Abstract
The mapping of high-dimensional olfactory stimuli onto the two-dimensional surface of the nasal sensory epithelium constitutes the first step in the neuronal encoding of olfactory input. We have used zebrafish as a model system to analyze the spatial distribution of odorant receptor molecules in the olfactory epithelium by quantitative in situ hybridization. To this end, we have cloned 10 very divergent zebrafish odorant receptor molecules by PCR. Individual genes are expressed in sparse olfactory receptor neurons. Analysis of the position of labeled cells in a simplified coordinate system revealed three concentric, albeit overlapping, expression domains for the four odorant receptors analyzed in detail. Such regionalized expression should result in a corresponding segregation of functional response properties. This might represent the first step of spatial encoding of olfactory input or be essential for the development of the olfactory system.
Keywords: sensory system, smell, teleost, in situ hybridization, expression zones
How different odors are distinguished and recognized in the olfactory nervous system is an intriguing question. The primary event of olfaction is the interaction of odorants with a large family of receptor molecules. These receptors have been cloned by an elegant approach based on their affiliation to the heptahelical family (1). With two possible exceptions (2, 3), the response spectra of these molecules to odorants are not known so far. Analysis of the spatial distribution of odorant receptor expression, nevertheless, has allowed investigators to infer some features of information processing in the olfactory epithelium (4). Individual odorant receptor molecules are expressed in a tiny percentage of receptor neurons, statistically excluding expression of more than one or, at most, a few of these receptor molecules in an individual neuron. Receptor neurons expressing individual odorant receptor molecules exhibit a scattered distribution (5–7). That is, a particular receptor specificity is not clustered on the sensory surface (as in the visual and auditory system), but distributed. Nevertheless, expression of odorant receptors in the mammalian olfactory epithelium is not random, but segregated into at least four zones (6, 7). Functional response properties of the epithelium are correspondingly segregated in mammalian (8) as well as fish species (9). It is therefore puzzling that a study on catfish odorant receptors suggested a homogeneous, non-zonal expression pattern (10).
In addition to the zonally restricted distribution of receptor specificities on the level of the epithelium, the axonal projection from the sensory surface toward the olfactory bulb is also segregated, at least in rat, where a rough quadrant-to-quadrant topology is found (11). These quadrants may be related to the zones of odorant receptor expression (12).
So far it is not known whether the unusual regulation of expression in mammalian species (disperse, but zonal) might have a function in olfactory coding or for the development of the olfactory system. If zonal expression of odorant receptors is essential for the organization of the olfactory system, it would be expected to be conserved in distantly related species. To test this expectation and to examine the basis for possible mechanisms governing the expression of odorant receptor molecules, we have quantitatively analyzed the spatial distribution of odorant receptor molecules in the zebrafish olfactory epithelium. Zebrafish, Danio rerio, is a lower vertebrate that possesses a presumably less complex, but nevertheless well-developed, sense of smell (13, 14).
We report the cloning of 10 odorant receptors from zebrafish and the quantitative analysis of their in situ hybridization patterns. We find a spatial segregation of odorant receptors in the zebrafish olfactory epithelium into at least three overlapping, but clearly distinct, expression domains.
MATERIALS AND METHODS
Zebrafish Strains and Maintenance.
The wild-type strain Tü was used for the generation of PCR fragment clones. In situ hybridizations were performed with the Tü and the Ab strains, their F1 progeny, and fish of heterogenous genetic background from a local pet shop. Adult fish aged between 6 months and 1.5 years were used. The strains were raised in an automated tank system (15).
PCR Amplification of Genomic DNA with Odorant Receptor-Specific Primers.
Each PCR used 50 ng of purified genomic DNA. An alignment of 74 members of the G protein-coupled receptor superfamily (16) served to identify highly conserved stretches of amino acids. Ambiguous amino acids and codon degeneracy were taken into account by introduction of inosines and degeneracy into 27- to 30-mers. Several different oligonucleotides were synthesized against the intracellular region adjacent to transmembrane domain III (5′ primer) and against the N-terminal half of transmembrane domain VII (3′ primer). The oligonucleotides with the highest yield were 5′-ATGGCITA(T/C)GA(T/C)(A/C)GITA(T/C)GTIGCIATITG-3′ (5′ primer) and 5′-(G/A)TTIC(G/T)IA(G/A)IGT(G/A)TAIAT(G/A)A(A/T)IGG(G/A)TTIA-3′ (3′ primer). PCR was performed with AmpliTaq polymerase (Perkin–Elmer) as follows: five cycles of 60 sec at 96°C, 2-min ramp, 2 min at 40–45°C, 1-min ramp, and 30 sec at 72°C; followed by 25 cycles of 30 sec at 94°C, 2-min ramp, 1 min at 45–55°C, 1-min ramp, and 30 sec at 72°C with 2-sec extension per cycle. Five percent of this reaction was further amplified with new reagents (booster PCR) as follows: 20 cycles of 30 sec at 94°C, 2-min ramp, 1 min at 45–55°C, 1-min ramp, and 30 sec at 72°C with 5-sec extension per cycle.
Cloning and Sequencing of PCR Products.
PCR products were blunted with T4 DNA polymerase and ligated into the SmaI restriction site of pBluescript II KS (Stratagene). E. coli strain DH5α (GIBCO/BRL) was chemically transformed and ≈1000 insert-containing clones identified by blue/white selection. Insert size was determined by analytical PCR with vector primers. About two-thirds of the clones fell within the 500- to 600-bp range expected for odorant receptor molecules by comparison with rat sequences (1). Further grouping was done by comparing restriction fragment patterns (HaeIII, PvuII, RsaI, and TaqI) for these clones. Fifty clones with substantially different sizing patterns were partially sequenced with the automated laser fluorescence (ALF) sequencing system (Pharmacia). Ten different sequences were obtained and sequenced in both directions. In four cases, the corresponding cDNA clones were obtained from zebrafish nasal epithelium and found to be completely identical (S.K., unpublished observation). The ten clones were named fZOR1-10 for fragment of zebrafish odorant receptor 1–10.
In Situ Hybridization with Odorant Receptor Probes.
For in situ hybridization, the epithelium was sectioned parallel to its base (cf. Fig. 1B). Cryostat sections (14 μm) of fresh-frozen adult zebrafish nasal epithelium embedded in TissueTec (Miles) were thaw-mounted onto Vectabond-coated slides (Vector), dried for 3 hr at 55°C, and fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS; pH 7.4) for 10 min at room temperature. Sections were permeabilized in 0.2 M HCl for 10 min, digested with 1 μg of proteinase K per ml (Boehringer Mannheim) in 0.1 M Tris·HCl (pH 8.0) for 7.5 min at 37°C, and treated with 5 mM acetanhydride in 0.1 M triethanolamine (pH 8.0) for 10 min. Between individual steps, slides were rinsed with PBS. Digoxigenylated riboprobes were used for hybridization at a concentration of 3–5 μg/ml. Sense riboprobe was used as negative control. No labeling was observed with sense probes. Most of the multiple cloning site of the pBluescript vector between the RNA polymerase promoter and the insert had to be removed for the synthesis of riboprobes, because it generated an unacceptably high background signal (F.W., unpublished observation; ref. 17). Hybridization was performed overnight at 60°C in 50% formamide, 5× Denhardt’s reagent, 5× standard saline citrate (SSC; 20× SSC is 3 M NaCl and 0.3 M sodium citrate, pH 7.0), 0.4 mg of proteinase K-treated torula yeast RNA per ml (Type VI, Sigma), and 0.1 mg of tRNA from bakers’ yeast per ml. Washes were as follows: 30 min with 50% formamide/2× SSC at 60°C, 1 hr with 0.2× SSC at 60°C, and 15 min with 0.2× SSC at room temperature. Detection of bound probe was performed essentially as described in ref. 18 using nitroblue tetrazolium chloride/5-bromo-4-chloro-3-indolyl-phosphate as color substrate.
Figure 1.
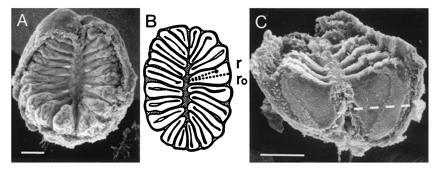
Spatial organization of the zebrafish nasal epithelium. (A) Scanning electron micrograph of an olfactory rosette of an adult zebrafish. The lamellae depart from a central (nonsensory) raphe. Long, regular, and dense, but sometimes patchy, cilia mark the nonsensory region of the epithelium. Olfactory receptor cells have fewer and shorter cilia and are homogenously distributed on the flat sides in the inner two-thirds of the lamellae (C). (Bar = 100 μm.) (B) Schematic representation of a horizontal cross-section through an olfactory rosette. The lamellae are cut perpendicular to their flat faces. This plane of section is used for all in situ hybridizations. Each lamella consists of two half-lamellae separated by extracellular matrix (gray). The lumen (interface to the water) is surrounded by a black line. The measurement of radial distances (r/ro) is visualized. (C) The dashed line is at relative height 0.60, the cutoff for analysis of radial distances. (Bar = 100 μm.)
The stringency of in situ hybridization employed here is ≈5°C below hybrid melting temperature, as determined in initial experiments. As a consequence of the high stringency, the probes presumably recognize only their cognate and very closely related genes. fZOR5 and fZOR9 both represent single genes, and fZOR6 and fZOR8 are members of several-gene subfamilies (F.W., unpublished observation).
Measuring the Distribution of Odorant Receptor-Positive Cells.
The distribution of receptor neurons labeled with a single odorant receptor probe was assessed in complete series of sections of nasal epithelium. Images of tissue sections were acquired with an Axiophot microscope (Zeiss) with Nomarski optics fitted with a color charged coupled device camera (Sony DXC-930p). Images were analyzed with National Institutes of Health image version 1.52. Labeled cells were evaluated with respect to radial position. Sections were counted from the top of the epithelium, beginning with the first section containing labeled cells and ending with the last such section. To account for differences in the overall size of epithelia, these z values were normalized to percent relative height (0% corresponding to the top). Radial position is determined as distance from the inner curve of sensory epithelium, measured along the lamella and divided by total length of the lamella (Fig. 1B). For presentation of data, only sections between 0% and 60% relative height were summed together, since the more basal sections contain disproportionally less sensory epithelium (Fig. 1C).
Statistical Analysis of Radial Distributions.
The observed radial distributions were analyzed using bootstrap methods (19). Though being computationally more demanding than usual statistical methods, bootstrap methods do not require any assumptions about the type of the underlying distribution. This feature is of particular advantage in view of the nonnormal character of the observed spatial distributions. Radial distributions for individual animals were characterized by their mean, variance, and skewness. For each of the receptors, the expectation values and their confidence intervals for mean, variance, and skewness were determined from their distribution in a set of 104 bootstrap samples (19). A bootstrap sample of an original data group of N points is obtained by randomly drawing N data points, with replacement, from those original data. Expectation values are determined from averaging over the corresponding sets of bootstrap samples. Confidence intervals were obtained from the 2.5 and 97.5 percentiles of the respective distributions in the set of bootstrap samples, giving a 95% confidence interval. In addition, the level of significance of the difference of radial distributions was obtained by permutation bootstrap tests (19, 20), using the difference of sample means and skewness as test quantities.
RESULTS
Cloning of Odorant Receptors.
Genomic DNA of a single zebrafish was amplified by PCR using primers homologous to rat odorant receptors and subsequently cloned in the plasmid vector pBluescript II KS. A family of 10 putative odorant receptor gene fragments was obtained. The deduced amino acid sequences are shown in Fig. 2. These receptors possess most of the highly conserved sequence motifs common to the superfamily of G protein-coupled receptors, and, furthermore, they exhibit several of the motifs characteristic of the family of odorant receptors but not conserved in other families (16). In a hydrophobicity plot, all 10 fragments show the expected four hydrophobic peaks—i.e., transmembrane domains IV, V, VI, and VII (data not shown). We conclude that these 10 clones belong to the group of odorant receptors within the family of G protein-coupled heptahelical receptors. The cloned family is highly divergent, with average amino acid homology of only 29% and minimal and maximal values of 20% and 46% (fZOR5 and fZOR8, respectively). Southern blot analysis (data not shown) indicates that each of the cloned fragments is a member of a different subfamily consisting of one to several genes.
Figure 2.
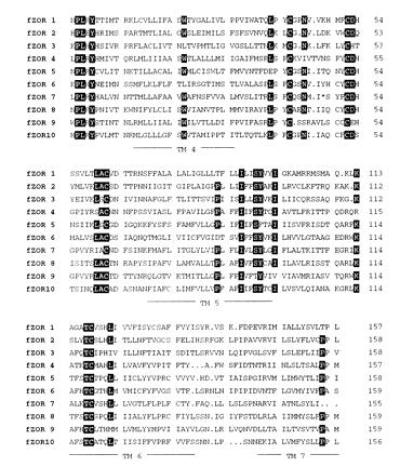
Deduced amino acid sequences of 10 zebrafish odorant receptor molecule fragments fZOR1-10. Amino acids are depicted in one-letter code. The asterisk in fZOR7 stands for a stop codon that is also present in the gene and may be due to a recent inactivation of the gene, since mRNA for fZOR7 is present at normal levels. Sequences were aligned using the UPMGAA algorithm (GCG). Dots indicate gaps inserted for optimal alignment. The predicted positions of transmembrane domains IV–VII are underlined. Amino acids conserved 80% or more are depicted by inverse letters.
Scattered Distribution of Odorant Receptor-Positive Olfactory Receptor Neurons.
We analyzed the expression of the gene family at the cellular level by in situ hybridization of sections of fresh-frozen nasal epithelium with fZOR1-10. All the receptors labeled a sparse population of cells that was restricted to the inner two-thirds of the lamellar length, the region of olfactory epithelium proper, as defined by backtracing from the olfactory bulb (21). This distribution is consistent with the interpretation that the ZOR family is expressed specifically in olfactory neurons and thereby supports the notion that fZOR1-10 represent odorant receptor molecules. Further support is derived from the cloning of several of the corresponding full-length molecules from a cDNA library of nasal epithelium (S.K., unpublished observation).
The frequency of labeled cells ranged between 100 and 500 cells per nasal epithelium. Interindividual variation was at least 2-fold and conceivably might be related to individual differences in perception thresholds (14). The numbers refer not necessarily to individual genes, since hybridization stringency, albeit high (see Materials and Methods), would still allow cross-hybridization to closely related subfamily members. The total number of olfactory receptor neurons is estimated to be 25,000—i.e., maximally 10% of all receptor neurons are labeled with our probes. The family size may therefore be several-fold larger, but perhaps not >100 genes. This estimate is in good accordance with data from Southern blot analysis (F.W., unpublished observation).
No significant differences in labeling frequency of an individual odorant receptor probe were seen between female and male epithelia or between left and right epithelia of one individual.
Separate Expression Domains for Odorant Receptors.
In initial observations, we noticed for several odorant receptors that in some sections labeled cells were arranged neatly in concentric rings around the median raphe (Fig. 3 B–D). The diameter of these rings was different for different odorant receptors. With some probes annular arrangement is not conspicuous (e.g., fZOR6 in Fig. 3A), but, nevertheless, in all individual epithelia examined, a preferred distance from the median raphe was observed. On closer inspection, we found that these annular patterns were more pronounced in the apical portion of the bowl-shaped olfactory epithelium, close to the opening of the organ, but often were found also in the basal sections.
Figure 3.
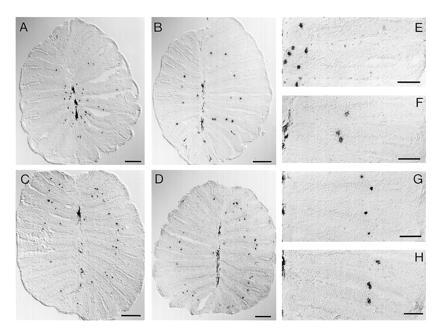
Spatial expression patterns of odorant receptor molecules. Horizontal tissue sections of fresh-frozen nasal epithelium were hybridized with digoxigenylated riboprobes of fZOR6 (A and E), fZOR9 (B and F), fZOR8 (C and G), and fZOR5 (D and H). For orientation, compare Fig. 1. In A–D, the complete sections are depicted (cf. Fig. 1B). (Bar = 100 μm.) (E–H) Photomicrographs taken at higher magnification. (Bar = 50 μm.) Melanophores in the median raphe are visible as elongated dark stain (left edge of micrograph). Labeled cells are small, roundish, and dark.
Next a statistical analysis of the annular expression pattern was performed for four different odorant receptor probes (fZOR5, fZOR6, fZOR8, and fZOR9). The position of labeled cells along the lamella was measured, and the relative radial position was determined as described in Materials and Methods. Distributions of fZOR5, fZOR6, and fZOR9 are found to be clearly distinct, whereas fZOR5 and fZOR8 show a very similar pattern (Fig. 4). fZOR6 occupies an inner ring, fZOR9 is found at an intermediate radial position, and fZOR5 and fZOR8 are preferentially found close to the outer margin of the sensory region [at radial coordinate 0.7, as defined by a careful morphological examination of the sections and analysis of retrogradely labeled receptor neuron distribution (21); and data not shown]. The radial coordinate for peak frequency is 0.15 for fZOR6, 0.45 for fZOR9, 0.50 for fZOR8, and 0.55 for fZOR5. The pattern of distribution is reproducible between animals for a given odorant receptor. As a further control, we have performed in situ hybridization with fZOR5 and fZOR9 on alternating sections of the same epithelia. Again, we observe clearly distinct preferred radii for these two receptors (data not shown). This pattern of distribution provides for spatial segregation in the sense that there exist clearly separate frequency peaks for three of the four odorant receptors (fZOR6, fZOR9, and fZOR5). fZOR5 and fZOR8 may share a domain, as discussed below. Nevertheless, each odorant receptor is expressed along the whole extent of the olfactory epithelium, albeit with lower frequency outside its preferred region.
Figure 4.
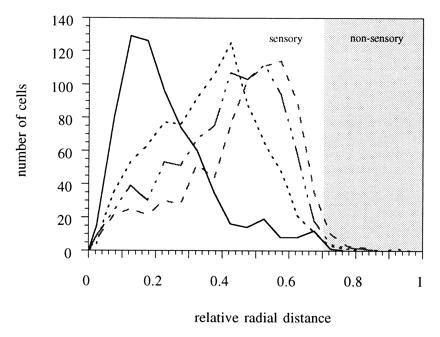
Radial distribution of cells labeled by in situ hybridization. Cells were labeled by in situ hybridization with fZOR6 (——), fZOR9 (···), fZOR8 (— · —), and fZOR5 (---). Their radial distances were determined within each section and normalized as described. The relative radii were summed for three to seven nasal epithelia per receptor, corresponding to 600–900 labeled cells, and the resulting data are presented with a bin size of 0.05. Data shown are all obtained from fish of genotype Ab/Tü. The annular expression seen here (a distinct maximum of labeled cells at a particular radial position) was also observed with fish from heterogenous genetic background.
To evaluate the significance of differences of the observed distributions, we performed bootstrap analysis for three parameters of the distribution, mean, variance, and skewness, as described in Materials and Methods. Table 1 depicts expectation values and confidence intervals thus obtained for all four experimentally measured distributions. Confidence intervals for distribution means for fZOR6, fZOR9, and fZOR5 do not overlap—i.e., these distributions are significantly different (P < 0.01 from permutation analysis). This conclusion is supported by the analysis of skewness of these three distributions, which is very cleary distinct for each of the three receptors (Table 1). Incidentally, the receptor expressed preferentially at intermediate radial positions (fZOR9) shows a quite symmetrical distribution, whereas those receptors expressed at the extreme innermost (fZOR6) and outermost (fZOR5) regions of the sensory epithelium are highly skewed in opposite directions. Presumably the discontinuation of the sensory region causes a steep drop in frequency of labeled cells. Therefore the skewness may also be a good indicator of radial position, besides the more obvious mean.
Table 1.
Statistical properties of radial distributions
| Receptor | Mean | Variance | Skewness |
|---|---|---|---|
| fZOR6 | 0.226 | 0.0181 | 1.08 |
| (0.206 to 0.241) | (0.0112 to 0.0244) | (0.818 to 1.30) | |
| fZOR9 | 0.354 | 0.0213 | −0.0608 |
| (0.342 to 0.366) | (0.0192 to 0.0236) | (−0.109 to −0.0290) | |
| fZOR8 | 0.414 | 0.0248 | −0.454 |
| (0.396 to 0.437) | (0.0226 to 0.0271) | (−0.770 to −0.210) | |
| fZOR5 | 0.456 | 0.0255 | −0.858 |
| (0.434 to 0.477) | (0.0227 to 0.0271) | (−1.08 to −0.635) |
The radial distribution of labeled cells was determined for individual epithelia for each of the four receptors fZOR6, fZOR9, fZOR8, and fZOR5. Mean, variance, and skewness of these distributions were obtained. Expectation values for these parameters, together with 95% confidence intervals (in parentheses), were obtained by bootstrap analysis. Nonoverlapping confidence intervals indicate a difference significant at P < 0.05. Note that values for the means differ from the frequency peaks seen in Fig. 4, a necessary consequence of the asymmetry of the observed distributions.
The distribution of the fourth receptor, fZOR8, is intermediate between that of fZOR9 and fZOR5, but closer to fZOR5. The variances for fZOR5 and fZOR8 are very similar. Mean and skewness for fZOR8 are significantly different from fZOR9 at the 99% level, but not from fZOR5 (see also Table 1). It is consistent with these findings (albeit not proven to be due to the considerable interindividual variation for fZOR8) that fZOR5 and fZOR8 share a common expression domain.
We have also analyzed possible male/female differences in preferred radial distribution, since pheromone receptors would be expected to be expressed in a sex-linked manner. For all four receptors, female and male epithelia were analyzed separately. We find no indication for sex-linked differences in the number and position of fuzzy expression zones. In all cases, interindividual variation within a sex was as great as between sexes (data not shown).
DISCUSSION
The neuronal representation of odors in the olfactory nervous system presents an intriguing problem. It is not possible to array olfactory stimuli linearly in one or two dimensions, as would be feasible for sensory stimuli in, for example, the visual and auditory systems. Therefore topological mapping is not the generic way of representing olfactory information. Indeed, cells expressing an individual odorant receptor are scattered on the olfactory epithelium, so that cellular neighborhood relationships on the sensory surface do not a priori represent stimulus similarities. Accordingly, the axonal projection of the receptor neurons to the olfactory bulb is not topological (11, 21, 22). It is not designed to retain neighborhood relationships, but, on the contrary, to sort out stimulus similarities by convergence of receptor neurons of alike specificity into one glomerulus (23, 24). However, a distinctive spatial segregation is superimposed on this pattern in the mammalian olfactory system. On a coarse level, the projection to the bulb does exhibit a region-to-region topology (11), and this zonal projection pattern is accompanied by a zonally restricted expression of odorant receptors in the olfactory epithelium (5–7).
The function of that global patterning is presently unknown. It has been suggested that zonal segregation of odorant receptor expression is essential for the differentiation of the complex olfactory system of higher vertebrates (6, 7, 25). Consistent with this idea, Ngai et al. (10) have reported a nonzonal expression pattern in catfish olfactory epithelium, fish presumably having less complex olfactory systems than higher vertebrates. We reasoned that a detailed statistical analysis of odorant receptor expression pattern in a fish species would help to decide whether spatially segregated expression of odorant receptors is indeed a peculiarity of mammals or a constitutive feature of the vertebrate olfactory system.
We have performed an in-depth analysis of the expression pattern of several odorant receptors in the olfactory epithelium of zebrafish. This species is currently investigated as a developmental model system for vertebrates. Zebrafish possess an olfactory system that is typical for teleosts (13) and seem to have average olfactory abilities (14). We have obtained PCR-derived clones of a family of 10 very divergent zebrafish odorant receptors. We find that all 10 odorant receptor molecules are expressed in sparsely scattered cells. This feature is similarly seen for catfish, mouse, and rat odorant receptor molecules (5–7, 10).
For a particular odorant receptor, we find a preferential accumulation of cells at a certain radial distance from the center of the epithelium. We observed three such preferred distances that were clearly distinct from one another (0.15, 0.45, and 0.55 relative radial distances for receptors fZOR6, fZOR9, and fZOR5, respectively). As the sensory area spans only the inner two-thirds of the olfactory lamella, these values cover the whole extent of it. The entire distributions for three of the four odorant receptors analyzed in detail are clearly distinct as well, albeit broadly overlapping. Statistical analysis has shown that the differences in distribution are highly significant. The forth odorant receptor (fZOR8) may share a domain with fZOR5. Each domain presumably accommodates several different receptors, since each of fZOR1-10 are expressed in the olfactory epithelium. Indeed, preliminary results indicate that an additional receptor (fZOR7) is also preferentially accumulated in the outermost domain.
Such annular expression, as reported here, has not been observed in the single other study on expression of fish odorant receptors (10). However, the serial reconstruction of sections attempted in that study is subject to artifacts caused by uneven distortion of sections. Indeed, we have found it necessary to express the position along lamellae as relative radial coordinate to correct for distortion of sections (and differences in absolute size between epithelia). Therefore it cannot be excluded that the effect may have been missed in the study of Ngai et al. (10). Since zebrafish has an olfactory system typical for teleosts in other respects (13, 14, 26), there is no reason to assume that expression in overlapping concentric domains might be a peculiarity of zebrafish.
Since the type of receptor expressed determines the ligand specificity, one would expect spatial nonhomogeneity of odorant receptor expression to result in spatial nonhomogeneity of physiological response characteristics. Indeed, a central/peripheral gradient not unlike the expression pattern observed in zebrafish has been noted for the electrical responsiveness of salmonid olfactory epithelium to odorants. Responses to bile acids are more pronounced in the peripheral sensory epithelium, whereas responses to amino acids are concentrated in the central regions (9).
Despite widely divergent morphologies of the sensory surface, the basic design of odorant receptor expression patterns is strikingly similar in fish and mammals. In both cases, the olfactory epithelium is subdivided with respect to receptor expression into few broad regions covering the whole extent of the sensory area. As in rodents, we find that members of different subfamilies are expressed in the same domain (6, 7). Fish expression domains, however, show substantially more overlap than rodent expression zones (6, 7). In fish as well as in mammals, the pattern is organized concentrically around a midline structure (7). Along the flow direction of the odorant carrier medium, the receptor expression pattern has a stripe-like appearance in both cases (6, 7).
These profound structural similarities could result from a common developmental origin (homology), or they could indicate a common function (analogy). Otherwise, the similarities would be incidental. As far as the function of spatial segregation of odorant receptor expression is unknown, a comparison can only be based on ontogenetic arguments. In rodents, expression zones are present from the very beginning of receptor expression. All zones develop synchronously and without influence of the olfactory bulb. Therefore they have been suggested to be patterned by spatial cues intrinsic to the olfactory epithelium (27). The fish olfactory rosette grows throughout the animal’s lifetime. Preliminary evidence suggests that the pattern of odorant receptor expression is virtually identical 75 days postfertilization and in 1.5-year-old fish, whereas the total length of the lamellae increases about 2-fold during that time. The dynamical stability of the pattern is consistent with the assumption that zebrafish expression domains, like rodent expression zones, are induced by spatial cues, rather than being a spatial record of a temporal regulatory process. Thus, both expression patterns presumably are established by similar developmental mechanisms. Therefore it is tempting to assume that teleost expression domains are the evolutionary ancestor of rodent expression zones. Because of their overlapping appearance, they might be termed fuzzy expression zones. The evolutionary conservation of the feature of spatial segregation of odorant receptor expression indicates an essential role in the organization of the vertebrate olfactory system.
What might be the advantage of spatial segregation of odorant receptor expression? It has been reasoned that zonal organization might be a strategy to simplify embryonic development (6, 7, 10). Zonal positional markers might provide external cues for a cell to select out of a large repertoire one or a few receptor genes for expression. Furthermore, zonal restriction of receptor expression might provide a presorting for axonal targeting, as cells expressing the same receptor converge onto common glomeruli in rat and mouse (23, 24). Because catfish seemed not to exhibit spatial segregation (10), these two mechanisms have been suggested to be evolutionary recent specializations of the complex mammalian olfactory system (6, 7), implying that no such mechanisms are needed to determine receptor gene expression and axon targeting in systems of the size of a single mammalian expression zone or the whole fish epithelium. Our data, however, do not support such a model. The expression pattern of odorant receptors is nonrandom even in systems of the complexity of the zebrafish nasal epithelium. Therefore, nonautonomous regulation of receptor expression and perhaps also presorting of axons might be constitutive mechanisms of the self-organization of the vertebrate olfactory system, instead of being mammalian specializations. That spatial segregation of receptor expression is related to axon targeting in the zebrafish is supported by preliminary results of tracing experiments, indicating that receptor neurons connected to one particular of the morhologically identifiable glomeruli (28) are distributed similarly to members of a particular expression domain (S.K., unpublished observation).
Alternatively, spatial segregation may be present for physiological reasons—e.g., because it might matter for the response properties of a particular olfactory neuron type to be in close proximity with only a subset of other types. This assumption would imply an interaction between neighboring olfactory receptor neurons. Biochemical as well as anatomical correlates for such lateral interactions at the level of the epithelium have been found in various systems (29–31).
In conclusion, we have shown that spatial segregation of odorant receptors observed in mammals is generalizable to the fish olfactory system. This evolutionary conservation supports an essential role for expression domains in the construction and/or function of the vertebrate olfactory system.
Acknowledgments
We thank Carmen Gitter for expert technical assistance and Friedrich Bonhoeffer for generous support. We also thank Reinhold Welker and Sybille Schwarz, who were involved in an early phase of this work in the cloning and characterization of the PCR-generated fragments. Andreas Rummrich helped with the analysis of sequences. Silke Argo provided in situ hybridization specimens of juvenile epithelia. Harald von Campenhausen and Jürgen Berger supplied the scanning electron micrographs. Christiane Nüsslein-Volhard generously consented to the use of her fish facility, and Mary Mullins kindly supplied partially inbred zebrafish strains.
Footnotes
References
- 1.Buck L, Axel R. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 2.Raming K, Krieger J, Strotmann J, Boekhoff I, Kubick S, Baumstark C, Breer H. Nature (London) 1993;361:353–356. doi: 10.1038/361353a0. [DOI] [PubMed] [Google Scholar]
- 3.Sengupta P, Chou J H, Bargman C I. Cell. 1996;84:899–909. doi: 10.1016/s0092-8674(00)81068-5. [DOI] [PubMed] [Google Scholar]
- 4.Sullivan S L, Ressler K J, Buck L B. Curr Opin Genet Dev. 1995;5:516–523. doi: 10.1016/0959-437x(95)90057-n. [DOI] [PubMed] [Google Scholar]
- 5.Strotmann J, Wanner I, Krieger J, Ramming K, Breer H. NeuroReport. 1992;3:1053–1056. doi: 10.1097/00001756-199212000-00005. [DOI] [PubMed] [Google Scholar]
- 6.Ressler K J, Sullivan S L, Buck L B. Cell. 1993;73:597–609. doi: 10.1016/0092-8674(93)90145-g. [DOI] [PubMed] [Google Scholar]
- 7.Vassar R, Ngai J, Axel R. Cell. 1993;74:309–318. doi: 10.1016/0092-8674(93)90422-m. [DOI] [PubMed] [Google Scholar]
- 8.Mackay-Sim A, Kesteven S. J Neurophysiol. 1994;71:150–160. doi: 10.1152/jn.1994.71.1.150. [DOI] [PubMed] [Google Scholar]
- 9.Thommesen G. Acta Physiol Scand. 1982;115:47–56. doi: 10.1111/j.1748-1716.1982.tb07044.x. [DOI] [PubMed] [Google Scholar]
- 10.Ngai J, Chess A, Dowling M M, Necles N, Macagno E R, Axel R. Cell. 1993;72:667–680. doi: 10.1016/0092-8674(93)90396-8. [DOI] [PubMed] [Google Scholar]
- 11.Astic L, Saucier D. Brain Res Bull. 1986;16:445–454. doi: 10.1016/0361-9230(86)90172-3. [DOI] [PubMed] [Google Scholar]
- 12.Strotmann J, Wanner I, Helfrich T, Beck A, Breer H. Cell Tissue Res. 1994;278:11–20. doi: 10.1007/BF00305773. [DOI] [PubMed] [Google Scholar]
- 13.Byrd C A, Brunjes P C. J Comp Neurol. 1995;358:247–259. doi: 10.1002/cne.903580207. [DOI] [PubMed] [Google Scholar]
- 14.Michel W C, Lubomudrov L M. J Comp Physiol A. 1995;177:191–199. doi: 10.1007/BF00225098. [DOI] [PubMed] [Google Scholar]
- 15.Mullins M C, Hammerschmidt M, Haffter P, Nüsslein-Volhard C. Curr Biol. 1994;4:189–202. doi: 10.1016/s0960-9822(00)00048-8. [DOI] [PubMed] [Google Scholar]
- 16.Probst W C, Snyder L A, Schuster D I, Brosius J, Sealfon S C. DNA Cell Biol. 1992;11:1–20. doi: 10.1089/dna.1992.11.1. [DOI] [PubMed] [Google Scholar]
- 17.Witkiewicz H, Bolander M E, Edwards D R. BioTechniques. 1993;14:458–463. [PubMed] [Google Scholar]
- 18.Schaeren-Wiemers N, Gerfin-Moser A. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- 19.Efron B, Tibshirani R J. An Introduction to the Bootstrap. London: Chapman & Hall; 1993. [Google Scholar]
- 20.Good P. Permutation Tests. Berlin: Springer; 1994. [Google Scholar]
- 21.Baier H, Rotter S, Korsching S. Proc Natl Acad Sci USA. 1994;91:11646–11650. doi: 10.1073/pnas.91.24.11646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riddle D R, Oakley B. J Comp Neurol. 1992;324:575–589. doi: 10.1002/cne.903240410. [DOI] [PubMed] [Google Scholar]
- 23.Vassar R, Chao S K, Sitcheran R, Nunez J M, Vosshall L B, Axel R. Cell. 1994;79:981–991. doi: 10.1016/0092-8674(94)90029-9. [DOI] [PubMed] [Google Scholar]
- 24.Ressler K J, Sullivan S L, Buck L B. Cell. 1994;79:1245–1255. doi: 10.1016/0092-8674(94)90015-9. [DOI] [PubMed] [Google Scholar]
- 25.Chess A, Simon I, Cedar H, Axel R. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- 26.Hansen A, Zeiske E. J Comp Neurol. 1993;333:289–300. doi: 10.1002/cne.903330213. [DOI] [PubMed] [Google Scholar]
- 27.Sullivan S L, Bohm S, Ressler K J, Horowitz L F, Buck L B. Neuron. 1995;15:779–789. doi: 10.1016/0896-6273(95)90170-1. [DOI] [PubMed] [Google Scholar]
- 28.Baier H, Korsching S. J Neurosci. 1994;14:219–230. doi: 10.1523/JNEUROSCI.14-01-00219.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Breer H, Shepherd G M. Trends Neurosci. 1993;16:5–9. doi: 10.1016/0166-2236(93)90040-s. [DOI] [PubMed] [Google Scholar]
- 30.Leinders-Zufall T, Shepherd G M, Zufall F. J Neurophysiol. 1995;74:1498–1508. doi: 10.1152/jn.1995.74.4.1498. [DOI] [PubMed] [Google Scholar]
- 31.Fishelson L. Anat Rec. 1995;243:403–412. doi: 10.1002/ar.1092430402. [DOI] [PubMed] [Google Scholar]


