBLASTX against more than 40,000 ESTs derived from eight apple EST libraries of different tissues, including fruit cortex (AALA), seeds (AAWA), preopened floral buds (AFBC), phloem (AAOA), xylem (AAZA), young expanding leaves (AELA), young shoots (AVBB), and root tips (ABPB). The prevalence of these transcripts among all other apple EST libraries was then compared with that in the two apple skin peel libraries in order to identify enhanced expression in apple fruit peel.
Gene Expression Analysis by RT-PCR
For a two-step quantitative RT-PCR, a known amount of DNase-treated total tomato fruit flesh or peel RNA was reverse transcribed using Invitrogen's SuperScript II reverse transcriptase kit. Platinum SYBR SuperMix (Invitrogen) RT-PCRs were tracked on an ABI 7300 instrument (Applied Biosystems). Each sample was amplified by PCR using the same amount of cDNA template in triplicate reactions. Gene-specific quantitative RT-PCR primer pairs for the 19 selected genes encoding peel-associated proteins were designed using the Primer Express software (Applied Biosystems). SlASR1 was used as an endogenous control. Following an initial step in the thermal cycler for 15 min at 95°C, PCR amplification proceeded for 40 cycles of 15 s at 95°C and 30 s at 60°C and was completed by melting curve analysis to confirm the specificity of PCR products. The baseline and threshold values were adjusted according to the manufacturer's instructions. Similar results were obtained from relative quantification of transcript abundance determined independently by the standard curve method described in Applied Biosystems User Bulletin 2 (http://www3.appliedbiosystems.com/cms/groups/mcb_support/documents/generaldocuments/cms_040980.pdf). Oligonucleotide sequences designed for the amplification of the transcripts presented in Figures 3 and 6 were as follows: 5′-TGACCATGGTTTTCAGCCAAT-3′ (SlCER1 forward) and 5′-TAACTCATAAGCCACCACATCAATG-3′ (SlCER1 reverse); 5′-ATGCAAAGCTGAGGAAATGTTG-3′ (SlSHN2 forward) and 5′-GATGTTTTTTGCCACACTCCAA-3′ (SlSHN2 reverse); 5′-GTAAAGATTGCAGTTGTGGAAGTGA-3′ (SlTHM27 forward) and 5′-TTCAAGCCCAAAAAGTCATAACC-3′ (SlTHM27 reverse); 5′-GCGAGCGCTAGTGCTGGTAT-3′ (SlMIXTA2 forward) and 5′-TAATATGTTGCGCATTTTCGAAA-3′ (SlMIXTA2 reverse); 5′-ACAAGAAGATCCACAAGGGAAAGT-3′ (SlCER6 forward) and 5′-CGGACCGATCGTAGTGATGTT-3′ (SlCER6 reverse); and 5′-GATCGTAGCTGGACCCTCTGC-3′ (SlCHS1 forward) and 5′-GTTTTTCACAAACCAACAGTTCTGAT-3′ (SlCHS1 reverse).
Nontargeted Profiling of Semipolar Compounds by UPLC-QTOF-MS
For the profiling of semipolar compounds, frozen tomato tissues were ground to a fine powder using an analytical mill (IKA; A11 basic) or mortar and pestle and extracted according to De Vos et al. (2007). Frozen tissue (500 mg) was weighed in a 15-mL polypropylene tube and extracted with methanol (the solid:liquid ratio was kept at 1:3, w/v). The mixture was sonicated for 20 min at room temperature, centrifuged (3,000g, 10 min), and filtered through a 0.22-μm polytetrafluoroethylene membrane filter (Acrodisc CR 13 mm; PALL) before injection to the UPLC-QTOF-MS instrument. Mass spectral analyses were carried out by the UPLC-QTOF instrument (Waters Premier QTOF), with the UPLC column connected online to a UV detector (measuring at 240 nm; Waters Acquity) and then to the MS detector. Separation of metabolites was performed on the 100- × 2.1-mm i.d., 1.7-μm UPLC BEH C18 column (Waters Acquity). The mobile phase consisted of 0.1% formic acid in acetonitrile:water (5:95, v/v; phase A) and 0.1% formic acid in acetonitrile (phase B). The linear gradient program was as follows: 100% to 72% A over 22 min, 72% to 60% A over 0.5 min, 60% to 0% A over 0.5 min, held at 100% B for a further 1.5 min, then returned to the initial conditions (100% A) in 0.5 min and conditioning at 100% A for 1 min. The flow rate was 0.3 mL min−1, and the column temperature was kept at 35°C. Masses of the eluted compounds were detected with a QTOF Premier MS instrument equipped with an ESI source (performed in ESI-positive and ESI-negative modes). The following settings were applied during the UPLC-MS runs: capillary voltage at 3.0 kV; cone voltage at 30 eV; collision energy at 3 eV; and argon was used as a collision gas. The m/z range was 50 to 1,500 D. The following settings were applied during the UPLC-MS/MS run: capillary spray at 3.0 kV; cone voltage at 30 eV; collision energies were 10 to 35 eV for positive mode and 15 to 40 eV for negative mode; and argon was used as a collision gas. The MS system was calibrated using sodium formate, and Leu enkephalin was used as the lock mass. A mixture of 15 standard compounds, injected after each batch of 10 tomato samples, was used for instrument quality control. MassLynx software version 4.1 (Waters) was used to control the instrument and calculate accurate masses. The UV spectra (200–600 nm) were acquired on a UPLC (Waters, Acquity) instrument equipped with an Acquity 2996 PDA under LC conditions as described above for the UPLC-QTOF analysis. Metabolites were identified using standard compounds by comparison of their retention times, UV spectra, and MS/MS fragments. When the corresponding standards were not available, compounds were putatively identified applying several steps. First, the elemental composition was selected according to the accurate masses and the isotopic pattern using MassLynx software. Then, the elemental composition obtained was searched against the KNApSAcK metabolite database (http://prime.psc.riken.jp/KNApSAcK; Shinbo et al., 2006), the Dictionary of Natural Products (Chapman & Hall/CRC), and the MOTO database (http://appliedbioinformatics.wur.nl/moto; Moco et al., 2006). When a suitable candidate was not found, more comprehensive chemical databases were searched using the SciFinder tool (SciFinder Scholar). Predicted Log D values for pH 3 (pH of the UPLC mobile phase), found using the SciFinder tool, were utilized for the retention time prediction in order to narrow the number of proposed structures. The interpretation of the observed UV and MS/MS spectra in comparison with those found in the literature (when possible) was the main tool for putative identification of metabolites.
GC-MS Analysis of Derivatized Extracts
We used a method modified from the one described by Roessner-Tunali et al. (2003). Tissue powder (100 mg) was extracted in 700 μL of methanol with 60 μL of internal standard (0.2 mg of ribitol in 1 mL of water). After mixing vigorously, the extract was sonicated in a bath sonicator for 20 min and centrifuged at 20,000g. Chloroform (375 μL) and water (750 μL) were added to the supernatant, and the mixture was vortexed and centrifuged. An aliquot of the upper methanol/water phase (400 μL) was taken and lyophilized. The dried material was redissolved and derivatized for 90 min at 37°C (in 40 μL of 20 mg mL−1 methoxyamine hydrochloride in pyridine), followed by a 30-min treatment with 70 μL of N-methyl-N(trimethylsilyl) trifluoroacetamide at 37°C and centrifugation. As tomato fruit contains high amounts of sugars, it was necessary to measure two different sample sizes in order to avoid the overloaded peaks of sugar and clearly detect compounds with lower abundance (after derivatization, each sample was divided into two portions). Sample volumes of 1 μL were injected into the GC column for sugar analysis, and sample volumes of 1.5 μL were injected for lower abundance metabolite detection. A retention time standard mixture (14 μg mL−1 each of n-dodecane, n-pentadecane, n-nonadecane, n-docosane, n-octacosane, n-dotriacontane, and n-hexatriacontane in pyridine) was injected after each set of six samples. The GC-MS system was composed of a COMBI PAL autosampler (CTC analytics), a Trace GC Ultra gas chromatograph equipped with a PTV injector, and a DSQ quadrupole mass spectrometer (ThermoElectron). GC was performed on a 30-m × 0.25-mm × 0.25-μm Zebron ZB-5ms MS column (Phenomenex). The PTV split technique was carried out as follows. Sugars were analyzed with the split 1:100, and the lower abundance compounds were analyzed with the split 1:10. PTV inlet temperature was set at 45°C, followed by the temperature program: hold at 45°C for 0.05 min, then raise to 70°C with ramp rate of 10°C s−1, hold at this temperature for 0.25 min, then followed by the transfer-to-column stage (raising to 270°C with ramp rate of 14.5°C s−1, hold at 270°C for 0.8 min), and followed by a cleaning stage (raising to 330°C with ramp rate of 10°C s−1, hold at 330°C for 10 min). The interface was heated to 300°C, and the ion source was adjusted to 250°C. Helium was used as the carrier gas at a flow rate of 1.2 mL min−1. The analysis was performed under the following temperature program: 1 min isothermal at 40°C, followed by a 15°C min−1 ramp to 320°C, and then holding this temperature for 4.5 min. Mass spectra were recorded at nine scans per second with an m/z 40 to 450 scanning range from 5 to 10 min and with an m/z 50 to 600 scanning range from 10 to 24 min. For the analysis of the low-abundance compounds, the filament was switched off from 12.95 to 13.60 min in order to prevent damage to the MS detector from the high concentration sugar compounds.
Both the reconstructed ion chromatograms and mass spectra were evaluated using the Xcalibur software version 1.4 (ThermoFinnigan). Compounds were identified by comparison of their retention index and mass spectra with those generated for authentic standards analyzed on our instrument. When the corresponding standards were not available, compounds were putatively identified by comparison of their retention index and mass spectra with those present in the mass spectra library of the Max-Planck-Institute for Plant Physiology in Golm, Germany (Q_MSRI_ID, http://csbdb.mpimp-golm.mpg.de/csbdb/gmd/msri/gmd_msri.html) and the commercial mass spectra library NIST05 (http://www.nist.gov/). The response values for metabolites resulting from the Xcalibur processing method were normalized to the ribitol internal standard.
Analysis of UPLC-QTOF-MS and GC-MS Data
The analysis of the raw UPLC-QTOF-MS data was performed using XCMS software (Smith et al., 2006) from the Bioconductor package (version 2.1) for the R statistical language (version 2.6.1). XCMS performs chromatogram alignment, mass signal detection, and peak integration. The XCMS set was constructed with the following parameters: fwhm = 10.8, step = 0.05, steps = 4, mzdiff = 0.07, snthresh = 8, and max = 1,000. Since injections of samples in the positive and negative ionization modes were performed in the separate injection sets, XCMS preprocessing was done for each ionization mode independently. The beginning of the chromatogram representing the column void volume and the last 3.5 min corresponding to column washing and equilibration were removed from analysis. To estimate the number of metabolites up-regulated in the peel in at least one of the developmental stages, statistical filtering was applied to the XCMS output. Peaks that were retained had to adhere to the following filters for each ripening stage: (1) there had to be at least a 2-fold difference between the means of the peel and the flesh repeats; and (2) the between-repeats variability had to be twice lower than the fold change between peel and flesh. The repeats variability criteria were calculated for each ripening stage according to the following formula and had to be greater than 2.
 |
To cluster together masses belonging to the same metabolite, a custom computer program implemented in MATLAB 7.3 was developed. The program accepts as an input the differential mass signals in both positive and negative ionization modes. The algorithm comprises two steps. First, based on a user-defined time shift, the retention time values are synchronized between the positive and negative modes. Next, using a greedy clustering process, the mass signals in a joined list of both modes are grouped according to the similarity in their abundance profiles across different samples and according to the proximity in their retention times. Pearson correlation is used as the distance measure.
Putatively assigned metabolites, analyzed by both UPLC-QTOF-MS (58 metabolites; Supplemental Table S6) and GC-MS (56 metabolites; Supplemental Table S7) were statistically treated using ANOVA. Two factors discriminated the metabolic samples: tissue type (peel versus flesh) and developmental stage (IG, MG, Br, Or, and Re). Therefore, a two-way ANOVA (P < 0.05) was chosen for the identification of differentially abundant metabolites between peel and flesh. Each metabolite subset consisted of 10 replicate triplets (three replicates for each sample). The RSD threshold score was determined according to the RSD 50% percentile of the whole data set. A single replicate outlier was removed from the triplet in case the deletion improved the RSD of the remaining replicate couple to below the RSD threshold. A whole triplet was removed in case the deletion of the outlier did not improve the RSD score. Differentiated metabolites with more than three deleted replicate triplets were removed from further analysis. Forty-five UPLC-QTOF-MS peel up-regulated metabolites and 25 GC-MS metabolites were retained following this preprocessing procedure. Average replicate values of the retained metabolites were used for further analysis.
Cuticular Wax and Cutin Analyses
For wax and cutin analysis, the cuticle was first extracted from manually dissected peels as described by Hovav et al. (2007). Cuticular waxes were extracted by immersing the peels (30–40 mg per sample) twice for 1 to 2 h into 5 mL of CHCl3 at room temperature. Both CHCl3 solutions were combined, and n-tetracosane was added as an internal standard. The solvent was removed under a gentle stream of nitrogen, and the remaining wax mixture was redissolved in 200 μL of CHCl3 and stored at 4°C until use. After the wax extraction, the remaining peels were depolymerized by the addition of 2 mL of BF3/methanol for 3 h at 70°C, and n-tetracosane was added as an internal standard. Then, after the addition of 2 mL of water, the cutin mixture was extracted three times with diethylether. The solvent of the combined extracts was removed under nitrogen, and the remaining cutin mixture was redissolved in 1,000 μL of CHCl3 and stored at 4°C until use. Fifty microliters of the wax or cutin samples was used for independent GC analyses. CHCl3 was evaporated from the samples under a nitrogen stream while heating to 50°C. Then, the wax or cutin mixtures were treated with bis-N,N-(trimethylsilyl)trifluoroacetamide (Sigma-Aldrich) in pyridine (30 min at 70°C) to transform all hydroxyl-containing compounds into the corresponding trimethylsilyl derivatives. The qualitative composition was studied with capillary GC (5890 II; Hewlett-Packard; 30-m DB-1, 0.32-mm i.d., film thickness = 1 μm; J&W Scientific) with He carrier gas inlet pressure for a constant flow of 1.4 mL min−1 and a mass spectrometric detector (5971; Hewlett-Packard). GC was carried out with temperature-programmed injection at 50°C, 2 min at 50°C, raised by 40°C min−1 to 200°C, held for 2 min at 200°C, raised by 3°C min−1 to 320°C, and held for 30 min at 320°C. The quantitative composition of the mixtures was studied using capillary GC-FID under the same GC conditions as described above, but the H2 carrier gas inlet pressure was programmed for a constant flow of 2 mL min−1. Single compounds were quantified against the internal standard by manual integration of peak areas.
Electron and Light Microscopy
Scanning electron microscopy was conducted as described by Panikashvili et al. (2007). For light microscopy, fruit tissues were fixed overnight in 2.5% paraformaldehyde solution and then washed for 30 min with 1% paraformaldehyde with 30% Suc. The tissues were embedded in 7% agarose LM-GQT (Laboratorios CONDA) and sliced using a Leica 2000 microtome with freezing. Sliced tissues were observed with an Olympus CLSM500 microscope.
Gene identifier numbers of all sequence data from this article can be found in Supplemental Tables S1 to S4.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Thirty gene expression profile clusters, composed of 574 nonredundant tomato peel-associated transcripts.
Supplemental Figure S2. Light microscopy images of stem peel cross sections reveals several layers of elongated collenchyma cells as well as occasional single parenchyma cells beneath the stem epidermis.
Supplemental Table S1. Gene annotations and putative pathways of the 574 nonredundant peel-associated transcripts included in the clustering analysis.
Supplemental Table S2. Peel/flesh expression ratios of transcripts presented in Table I (log2).
Supplemental Table S3. Affymetrix identifiers and The Institute for Genomic Research TCs/GenBank accession numbers of 140 transcripts that were found to be enriched (2-fold and higher) in tomato stem epidermis versus whole stem.
Supplemental Table S4. Peel-associated tomato transcripts (fruit and/or stem) of which homologs/orthologs are also enriched in epidermis-related tissue(s) of at least one other organism.
Supplemental Table S5. Distribution of differential mass signals detected by UPLC-QTOF-MS (in ESI-negative [−] and ESI-positive [+] modes) at different stages of fruit development.
Supplemental Table S6. List of putative metabolites identified in tomato fruit peel and flesh by UPLC-QTOF-MS (in ESI-negative [−] and ESI-positive [+] modes) at different stages of fruit development.
Supplemental Table S7. List of compounds detected in tomato fruit by GC-MS analysis.
Supplementary Material
Acknowledgments
We are grateful to Arye Tishbee for operating the LC-MS instrument, Dr. Eugenia Klein for her support with scanning electron microscopy, Sergey Malitsky for his assistance with metabolic profiling, and Hadar Less for his help with array data analysis. We also thank Ric de Vos for his hospitality in Plant Research International and assistance with the LC-MS analysis and the Tomato Genetics Resource Center for tomato seeds.
The work was supported by the William Z. and Eda Bess Novick Young Scientist Fund, Mrs. Louise Gartner, Dallas, TX, the Y. Leon Benoziyo Institute for Molecular Medicine, the European Union project META-PHOR (contract no. FOODCT–2006–036220), the Agriculture and Agri-Food Canada/Binational Agricultural Research and Development Fund (research project no. C–9105–06), and the Minerva Foundation. Work by R.J. and Z.W. was supported by a Special Research Opportunity Grant from the Natural Sciences and Engineering Research Council of Canada. A. Aharoni is an incumbent of the Adolfo and Evelyn Blum Career Development Chair.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Asaph Aharoni (asaph.aharoni@weizmann.ac.il).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Aharoni A, Dixit S, Jetter R, Thoenes E, van Arkel G, Pereira A (2004) The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16 2463–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alba R, Fei Z, Payton P, Liu Y, Moore SL, Debbie P, Cohn J, D'Ascenzo M, Gordon JS, Rose JK, et al (2004) ESTs, cDNA microarrays, and gene expression profiling: tools for dissecting plant physiology and development. Plant J 39 697–714 [DOI] [PubMed] [Google Scholar]
- Alba R, Payton P, Fei Z, McQuinn R, Debbie P, Martin GB, Tanksley SD, Giovannoni JJ (2005) Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. Plant Cell 17 2954–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida DPF, Huber DJ (2001) Transient increase in locular pressure and occlusion of endocarpic apertures in ripening tomato fruit. J Plant Physiol 158 199–203 [Google Scholar]
- Andrews J, Adams SR, Burton KS, Edmondson RN (2002) Partial purification of tomato fruit peroxidase and its effect on the mechanical properties of tomato fruit skin. J Exp Bot 53 2393–2399 [DOI] [PubMed] [Google Scholar]
- Andrews J, Malone M, Thompson DS, Ho LC, Burton KS (2000) Peroxidase isozyme patterns in the skin of maturing tomato fruit. Plant Cell Environ 23 415–422 [Google Scholar]
- Andrews PK, Fahy DA, Foyer CH (2004) Relationships between fruit exocarp antioxidants in the tomato (Lycopersicon esculentum) high pigment-1 mutant during development. Physiol Plant 120 519–528 [DOI] [PubMed] [Google Scholar]
- Arpat AB, Waugh M, Sullivan JP, Gonzales M, Frisch D, Main D, Wood T, Leslie A, Wing RA, Wilkins TA (2004) Functional genomics of cell elongation in developing cotton fibers. Plant Mol Biol 54 911–929 [DOI] [PubMed] [Google Scholar]
- Avila J, Nieto C, Canas L, Benito MJ, Paz-Ares J (1993) Petunia hybrida genes related to the maize regulatory C1 gene and to animal myb proto-oncogenes. Plant J 3 553–562 [DOI] [PubMed] [Google Scholar]
- Baker EA (1982) Chemistry and morphology of plant epicuticular waxes. In DF Cutler, KL Alvin, CE Price, eds, The Plant Cuticle. Academic Press, London, pp 139–165
- Bargel H, Neinhuis C (2004) Altered tomato (Lycopersicon esculentum Mill.) fruit cuticle biomechanics of a pleiotropic non ripening mutant. J Plant Growth Regul 23 61–75 [Google Scholar]
- Bargel H, Neinhuis C (2005) Tomato (Lycopersicon esculentum Mill.) fruit growth and ripening as related to the biomechanical properties of fruit skin and isolated cuticle. J Exp Bot 56 1049–1060 [DOI] [PubMed] [Google Scholar]
- Barleben L, Panjikar S, Ruppert M, Koepke J, Stockigt J (2007) Molecular architecture of strictosidine glucosidase: the gateway to the biosynthesis of the monoterpenoid indole alkaloid family. Plant Cell 19 2886–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer S, Schulte E, Thier HP (2004. a) Composition of the surface wax from tomatoes. I. Identification of the components by GC/MS. Eur Food Res Technol 219 223–228 [Google Scholar]
- Bauer S, Schulte E, Thier HP (2004. b) Composition of the surface wax from tomatoes. II. Quantification of the components at the ripe red stage and during ripening. Eur Food Res Technol 219 487–491 [Google Scholar]
- Bird D, Beisson F, Brigham A, Shin J, Greer S, Jetter R, Kunst L, Wu XW, Yephremov A, Samuels L (2007) Characterization of Arabidopsis ABCG11/WBC11, an ATP binding cassette (ABC) transporter that is required for cuticular lipid secretion. Plant J 52 485–498 [DOI] [PubMed] [Google Scholar]
- Blee E, Schuber F (1993) Biosynthesis of cutin monomers: involvement of a lipoxygenase peroxygenase pathway. Plant J 4 113–123 [Google Scholar]
- Bors W, Wachtveitl J, Saran M (1989) The mechanism of cytochrome C reduction by alkyl radicals: evidence for multiple reaction pathways. Free Radic Res Commun 6 251–256 [DOI] [PubMed] [Google Scholar]
- Bovy A, de Vos R, Kemper M, Schijlen E, Almenar Pertejo M, Muir S, Collins G, Robinson S, Verhoeyen M, Hughes S, et al (2002) High-flavonol tomatoes resulting from the heterologous expression of the maize transcription factor genes LC and C1. Plant Cell 14 2509–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broun P, Poindexter P, Osborne E, Jiang CZ, Riechmann JL (2004) WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc Natl Acad Sci USA 101 4706–4711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrari F, Baxter C, Usadel B, Urbanczyk-Wochniak E, Zanor M-I, Nunes-Nesi A, Nikiforova V, Centero D, Ratzka A, Pauly M, et al (2006) Integrated analysis of metabolite and transcript levels reveals the metabolic shifts that underlie tomato fruit development and highlight regulatory aspects of metabolic network behavior. Plant Physiol 142 1380–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Goodwin SM, Boroff VL, Liu X, Jenks MA (2003) Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and wax production. Plant Cell 15 1170–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SW, Lee SK, Kim EO, Oh JH, Yoon KS, Parris N, Hicks KB, Moreau RA (2007) Antioxidant and antimelanogenic activities of polyamine conjugates from corn bran and related hydroxycinnamic acids. J Agric Food Chem 55 3920–3925 [DOI] [PubMed] [Google Scholar]
- Davies KM, Schwinn KE (2003) Transcriptional regulation of secondary metabolism. Funct Plant Biol 30 913–925 [DOI] [PubMed] [Google Scholar]
- De Vos RCH, Moco S, Lommen A, Keurentjes JJB, Bino RJ, Hall RD (2007) Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat Protocols 2 778–791 [DOI] [PubMed] [Google Scholar]
- Dräger B (2006) Tropinone reductases, enzymes at the branch point of tropane alkaloid metabolism. Phytochemistry 67 327–337 [DOI] [PubMed] [Google Scholar]
- Duggar BM (1913) Lycopersicin, the red pigment of the tomato, and the effects of conditions upon its development. Wash Univ Studies 1 22–45 [Google Scholar]
- Eigenbrode SD, Espelie KE (1995) Effects of plant epicuticular lipids on insect herbivores. Annu Rev Entomol 40 171–194 [Google Scholar]
- Fei Z, Tang X, Alba RM, White JA, Ronning CM, Martin GB, Tanksley SD, Giovannoni JJ (2004) Comprehensive EST analysis of tomato and comparative genomics of fruit ripening. Plant J 40 47–59 [DOI] [PubMed] [Google Scholar]
- Fernie AR, Trethewey RN, Krotzky AJ, Willmitzer L (2004) Metabolite profiling: from diagnostics to systems biology. Nat Rev Mol Cell Biol 5 763–769 [DOI] [PubMed] [Google Scholar]
- Friedman M (2002) Tomato glycoalkaloids: role in the plant and in the diet. J Agric Food Chem 50 5751–5780 [DOI] [PubMed] [Google Scholar]
- Fry SC, Dumville JC, Miller JG (2001) Fingerprinting of polysaccharides attacked by hydroxyl radicals in vitro and in the cell walls of ripening pear fruit. Biochem J 357 729–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara Y, Takaki A, Uehara Y, Ikeda T, Okawa M, Yamauchi K, Ono M, Yoshimitsu H, Nohara T (2004) Tomato steroidal alkaloid glycosides, esculeosides A and B, from ripe fruits. Tetrahedron 60 4915–4920 [Google Scholar]
- Gillaspy G, Ben-David H, Gruissem W (1993) Fruits: a developmental perspective. Plant Cell 5 1439–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni JJ (2007) Fruit ripening mutants yield insights into ripening control. Curr Opin Plant Biol 10 283–289 [DOI] [PubMed] [Google Scholar]
- Giovannoni JJ, Noensie EN, Ruezinsky DM, Lu X, Tracy SL, Ganal MW, Martin GB, Pillen K, Alpert K, Tanksley SD (1995) Molecular genetic analysis of the ripening-inhibitor and non-ripening loci of tomato: a first step in genetic map-based cloning of fruit ripening genes. Mol Gen Genet 248 195–206 [DOI] [PubMed] [Google Scholar]
- Glover BJ, Perez-Rodriguez M, Martin C (1998) Development of several epidermal cell types can be specified by the same MYB-related plant transcription factor. Development 125 3497–3508 [DOI] [PubMed] [Google Scholar]
- Graca J, Schreiber L, Rodrigues J, Pereira H (2002) Glycerol and glyceryl esters of omega-hydroxyacids in cutins. Phytochemistry 61 205–215 [DOI] [PubMed] [Google Scholar]
- Heredia A (2003) Biophysical and biochemical characteristics of cutin, a plant barrier biopolymer. Biochim Biophys Acta 1620 1–7 [DOI] [PubMed] [Google Scholar]
- Hovav R, Chehanovsky N, Moy M, Jetter R, Schaffer AA (2007) The identification of a gene (Cwp1), silenced during Solanum evolution, which causes cuticle microfissuring and dehydration when expressed in tomato fruit. Plant J 52 627–639 [DOI] [PubMed] [Google Scholar]
- Hunt GM, Baker EA (1980) Phenolic constituents of tomato fruit cuticles. Phytochemistry 19 1415–1419 [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffé FW, Tattersall A, Glover BJ (2007) A truncated MYB transcription factor from Antirrhinum majus regulates epidermal cell outgrowth. J Exp Bot 58 1515–1524 [DOI] [PubMed] [Google Scholar]
- Jetter R, Kunst L, Samuels AL (2006) Composition of plant cuticular waxes. In M Riederer, C Muller, eds, Biology of the Plant Cuticle. Blackwell, Sheffield, UK, pp 145–181
- Jin HL, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19 6150–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowska M, Zimowski J, Paczkowski C, Wojciechowski ZA (2005) The formation of sugar chains in triterpenoid saponins and glycoalkaloids. Phytochem Rev 4 237–257 [Google Scholar]
- Kannangara R, Branigan C, Liu Y, Penfield T, Rao V, Mouille G, Hofte H, Pauly M, Riechmann JL, Broun P (2007) The transcription factor WIN1/SHN1 regulates cutin biosynthesis in Arabidopsis thaliana. Plant Cell 19 1278–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstiens G (1996. a) Cuticular water permeability and its physiological significance. J Exp Bot 47 1813–1832 [Google Scholar]
- Kerstiens G (1996. b) Signalling across the divide: a wider perspective of cuticular structure-function relationships. Trends Plant Sci 1 125–129 [Google Scholar]
- Kolattukudy PE (1980) Biopolyester membranes of plants: cutin and suberin. Science 208 990–1000 [DOI] [PubMed] [Google Scholar]
- Kolattukudy PE (1981) Structure, biosynthesis, and biodegradation of cutin and suberin. Annu Rev Plant Physiol Plant Mol Biol 32 539–567 [Google Scholar]
- Kolattukudy PE (2001) Polyesters in higher plants. Adv Biochem Eng Biotechnol 71 1–49 [DOI] [PubMed] [Google Scholar]
- Kranz HD, Denekamp M, Greco R, Jin H, Leyva A, Meissner RC, Petroni K, Urzainqui A, Bevan M, Martin C, et al (1998) Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J 16 263–276 [DOI] [PubMed] [Google Scholar]
- Krolikowski KA, Victor JL, Wagler TN, Lolle SJ, Pruitt RE (2003) Isolation and characterization of the Arabidopsis organ fusion gene HOTHEAD. Plant J 35 501–511 [DOI] [PubMed] [Google Scholar]
- Kurdyukov S, Faust A, Trenkamp S, Bar S, Franke R, Efremova N, Tietjen K, Schreiber L, Saedler H, Yephremov A (2006) Genetic and biochemical evidence for involvement of HOTHEAD in the biosynthesis of long-chain alpha,omega-dicarboxylic fatty acids and formation of extracellular matrix. Planta 224 315–329 [DOI] [PubMed] [Google Scholar]
- Leide J, Hildebrandt U, Reussing K, Riederer M, Vogg G (2007) The developmental pattern of tomato fruit wax accumulation and its impact on cuticular transpiration barrier properties: effects of a deficiency in a beta-ketoacyl-coenzyme A synthase (LeCER6). Plant Physiol 144 1667–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire-Chamley M, Petit J, Garcia V, Just D, Baldet P, Germain V, Fagard M, Mouassite M, Cheniclet C, Rothan C (2005) Changes in transcriptional profiles are associated with early fruit tissue specialization in tomato. Plant Physiol 139 750–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Beisson F, Koo AJ, Molina I, Pollard M, Ohlrogge J (2007) Identification of acyltransferases required for cutin biosynthesis and production of cutin with suberin-like monomers. Proc Natl Acad Sci USA 104 18339–18344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Hamilton WDO, Merryweather A (1996) Cloning and initial characterization of 14 myb-related cDNAs from tomato (Lycopersicon esculentum cv Ailsa Craig). Plant Mol Biol 30 1009–1020 [DOI] [PubMed] [Google Scholar]
- Lindstrom EW (1925) Inheritance in tomatoes. Genetics 10 305–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Casado G, Matas AJ, Domínguez E, Cuartero J, Heredia A (2007) Biomechanics of isolated tomato (Solanum lycopersicum L.) fruit cuticles: the role of the cutin matrix and polysaccharides. J Exp Bot 58 3875–3883 [DOI] [PubMed] [Google Scholar]
- Luo B, Xue XY, Hu WL, Wang LJ, Chen XY (2007) An ABC transporter gene of Arabidopsis thaliana, AtWBC11, is involved in cuticle development and prevention of organ fusion. Plant Cell Physiol 48 1790–1802 [DOI] [PubMed] [Google Scholar]
- Luque P, Bruque S, Heredia A (1995) Water permeability of isolated cuticular membranes: a structural analysis. Arch Biochem Biophys 317 417–422 [DOI] [PubMed] [Google Scholar]
- Mattanovich D, Borth N (2006) Applications of cell sorting in biotechnology. Microb Cell Fact 5 12–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner R, Jacobson Y, Melamed S, Levyatuv S, Shalev G, Ashri A, Elkind Y, Levy A (1997) A new model system for tomato genetics. Plant J 12 1465–1472 [Google Scholar]
- Millar AA, Clemens S, Zachgo S, Giblin EM, Taylor DC, Kunst L (1999) CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell 11 825–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moco S, Bino RJ, Vorst O, Verhoeven HA, de Groot J, van Beek TA, Vervoort J, de Vos CH (2006) A liquid chromatography mass spectrometry based metabolome database for tomato. Plant Physiol 141 1205–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moctezuma E, Smith DL, Gross KC (2003) Antisense suppression of a beta-galactosidase gene (TBG6) in tomato increases fruit cracking. J Exp Bot 54 2025–2033 [DOI] [PubMed] [Google Scholar]
- Montgomery J, Goldman S, Deikman J, Margossian L, Fischer RL (1993) Identification of an ethylene-responsive region in the promoter of a fruit ripening gene. Proc Natl Acad Sci USA 90 5939–5943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore S, Payton P, Wright M, Tanksley S, Giovannoni J (2005) Utilization of tomato microarrays for comparative gene expression analysis in the Solanaceae. J Exp Bot 56 2885–2895 [DOI] [PubMed] [Google Scholar]
- Muir SR, Collins GJ, Robinson S, Hughes S, Bovy A, Ric De Vos CH, van Tunen AJ, Verhoeyen ME (2001) Overexpression of petunia chalcone isomerase in tomato results in fruit containing increased levels of flavonols. Nat Biotechnol 19 470–474 [DOI] [PubMed] [Google Scholar]
- Nakazono M, Qiu F, Borsuk LA, Schnable PS (2003) Laser-capture microdissection, a tool for the global analysis of gene expression in specific plant cell types: identification of genes expressed differentially in epidermal cells or vascular tissues of maize. Plant Cell 15 583–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C (2006) Unraveling the complex network of cuticular structure and function. Curr Opin Plant Biol 9 281–287 [DOI] [PubMed] [Google Scholar]
- Niggeweg R, Michael AJ, Martin C (2004) Engineering plants with increased levels of the antioxidant chlorogenic acid. Nat Biotechnol 22 746–754 [DOI] [PubMed] [Google Scholar]
- Noda K, Glover BJ, Linstead P, Martin C (1994) Flower color intensity depends on specialized cell-shape controlled by a Myb-related transcription factor. Nature 369 661–664 [DOI] [PubMed] [Google Scholar]
- Panikashvili D, Savaldi-Goldstein S, Mandel T, Yifhar T, Franke RB, Hofer R, Schreiber L, Chory J, Aharoni A (2007) The Arabidopsis DESPERADO/AtWBC11 transporter is required for cutin and wax secretion. Plant Physiol 145 1345–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Rodriguez M, Jaffe FW, Butelli E, Glover BJ, Martin C (2005) Development of three different cell types is associated with the activity of a specific MYB transcription factor in the ventral petal of Antirrhinum majus flowers. Development 132 359–370 [DOI] [PubMed] [Google Scholar]
- Pighin JA, Zheng H, Balakshin LJ, Goodman IP, Western TL, Jetter R, Kunst L, Samuels AL (2004) Plant cuticular lipid export requires an ABC transporter. Science 306 702–704 [DOI] [PubMed] [Google Scholar]
- Qin YM, Hu CY, Pang Y, Kastaniotis AJ, Hiltunen JK, Zhu YX (2007. a) Saturated very-long-chain fatty acids promote cotton fiber and Arabidopsis cell elongation by activating ethylene biosynthesis. Plant Cell 19 3692–3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin YM, Pujol FM, Hu CY, Feng JX, Kastaniotis AJ, Hiltunen JK, Zhu YX (2007. b) Genetic and biochemical studies in yeast reveal that the cotton fibre-specific GhCER6 gene functions in fatty acid elongation. J Exp Bot 58 473–481 [DOI] [PubMed] [Google Scholar]
- Reina JJ, Heredia A (2001) Plant cutin biosynthesis: the involvement of a new acyltransferase. Trends Plant Sci 6 296. [DOI] [PubMed] [Google Scholar]
- Rick CM, Uhlig JW, Jones AD (1994) High alpha-tomatine content in ripe fruit of Andean Lycopersicon esculentum var. cerasiforme: developmental and genetic aspects. Proc Natl Acad Sci USA 91 12877–12881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riederer M, Burghardt M (2006) Cuticular transpiration. In M Riederer, C Muller, eds, Biology of the Plant Cuticle. Blackwell Publishing, Oxford, UK, pp 292–311
- Riederer M, Schreiber L (2001) Protecting against water loss: analysis of the barrier properties of plant cuticles. J Exp Bot 52 2023–2032 [DOI] [PubMed] [Google Scholar]
- Roessner-Tunali U, Hegemann B, Lytovchenko A, Carrari F, Bruedigam C, Granot D, Fernie AR (2003) Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol 133 84–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JKC, Bashir S, Giovannoni JJ, Jahn MM, Saravanan RS (2004) Tackling the plant proteome: practical approaches, hurdles and experimental tools. Plant J 39 715–733 [DOI] [PubMed] [Google Scholar]
- Rowland O, Zheng HQ, Hepworth SR, Lam P, Jetter R, Kunst L (2006) CER4 encodes an alcohol-forming fatty acyl-coenzyme A reductase involved in cuticular wax production in Arabidopsis. Plant Physiol 142 866–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saladie M, Matas AJ, Isaacson T, Jenks MA, Goodwin SM, Niklas KJ, Xiaolin R, Labavitch JM, Shackel KA, Fernie AR, et al (2007) A reevaluation of the key factors that influence tomato fruit softening and integrity. Plant Physiol 144 1012–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel Yang S, Cheung F, Lee JJ, Ha M, Wei NE, Sze SH, Stelly DM, Thaxton P, Triplett B, Town CD, et al (2006) Accumulation of genome-specific transcripts, transcription factors and phytohormonal regulators during early stages of fiber cell development in allotetraploid cotton. Plant J 47 761–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels L, Jetter R, Kunst L (2005) First steps in understanding the export of lipids to the plant cuticle. Plant Biosyst 139 65–68 [Google Scholar]
- Schijlen EG, de Vos CH, Martens S, Jonker HH, Rosin FM, Molthoff JW, Tikunov YM, Angenent GC, van Tunen AJ, Bovy AG (2007) RNA interference silencing of chalcone synthase, the first step in the flavonoid biosynthesis pathway, leads to parthenocarpic tomato fruits. Plant Physiol 144 1520–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnurr J, Shockey J, Browse J (2004) The acyl-CoA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. Plant Cell 16 629–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonherr J (1976) Naphthalene acetic acid permeability of Citrus leaf cuticle. Biochem Physiol Pflanz 170 309–319 [Google Scholar]
- Sharples SC, Fry SC (2007) Radioisotope ratios discriminate between competing pathways of cell wall polysaccharide and RNA biosynthesis in living plant cells. Plant J 52 252–262 [DOI] [PubMed] [Google Scholar]
- Shi YH, Zhu SW, Mao XZ, Feng JX, Qin YM, Zhang L, Cheng J, Wei LP, Wang ZY, Zhu YX (2006) Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell 18 651–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinbo Y, Nakamura Y, Altaf-Ul-Amin M, Asahi H, Kurokawa K, Arita M, Saito K, Ohta D, Shibata D, Kanaya S (2006) KNApSAcK: a comprehensive species-metabolite relationship database. In K Saito, RA Dixon, L Willmitzer, eds, Biotechnology in Agriculture and Forestry, Plant Metabolomics, Vol 57. Springer-Verlag, Berlin, pp 165–181
- Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G (2006) XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem 78 779–787 [DOI] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4 447–456 [DOI] [PubMed] [Google Scholar]
- Suh MC, Samuels AL, Jetter R, Kunst L, Pollard M, Ohlrogge J, Beisson F (2005) Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiol 139 1649–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamagnone L, Merida A, Parr A, Mackay S, Culianez-Macia FA, Roberts K, Martin C (1998) The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 10 135–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DS (2001) Extensiometric determination of the rheological properties of the epidermis of growing tomato fruit. J Exp Bot 52 1291–1301 [PubMed] [Google Scholar]
- Thompson DS, Davies WJ, Ho LC (1998) Regulation of tomato fruit growth by epidermal cell wall enzymes. Plant Cell Environ 21 589–599 [Google Scholar]
- Torres CA, Andrews PK (2006) Developmental changes in antioxidant metabolites, enzymes, and pigments in fruit exocarp of four tomato (Lycopersicon esculentum Mill.) genotypes: beta-carotene, high pigment-1, ripening inhibitor, and ‘Rutgers’. Plant Physiol Biochem 44 806–818 [DOI] [PubMed] [Google Scholar]
- Torres CA, Andrews PK, Davies NM (2006) Physiological and biochemical responses of fruit exocarp of tomato (Lycopersicon esculentum Mill.) mutants to natural photo-oxidative conditions. J Exp Bot 57 1933–1947 [DOI] [PubMed] [Google Scholar]
- Ukitsu H, Kuromori T, Toyooka K, Goto Y, Matsuoka K, Sakuradani E, Shimizu S, Kamiya A, Imura Y, Yuguchi M, et al (2007) Cytological and biochemical analysis of cof1, an Arabidopsis mutant of an ABC transporter gene. Plant Cell Physiol 48 1524–1533 [DOI] [PubMed] [Google Scholar]
- Vanengelen FA, Molthoff JW, Conner AJ, Nap JP, Pereira A, Stiekema WJ (1995) Pbinplus: an improved plant transformation vector based on Pbin19. Transgenic Res 4 288–290 [DOI] [PubMed] [Google Scholar]
- van Houwelingen A, Souer E, Spelt K, Kloos D, Mol J, Koes R (1998) Analysis of flower pigmentation mutants generated by random transposon mutagenesis in Petunia hybrida. Plant J 13 39–50 [DOI] [PubMed] [Google Scholar]
- Verhoeyen ME, Bovy A, Collins G, Muir S, Robinson S, de Vos CH, Colliver S (2002) Increasing antioxidant levels in tomatoes through modification of the flavonoid biosynthetic pathway. J Exp Bot 53 2099–2106 [DOI] [PubMed] [Google Scholar]
- Verwoerd TC, Dekker BM, Hoekema A (1989) A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res 17 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogg G, Fischer S, Leide J, Emmanuel E, Jetter R, Levy AA, Riederer M (2004) Tomato fruit cuticular waxes and their effects on transpiration barrier properties: functional characterization of a mutant deficient in a very-long-chain fatty acid beta-ketoacyl-CoA synthase. J Exp Bot 55 1401–1410 [DOI] [PubMed] [Google Scholar]
- Wellesen K, Durst F, Pinot F, Benveniste I, Nettesheim K, Wisman E, Steiner-Lange S, Saedler H, Yephremov A (2001) Functional analysis of the LACERATA gene of Arabidopsis provides evidence for different roles of fatty acid omega-hydroxylation in development. Proc Natl Acad Sci USA 98 9694–9699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney KD (2005) Linking frugivores to the dynamics of a fruit color polymorphism. Am J Bot 92 859–867 [DOI] [PubMed] [Google Scholar]
- Xu R, Fazio GC, Matsuda SPT (2004) On the origins of triterpenoid skeletal diversity. Phytochemistry 65 261–291 [DOI] [PubMed] [Google Scholar]
- Yazaki K (2006) ABC transporters involved in the transport of plant secondary metabolites. FEBS Lett 580 1183–1191 [DOI] [PubMed] [Google Scholar]
- Zhao J, Zhang W, Zhao Y, Gong X, Guo L, Zhu G, Wang X, Gong Z, Schumaker KS, Guo Y (2007) SAD2, an importin-like protein, is required for UV-B response in Arabidopsis by mediating MYB4 nuclear trafficking. Plant Cell 19 3805–3818 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.