Abstract
CLC Cl−/H+ exchangers are homodimers with Cl−-binding and H+-coupling residues contained within each subunit. It is not known whether the transport mechanism requires conformational rearrangement between subunits or whether each subunit operates as a separate exchanger. We designed various cysteine substitution mutants on a cysteine-less background of CLC-ec1, a bacterial CLC exchanger of known structure, with the aim of covalently linking the subunits. The constructs were cross-linked in air or with exogenous oxidant, and the cross-linked proteins were reconstituted to assess their function. In addition to conventional disulfides, a cysteine–lysine cross-bridge was formed with I2 as an oxidant. The constructs, all of which contained one, two, or four cross-bridges, were functionally active and kinetically competent with respect to Cl− turnover rate, Cl−/H+ exchange stoichiometry, and H+ pumping driven by a Cl− gradient. These results imply that large quaternary rearrangements, such as those known to occur for “common gating” in CLC channels, are not necessary for the ion transport cycle and that it is therefore likely that the transport mechanism is carried out by the subunits working individually, as with “fast gating” of the CLC channels.
Keywords: disulfide, oxidation, sulfenamide, antiporter, exchanger
The CLC Cl−-transporting proteins present an unusual circumstance in membrane biochemistry: a single protein family that is split into two mechanistically antithetical subtypes, channels and pumps (1–3). The Cl− channels are two-pore homodimers (4), with each subunit containing a gated, selective aqueous pore through which Cl− passively diffuses down its electrochemical gradient. The Cl− pumps, also homodimers, are H+-coupled exchange transporters that obligatorily swap Cl− for H+ on opposite sides of the membrane, with a stoichiometry of two anions for each proton (1, 5); these exchangers can use the free energy stored in a proton gradient to pump Cl− thermodynamically uphill, or vice versa. Despite their dissimilar ways of moving Cl− ions, all CLC proteins are built on the same basic structural plan. This conclusion emerges from sequence conservation across the CLC family, the x-ray crystal structure (6, 7) of a bacterial exchanger-type CLC (the only CLC structure known), and the functional behaviors of the channels, which may be variously rationalized in terms of the transporter's structure (3, 8, 9).
Having been studied for many years by high-resolution electrophysiological methods, CLC channels are understood quite well at the functional level. Two types of conformational changes (“gating processes”) control channel activity. The pores, each contained wholly within a single subunit, open and close independently on a millisecond timescale via “fast gating,” which involves sidechain rotation of a conserved glutamate residue (7). In striking contrast, the “common gating” process acts cooperatively on both pores, opening or occluding them simultaneously. Because the pores occupy physically distant regions in separate subunits, common gating must involve rearrangement of quaternary structure, i.e., some sort of communication between the subunits; indeed, recent work using fluorescently labeled CLC channels has revealed subunit movements as large as ≈20 Å associated with common gating (10).
Much less is known about the mechanism of the Cl−/H+ exchangers. The question that concerns us here is whether the subunits act independently or cooperatively, either of which possibility may be supported by plausible but inconclusive arguments. For instance, independent transport within each subunit is softly implied by the bacterial transporter structure alone, which shows each subunit carrying its own Cl−-binding region located far from the subunit–subunit interface and from its twin in the other subunit. Moreover, the conserved glutamates corresponding to the fast gate in the channel subclass also are located in this Cl−-binding area and are crucially involved in coupling H+ to the movement of Cl− (1). In addition, the turnover rate of Cl−/H+ exchange (11) is in the same range as the kinetics of fast gating in the channels. On the other hand, coordinated countertransport of Cl− and H+ requires a cycle of conformational changes that must be more elaborate than rotation of a single glutamate sidechain; conformations should therefore exist that have not yet been seen in crystal structures, and these could in principle involve movements of the subunits with respect to each other.
In this study, we ask whether substantial intersubunit movements are required for Cl−/H+ exchange in CLC-ec1, a CLC exchange transporter from Escherichia coli. We attack this question by constraining movement of the subunits through covalent cross-links designed from the known structure of the protein, an approach that has been similarly used to argue that substrate transport by GltT, a trimeric glutamate transporter, occurs without quaternary rearrangement (12). By finding that Cl−/H+ exchange is preserved in proteins cross-linked across the dimer interface at various points and straitjacketed by multiple cross-links, we conclude that transport is carried out in parallel by each subunit of the homodimer.
Results
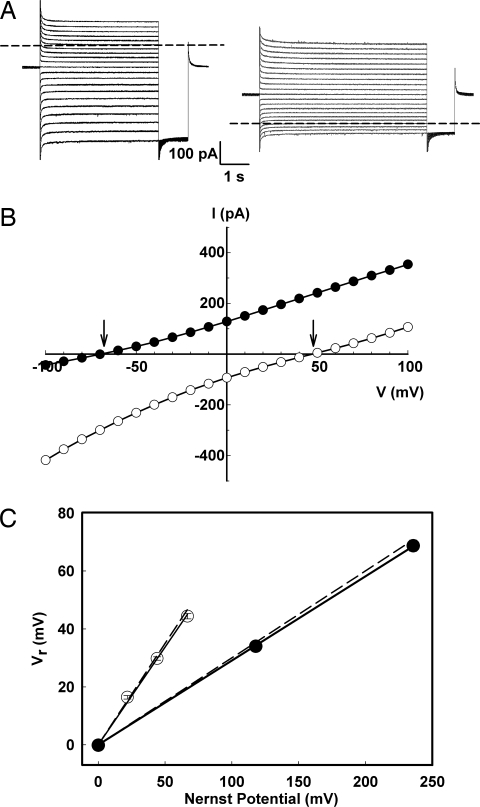
To obtain a clean background on which to introduce cysteine mutations, we substituted the three native cysteines in CLC-ec1 with innocuous side chains. This “Cys-less” mutant, C85A/C302A/C347S, is readily overexpressed, purified, and reconstituted. The construct also is well coupled in Cl−/H+ exchange, as shown by analysis of currents mediated by Cys-less incorporated into planar lipid bilayers (Fig. 1), using solutions with various Cl− or pH gradients. Large currents are readily observed, with reversal potentials determined from current–voltage curves quantitatively demanded of a 2Cl−/1H+ exchange stoichiometry, according to
where r = H+/Cl− stoichiometry and ECl and EH represent Nernst equilibrium potentials of each ion. This result precisely parallels wild-type behavior (1, 13), and the proton-pumping and unitary Cl− transport rates of Cys-less also are similar to wild type (see below). Cys-less is thus certified as a mechanistically valid background protein on which to engineer covalent cross-links.
Fig. 1.
Cl−/H+ coupling in the Cys-less transporter. Cys-less CLC-ec1 was inserted into planar lipid bilayers, and the resulting currents were recorded under voltage-clamp conditions. (A) Representative families of currents, with voltages from −100 to 100 mV in 10-mV steps, with either Cl− (300 mM/17 mM) (Left) or pH gradient (3.0/7.0) (Right) across the bilayer. Dashed lines mark zero-current level. (B) Current–voltage curves measured 200 ms before the end of the pulse interval for the gradient of Cl− (open points) or H+ (filled points). Reversal potentials are marked by arrows. (C) Variation of reversal potential, Vr, with Cl− or pH gradients. For Cl− gradients at symmetrical pH 3.0 (filled points), one side of the membrane was held at 300 mM KCl while KCl was varied on the opposite side. For pH gradients at symmetrical 300 mM KCl (open points), one side of the membrane was held at pH 3.0 while the opposite-side pH was varied. Gradients are reported as Nernst equilibrium potentials of each ion. Dashed lines represent the previously published measurements for wild-type protein (1).
Intersubunit Disulfide Cross-Links.
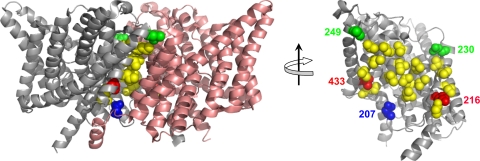
The dimer subunit interface in CLC-ec1 is roughly trapezoidal, ≈1,200 Å2 in area (Fig. 2). Most of the residues buried in the interface are nonpolar. Twelve pairs of residues whose side chains closely appose each other across the interface were chosen as candidates for cross-linking (Table 1). We tested these pairs individually by double-cysteine substitution, expecting from the twofold symmetry of the homodimer two covalent cross-links for each under oxidizing conditions. Residues tested are indicated by colored, space-filled side chains in Fig. 2. Fig. 2 also indicates (yellow residues) positions that, despite their proximity, fail to cross-link spontaneously and link incompletely, if at all, upon addition of oxidant; most of these are deeply buried in the interface, whereas those that cross-link spontaneously during protein purification are located on its edge.
Fig. 2.
Design of intersubunit cross-links. (Left) “Side view” of the dimeric transporter, with extracellular side up. (Right) “Head-on” cross-interface view of a single subunit obtained after removing one subunit and rotating the other by 90o, as indicated. Wild-type positions that, when cysteine-substituted, fail to form cross-links well are shown in yellow. Positions capable of fully cross-linking with a partner on the opposite subunit are shown in blue (207/207), green (230/249), and red (216/433).
Table 1.
Cross-linking characteristics of cysteine-substitution mutants
| Mutants | Cβ–Cβ distance, Å | Extent of cross-linking |
|---|---|---|
| Disulfide | ||
| D29C/R403C | 5.1 | Partial |
| L194C/L410C | 6.9 | Minimal |
| L194C/L422C | 7.0 | Minimal |
| I197C/L406C | 6.5 | Minimal |
| L198C/L198C | 7.9 | Minimal |
| I201C/I201C | 6.5 | Partial |
| K216C/T433C | 5.7 | Partial |
| I220C/L430C | 6.5 | Minimal |
| I223C/I426C | 5.7 | Minimal |
| T226C/L423C | 6.5 | Minimal |
| R230C/L249C | 5.9 | Complete |
| Q207C/Q207C | 17.8 | Complete |
| Sulfenamide | ||
| K216/T433C | 5.7 | Complete |
Cysteine-substitution mutants were scored for cross-linking in the presence of added oxidant, as observed by SDS/PAGE mobility.
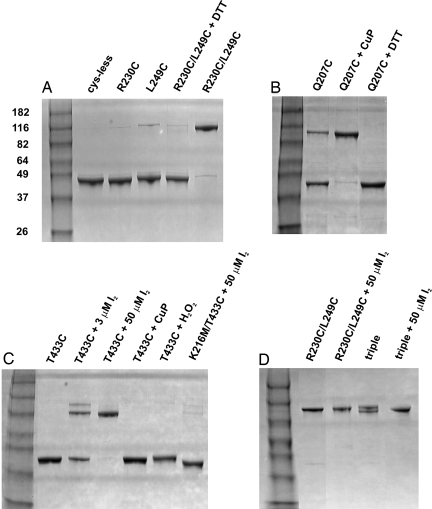
Only two of these tested substitutions, the R230C/L249C double mutant and the Q207C single mutant, could be quantitatively cross-linked, as indicated by approximate doubling of apparent size by SDS/PAGE analysis (Fig. 3). Cross-linking is prevented by including reducing agents during protein purification, but reversion of the cross-linked bands to monomer requires unusually harsh conditions (100 mM DTT at 50°C; data not shown). The 230C/249C pair is already fully cross-linked upon purification (4–6 h after disruption of the bacteria used for protein expression), as indicated by a complete absence of monomer on SDS/PAGE and low free cysteine by quantitative thiol analysis [supporting information (SI) Table 2]. These side chains lie close together (≈6 Å) across the interface; therefore, robust cross-linking is unsurprising.
Fig. 3.
Analysis of cross-linking by SDS/PAGE. The indicated CLC-ec1 variants were run on 10–12% SDS gels under nonreducing conditions. Lanes marked “DTT” are samples in which reducing conditions were maintained throughout the protein preparation. (A) Spontaneous cross-linking of 230C/249C. The molecular masses of standards are indicated in kilodaltons. (B) Spontaneous partial cross-linking for 207C driven to completion by 50 μM CuP. (C) Cysteine–lysine cross-linking. Shown are partial cross-linking of 433C at a concentration of I2 (3 μM) slightly below the protein concentration (4 μM) and complete cross-linking by excess I2. For the negative control lanes, CuP or H2O2 was used as an oxidant, and K216M was used as the mutant. (D) Straitjacketing by multiple cross-links in the triple mutant 230C/249C/433/C.
In contrast, cross-linking of 207C was unexpected, because this position dwells far off the dimer's twofold axis in the wild-type crystal structure, separated by ≈18 Å from its identical twin. Nevertheless, cross-linking proceeds rapidly to completion with added oxidant (Fig. 3B), a result suggesting that the cytoplasmic loop connecting transmembrane helices H and I, on which this residue lies, although well ordered in wild-type crystals (7), is trapped by disulfide formation during infrequent thermal fluctuations. To follow up with this idea, we determined the x-ray crystal structure of the fully cross-linked 207C–207C protein at 3.1-Å resolution (SI Fig. 8 and SI Table 3). Electron density is of good quality throughout, as illustrated for the extensively studied Cl−-binding region and for transmembrane helices H and I, which flank the cross-linked loop (residues 205–215). However, the H–I loop itself is invisible and is indeed the only segment of the 200-kDa asymmetric unit, aside from the N and C termini, with such disconnected backbone electron density. The crystal structure thus suggests that a trapped 207C–207C cross-bridge pulls this transmembrane connector off its natural docking sites on the membrane-embedded protein, disordering it locally without disrupting the transporter's overall structure.
Intersubunit Cysteine–Lysine Cross-Links.
Among the cysteine-substituted pairs that fail to cross-link well is 433C/216C (red in Fig. 2). We were therefore astonished to observe in experiments conceived as negative controls that the single cysteine replacement 433C rapidly and fully cross-links in the presence of I2 (Fig. 3C). This result was unexpected because, in the wild-type structure, the symmetry-related residues at this position are ≈30 Å apart, separated from each other by the entire span of the dimer interface. Because disulfide bond formation between these cysteines would require a gross quaternary rearrangement, it is tempting to infer that this cross-link heralds a novel, as yet uncrystallized conformation involved in the transport cycle. However, such an inference is inconsistent with the detailed nature of this cross-link in different oxidation conditions. At low concentration of I2 (3 μM, slightly substoichiometric to protein), much of the transporter remains monomeric, and the cross-linked protein shows up as a doublet (Fig. 3C). This result is incompatible with a single covalent linkage but is precisely as expected if cysteine oxidation leads somehow to a double cross-link. Such an idea predicts that weak oxidation conditions should produce all three possible forms of the protein, with zero, one, and two cross-links. The two latter forms should run at approximately twice the molecular weight of the former, but the doubly linked dimer, being the more compact, should have slightly higher mobility than the singly linked dimer. This explanation is further validated by using excess I2 (50 μM), where all of the protein collapses to a single cross-linked band running at the higher-mobility position. These results show that the cross-link in which 433C participates cannot be the result of a disulfide bond between the two cysteine residues. Instead, the dimer is linked covalently at two positions across the interface. We also note that I2 is specifically required here; neither H2O2 nor CuP, two strong oxidants commonly used to promote disulfide formation in proteins, produces this cross-link (Fig. 3C).
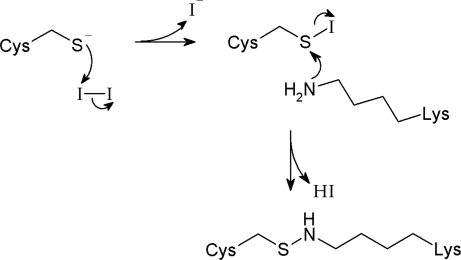
Inspection of the crystal structure reveals a chemically reactive residue located close to 433C in the opposite subunit: K216, such that its ε-amino group can be easily positioned within 4 Å of a substituted cysteine sulfur atom. This adventitious juxtaposition raises the possibility that the primary amine attacks the I2-oxidized sulfur, an activated sulfenyl iodide intermediate, to form a sulfenamide bond, according to the reaction of Fig. 4. To test the existence of a cysteine–lysine cross-bridge, we defanged the lysine by mutagenesis and challenged the double mutant T433C/K216M with I2 (Fig. 3C). Removal of the amino group abolishes the intersubunit cross-link, even under strong oxidizing conditions. Taken altogether, the results of Fig. 3C argue that I2 treatment of 433C links the dimer with two covalent cross-bridges via oxidative amination of both cysteines, at positions near the edges of the subunit interface (Fig. 2, red). As we have been unable to obtain reliable mass spectrometric data on peptide fragments after oxidation, we do not know whether the cross-link is a sulfenamide, as in Fig. 4, or a higher oxidation state such as a sulfinamide or sulfonamide.
Fig. 4.
Mechanism for Cys–Lys cross-link by I2-driven oxidative amination.
Straitjacketing the Interface with Multiple Cross-Links.
To summarize the biochemical results above, three classes of covalent cross-links worth further study have been identified. One of these, 207C–207C, is a single disulfide located near the cytoplasmic side of the interface on a linker connecting two transmembrane helices; this cross-link between a symmetry-related pair of residues locally distorts the protein backbone. Another covalent dimer arises from a double-cysteine substitution that spontaneously forms two disulfide bonds at position 230 on one subunit and position 249 on the other. Finally, a single cysteine substitution, 433C, produces an unusual double cysteine–lysine cross-link in the presence of I2.
The double-disulfide and double-sulfenamide cross-links are located at the four corners of the approximately flat subunit interface (Fig. 2). This intriguing geometry, reminiscent of welds bonding structural plates, propelled us to combine the cross-links in the same transporter in hopes of constructing a severely straitjacketed interface, in analogy with experiments constraining conformational movements of rhodopsin (14). The triple mutant required for this construction, 230C/249C/433C, expresses at high level and is well behaved biochemically. As with the 230C/249C double mutant, this “triple” spontaneously cross-links during purification (Fig. 3D). However, in contrast to the double mutant's behavior, a doublet band appears spontaneously. The lower-mobility band of this doublet runs at the same position as the fully cross-linked double-disulfide mutant. We consider that the higher-mobility band represents spontaneous cysteine–lysine cross-linking in air for two reasons. First, the free thiol content observed immediately on purification (SI Table 2), indicates that 20–30% of the cysteine at position 433 has already reacted. Second, treatment with excess I2 collapses the doublet to the high-mobility position. We cannot rigorously claim that this final product represents two cysteine–lysine links rather than one, because the gel's resolution may not be fine enough to discriminate among all these multiply linked species. We argue below, however, that the evidence strongly suggests that both of these bridges form and that, after I2 oxidation, the triple mutant is straitjacketed by all four intersubunit covalent links.
Functional Competence of Cross-Linked Transporters.
The four types of covalent dimers may be purified to homogeneity in milligram quantities. The various covalent attachments will, each in its own way, constrain relative motions of the subunits during the transport cycle. We therefore examined basic functional properties of these linked dimers to assess whether such constraints impair ion transport. In all experiments, the covalent dimers were reconstituted into liposomes for either ion flux measurements or electrophysiological recording in planar lipid bilayer membranes.
Cl−-driven H+ pumping.
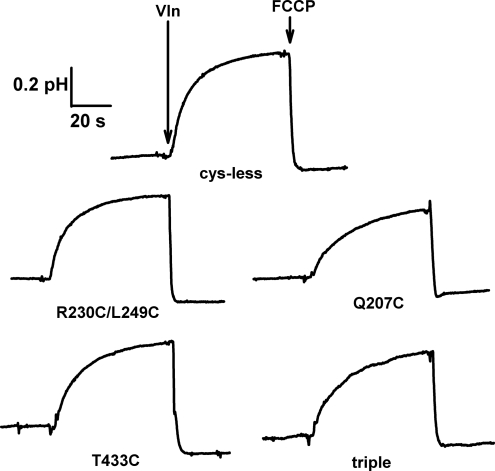
By definition, all secondary active transporters drive the uphill transport of one substrate by feeding off of the electrochemical gradient of the other. To gauge this capability for CLC-ec1 variants, we set up a Cl− gradient across the liposome membrane (300 mM inside and 10 mM outside) and followed the resulting uphill H+ influx by recording the pH rise of the liposome suspension (1, 13). Transport was initiated by setting the liposome transmembrane voltage to zero with the K+-specific ionophore valinomycin (Vln), which dissipates the charge imbalance arising from Cl−/H+ antiport; the accumulated proton gradient was subsequently collapsed with a weak-acid uncoupler [trifluoromethoxy carbonyl cyanide phenylhydrazone (FCCP)]. As shown in Fig. 5, all constructs pump protons with similar rates and extents, as for pumping by wild type protein (1). The singly linked 207C dimer is slightly less active than the other proteins, but its basic proton-uptake function is clearly intact. Signals of this magnitude represent a 2- to 3-unit pH gradient built up by the transporter (1).
Fig. 5.
Proton pumping by cross-linked transporters. CLC Cys-less and cross-linked variants indicated were reconstituted into liposomes and tested for Cl−-driven H+ pumping. Upward deflection indicates pH rise of the liposome suspension accompanying uphill proton influx.
Exchange stoichiometry.
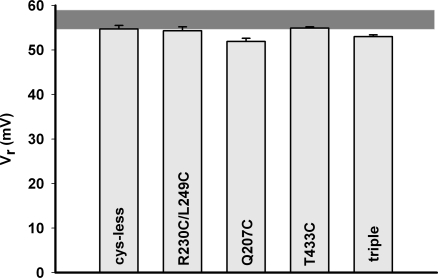
Although the proton-pumping traces in Fig. 5 qualitatively verify proper Cl−/H+ exchange in the linked dimers, they do not provide a measurement of the exchange stoichiometry, which for wild type protein is 2 Cl−/1 H+. This parameter is most conveniently assayed by electrical recording in planar bilayers, where the reversal potential in a 3-unit pH gradient is measured. All proteins give Cl−-selective currents, as in Fig. 1, from which reversal potentials are readily determined. All reverse at 53–57 mV (Fig. 6), similar to wild-type CLC-ec1, and close to the thermodynamic expectation (58.5 mV) for strict 2/1 exchange (Eq. 1). The reversal potential (51 mV) of the “strained” dimer 207C–207C is slightly lower, but this value still represents respectably tight exchange coupling, especially when compared with variously uncoupled mutants previously described (11, 15, 16). We conclude, then, that the normal mechanism of Cl−/H+ transport is unperturbed in the covalent–dimer constructs.
Fig. 6.
Cl−/H+ coupling of cross-linked transporters. Reversal potentials were determined on the indicated CLC exchangers under symmetrical Cl− pH gradients (4.0/7.0), as in Fig. 1. Each point represents the average ± SE of at least five independent measurements. The gray horizontal bar represents the range of values found for wild-type protein.
Unitary turnover rate.
The reversal potential, a null-point measurement, tells us nothing about the absolute ion-transport rate catalyzed by the exchanger. To measure this rate, we followed passive Cl− efflux under Poisson dilution conditions, i.e., at protein concentrations so low that most of the transporting liposomes contain only a single copy of the transporter and the remaining liposomes have no protein at all (11). Reconstituted liposomes loaded with high Cl− were suspended in low-Cl− solution, and KCl efflux was followed under conditions in which Cl− transport is rate-limiting. Time courses of Cl− appearance in the liposome suspension are shown in Fig. 7A for all of the proteins tested. Upon adding K+ and H+ ionophores, Cl− begins to increase in the suspension as efflux proceeds (except with the protein-free liposome controls; Fig. 7A, lowest trace). After a few minutes, Cl− release approaches completion, and detergent is added to disrupt all liposomes and thereby mark the entire trapped intraliposomal volume. The portion of the efflux that requires detergent for release, labeled fo in Fig. 7A, reflects the protein-free liposomes and serves as a quantitative indicator of the fraction of protein that is functionally reconstituted (11, 17). The striking result of Fig. 7A is that all these CLC-ec1 variants give identical fo values and similar unitary transport rates, on the order of 2,000 s−1, about half the rate of wild-type protein (11). Rates of the covalent dimers are 50–85% of the Cys-less rate, with the straitjacketed triple mutant being the slowest, 1,700 s−1. Thus, all cross-linked mutants support a kinetically competent transport cycle.
Fig. 7.
Unitary Cl− turnover rate. (A) Traces from Cl− electrode show the release of Cl− into the liposome suspension. Arrows mark the addition of Vln/FCCP at the beginning of the experiment and 50 mM octylglucose at the end. Raw traces are shown (from the top to the bottom traces) for Cys-less, 433C, 230C/249C, 207C/207C, triple, and control with no protein reconstituted. All mutants were assayed after full cross-linking. (B) Turnover rates γ (filled bars) and fo (open bars) for the indicated CLCs, each representing mean ± SE of five measurements.
Discussion
Studies on CLC channels suggest that a large conformational rearrangement of the subunits is directly involved in the common gating process (10, 18), but it is not known whether the CLC transporters require similar quaternary movements to move their substrate ions across biological membranes. An extreme possibility of subunit cooperation, for example, might envision an alternating cycle in which one subunit transports Cl− while the other adopts a different conformation to move H+ in the opposite direction. At the other extreme, communication between subunits might be so weak that each functions independently as a fully fledged transporter, a circumstance that would be strikingly analogous to fast gating in the CLC channels. This situation may be argued to apply to the CLC-ec1 transporter, because this protein lacks the C-terminal domain where the common gating movements occur (10). Nevertheless, this fundamental question has not been previously settled and so motivates this work. Because quaternary rearrangements would be manifested throughout the dimer interface, we sought to build multiple covalent links constraining relative movements of the subunits and to assess the functional consequences of these constraints.
Guided by the x-ray crystal structure of CLC-ec1, we engineered intersubunit cross-bridges by substituting cysteine at pairs of side chains close together on opposite sides of the dimer interface. Cross-link formation in various oxidation conditions was detected on SDS gels. Only about one-third of the tested pairs form robust cross-links, and these are all found near the edges of the interface, where water is close at hand to favor thiol deprotonation. Cross-links form poorly if at all with cysteine-substitutions in the interfacial interior, as expected from the known low reactivity of thiols with electrophiles in nonpolar protein environments (19).
Three pairs of residues were analyzed further. First, a symmetry-related pair (207C–207C) in a 10-residue loop connecting two transmembrane helices forms a single cross-link, but to do so the twin cysteines must move ≈10 Å from their positions in the wild-type crystal structure. This movement distorts the protein, but only locally in the connector-loops, which become disordered in the crystal structure of the cross-linked dimer. In the wild-type protein, electron density is well-defined for this ≈20 Å of extended backbone (7), so this disorder, although paradoxically a result of constraining the polypeptide chains, probably arises from the loss of favorable interactions with nearby protein groups brought about by the cross-link. It is also noteworthy that only two residues away from the beginning of the H–I loop is E203, whose carboxyl group is directly involved in proton movement (15); this critical glutamate is well ordered in the crystal structure of the cross-linked protein, and the tight Cl−/H+ coupling prevailing after cross-linking shows that despite its proximity to the disordered region, E203 is fulfilling its normal proton-transfer role.
A second pair of substituted cysteines (230C near the extracellular end of one transmembrane helix and 249C at the extracellular beginning of the next) lie close together across the subunit interface. These nonequivalent residues form two symmetry-related disulfide bonds separated by ≈35 Å at the corners of the interface's extracellular edge. In the wild-type protein, the paired side chains are in close proximity across the interface, and so cross-linking here is not expected to distort the protein. Indeed, these cysteines react to completion without added oxidant, such that the homodimer emerges from purification already fully cross-linked.
Finally, we stumbled upon an unusual type of cross-bridge in which a substituted cysteine (433C) covalently bonds to a nearby lysine, K216. The two resulting symmetry-related cross-links, ≈30 Å apart on the cytoplasmic edge of the interface, require added oxidant to form, with I2 being the most effective. We hasten to acknowledge that the detailed chemical character of this cross-link remains unproven. Attempts to identify cross-linked products after oxidation by mass spectrometry were unsuccessful, possibly due to incomplete trypsin fragmentation of the hydrophobic protein and to promiscuous, heterogeneous iodination at many residues (mostly surface-exposed aromatics). Nevertheless, the case for a cysteine–lysine linkage is strong. Under partial oxidation conditions, a single cysteine per monomer produces a double cross-link, as indicated by two distinguishable cross-linked species of slightly different mobilities. This cysteine's partner must therefore be a chemically reactive group that is not a thiol; the primary amino group of K216, in van der Waals contact with the 433 sidechain across the interface, offers itself as an obvious culprit. When this amino group is removed by decapitation to methionine, cross-linking fails. Moreover, the mechanism proposed for this reaction, I2 oxidation of a thiolate to a sulfenyl halide followed by nucleophilic attack by an amine to form a sulfenamide, makes chemical sense and is known in the synthetic–organic literature (20–22). It is still unclear whether this cross-link remains a sulfenamide or proceeds to higher oxidation states. But uncertainty on that detail does not undermine our purpose here, which is to covalently link the dimer. Oxidative cysteine amination by I2 is undocumented in proteins as far as we are aware, but similar cysteine–lysine cross-links have been observed in peptides and proteins, with biologically generated hypochlorous acid as oxidant (23, 24).
The 230C/249C disulfides and 433C/K216 sulfenamides were then combined in a “straitjacketed transporter” to simultaneously pinion the dimer at the four corners of the interface. A combination of SDS-gel behavior and quantitative thiol analysis shows that oxidizing this construct achieves at least three of these cross-links, the two disulfides and at least one sulfenamide, but our data do not rigorously demonstrate the final sulfenamide. It is very likely, however, that this fourth bond is in fact formed under the strong oxidation conditions used, in light of the natural proximity of the cysteine and lysine side chains, the susceptibility of sulfenyl iodide to nucleophilic attack, and the demonstrated ease of completing this reaction in the 433C single-cysteine protein.
The four covalent dimers were examined for functional proficiency. All perform H+-coupled Cl− antiport with characteristics close to wild-type and Cys-less proteins. They all pump protons uphill driven by downhill Cl− movement, with Cl−/H+ exchange stoichiometry close to 2. Absolute single-transporter turnover rates of the covalent dimers are close to that of the Cys-less control, and the straitjacketed transporter moves ions at fully half the rate of the unconstrained control. These results imply that whatever the cycle of conformational changes driving coordinated ion transport—and this is largely unknown—any functionally relevant movements of the subunits relative to each other must be small. Accordingly, we propose that the Cl−/H+ exchange mechanism is contained within each individual subunit; this inference points out a further close mechanistic correspondence between the CLC transporters and fast gating in the CLC channels (3). We cannot make the quantitative arguments required to claim strict independence for the transporter subunits, as applies to the channels, but we do propose that any conformational cross-talk between subunits in the transport cycle is weak. To a first approximation, then, the CLC-ec1 homodimer appears to be two Cl−/H+ exchange transporters simply glued together and working in parallel. The turnover rate of the wild-type protein was recently clocked at 4,200 Cl− ions per second per homodimer (11); accordingly, we assign half this unitary rate to the single transporting unit, the individual subunit of a “double-barreled” transporter.
Methods
Expression and purification of CLC-ec1, His-tag cleavage, functional reconstitution, electrical recordings, and liposome flux measurements were performed as documented in detail (11, 25), except where variations are specifically noted. Point mutations introduced by conventional PCR methods were confirmed by sequencing. For preparing CLC-ec1 under reducing conditions, 2 mM Tris[2-carboxylethyl] phosphine was included in all purification steps, and 20 mM DTT was added to the final protein sample to prevent spontaneous oxidation. SDS/PAGE gels (10–12%) were visualized by Coomassie blue. All procedures were carried out at room temperature, 21–23°C.
Crystallization of disulfide-cross-linked CLC-ec1(Q207C) in complex with a FAB fragment was carried out as described (13). To obtain acceptable crystal quality, this cysteine mutant was made on the wild-type protein, not on the cysteine-free construct used for other experiments here. Examination of the structure shows that positions of the three native cysteines are unaffected by the introduction of this additional cysteine. Diffraction data were collected at National Synchrotron Light Source, and the HKL program was used for spot integration and scaling. The structure was solved by molecular replacement in the CCP4 suite by using the wild-type protein [Protein Data Bank (PDB) ID code 1OTS] as search model, was refined in CCP4 with REFMAC5, and was adjusted manually for proper stereochemistry in COOT. Coordinates and structure factors are deposited in the PDB (ID code 2R9H)
For cross-linking in the presence of I2, it was necessary to remove all traces of imidazole (26). After elution from the Co-affinity column with 400 mM imidazole, the protein was chromatographed by FPLC on a Superdex 200 in cross-link buffer [CB; 100 mM NaCl, 20 mM Tris·HCl (pH 7.5), 10 mM decylmaltoside]. Protein concentration was adjusted to 0.2 mg/ml (3.8 μM monomer) before addition of I2 from a freshly prepared 20 mM solution in ethanol (final concentrations: 3 μM for partial cross-linking, 50 μM for complete reaction). Reaction was allowed to proceed for 15 min, and excess I2 was removed by repurifying the protein with FPLC as above. For cross-linking by Cu-phenanthroline (CuP), stock solutions of 40 mM aqueous CuCl2 and 120 mM 1,10-phenanthroline (in dimethyl sulfoxide) were mixed in equal parts, and the mixture was added to protein eluted from the Co-affinity column (50 μM final concentration). After an hour of incubation, the protein was purified by FPLC in CB as above.
Free thiol was quantified colorimetrically (27). Protein was purified under reducing conditions to prevent spontaneous cysteine oxidation. Spontaneous disulfide formation in 230C/249C constructs was initiated by removal of reducing agents with FPLC in CB; before the thiol assay, the sample buffer was exchanged by gel filtration for 100 mM Tris·HCl, 10 mM decylmaltoside, 3 mM EDTA (pH 8.0), and protein concentration was adjusted to 1 mg/ml (19 μM monomer). Dry urea (360 mg/ml) was then added to unfold the protein, followed by 0.2 mM dithio-bis(2-nitrobenzoic acid). Thiol content was determined from absorbance at 412 nm after 1 h of reaction, with correction for background determined in parallel on Cys-less protein.
Proton pumping was carried out with liposomes reconstituted at 20 mg/ml lipid and 2 μg of protein per milligram of lipid and was loaded with 300 mM KCl and 75 mM glutamic acid-NaOH (pH 4.8). A 100-μl sample was centrifuged through a 1.5-ml Sephadex G-50 column equilibrated with pump buffer [PB; 290 mM K-isethionate, 10 mM KCl, 2 mM glutamic acid-NaOH (pH 5.2)] and then diluted into a final volume of 2.0 ml PB in a stirred cell, where pH was followed. Proton uptake was initiated with 1 μM Vln and, after ≈1 min, was collapsed with 1–2 μM FCCP.
Absolute unitary transport rate was determined by using a Poisson dilution method (11). Briefly, immediately before the Cl− flux assay a 100-μl sample of reconstituted liposomes [0.2 μg of protein per milligram of lipid, loaded with 300 mM KCl, 25 mM citric acid-NaOH (pH 4.5)] was spun through a 1.5-ml Sephadex column equilibrated with 300 mM K-isethionate, 1 mM KCl, 25 mM citrate-NaOH (pH 4.5) and diluted into 1.9 ml of this solution in a stirred cell at 25°C. Cl− efflux was initiated by 1 μM of Vln/FCCP, and Cl− concentration was continuously recorded with a Ag/AgCl electrode; the reaction was ended by addition of 50 mM octyl glucoside. Transport rate was calculated from the initial rate of Cl− release (ions per second), determined in each experiment by calibrating the stirred cell with known KCl additions.
Planar bilayer recordings were carried out as described in detail (25) in conditions using a Cl− gradient (300 mM/17–300 mM KCl) at symmetrical pH (3.0 or 4.0) or a pH gradient (as indicated) at symmetrical 300 mM Cl−, buffered with 5 mM histidine/5 mM glutamic acid and adjusted to the desired pH with KOH. After insertion of transporters, both sides of the membrane were extensively perfused with fresh solution.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Josh Johnson and Jeff Agar for mass spectrometry; Barry Snider for advice on sulfenamide chemistry; and the staff at beamline X29A, Brookhaven National Synchrotron Light Source. This work was supported in part by National Institutes of Health Grant GM-31768 and by a Howard Hughes Medical Institute graduate fellowship (to W.N.).
Footnotes
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on May 1, 2007.
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2R9H).
This article contains supporting information online at www.pnas.org/cgi/content/full/0708639104/DC1.
References
- 1.Accardi A, Miller C. Nature. 2004;427:803–807. doi: 10.1038/nature02314. [DOI] [PubMed] [Google Scholar]
- 2.Jentsch TJ, Neagoe I, Scheel O. Curr Opin Neurobiol. 2005;15:319–325. doi: 10.1016/j.conb.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Miller C. Nature. 2006;440:484–489. doi: 10.1038/nature04713. [DOI] [PubMed] [Google Scholar]
- 4.Middleton RE, Pheasant DJ, Miller C. Biochemistry. 1994;33:13189–13198. doi: 10.1021/bi00249a005. [DOI] [PubMed] [Google Scholar]
- 5.DeAngeli A, Monachello D, Ephritikhine G, Frachisse JM, Thomine S, Gambale F, Barbier-Brygoo H. Nature. 2006;442:939–942. doi: 10.1038/nature05013. [DOI] [PubMed] [Google Scholar]
- 6.Dutzler R, Campbell EB, Cadene M, Chait BT, MacKinnon R. Nature. 2002;415:287–294. doi: 10.1038/415287a. [DOI] [PubMed] [Google Scholar]
- 7.Dutzler R, Campbell EB, MacKinnon R. Science. 2003;300:108–112. doi: 10.1126/science.1082708. [DOI] [PubMed] [Google Scholar]
- 8.Lin CW, Chen TY. J Gen Physiol. 2003;122:147–159. doi: 10.1085/jgp.200308845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engh AM, Maduke M. J Gen Physiol. 2005;125:601–617. doi: 10.1085/jgp.200509258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bykova EA, Zhang XD, Chen TY, Zheng J. Nat Struct Mol Biol. 2006;13:1115–1119. doi: 10.1038/nsmb1176. [DOI] [PubMed] [Google Scholar]
- 11.Walden M, Accardi A, Wu F, Xu C, Williams C, Miller C. J Gen Physiol. 2007;129:317–329. doi: 10.1085/jgp.200709756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Groeneveld M, Slotboom DJ. J Mol Biol. 2007;372:565–570. doi: 10.1016/j.jmb.2007.06.067. [DOI] [PubMed] [Google Scholar]
- 13.Nguitragool W, Miller C. J Mol Biol. 2006;362:682–690. doi: 10.1016/j.jmb.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 14.Struthers M, Yu H, Oprian DD. Biochemistry. 2000;39:7938–7942. doi: 10.1021/bi000771f. [DOI] [PubMed] [Google Scholar]
- 15.Accardi A, Walden M, Nguitragool W, Jayaram H, Williams C, Miller C. J Gen Physiol. 2005;126:563–570. doi: 10.1085/jgp.200509417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Accardi A, Lobet S, Williams C, Miller C, Dutzler R. J Mol Biol. 2006;362:691–699. doi: 10.1016/j.jmb.2006.07.081. [DOI] [PubMed] [Google Scholar]
- 17.Maduke M, Pheasant DJ, Miller C. J Gen Physiol. 1999;114:713–722. doi: 10.1085/jgp.114.5.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller C, White MM. Proc Natl Acad Sci USA. 1984;81:2772–2775. doi: 10.1073/pnas.81.9.2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, Xu Q, Cortes DM, Perozo E, Laskey A, Karlin A. Proc Natl Acad Sci USA. 2002;99:11605–11610. doi: 10.1073/pnas.192439299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis FA, Friedman AJ, Kluger EW, Skibo EB, Fretz ER, Milicia AP, LeMasters WC. J Org Chem. 1977;42:967–972. [Google Scholar]
- 21.Koval IV. Uspekhi Khim. 1996;65:452–473. [Google Scholar]
- 22.Goto K, Yamamoto G, Tan B, Okazaki R. TetrLett. 2001;42:4875–4877. [Google Scholar]
- 23.Raftery MJ, Yang Z, Valenzuela SM, Geczy CL. J Biol Chem. 2001;276:33393–33401. doi: 10.1074/jbc.M101566200. [DOI] [PubMed] [Google Scholar]
- 24.Fu X, Mueller DM, Heinecke JW. Biochemistry. 2002;41:1293–1301. doi: 10.1021/bi015777z. [DOI] [PubMed] [Google Scholar]
- 25.Accardi A, Kolmakova-Partensky L, Williams C, Miller C. J Gen Physiol. 2004;123:109–119. doi: 10.1085/jgp.200308935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolff J, Covelli I. Eur J Biochem. 1969;9:371–377. doi: 10.1111/j.1432-1033.1969.tb00618.x. [DOI] [PubMed] [Google Scholar]
- 27.Ellman GL. Arch Biochem Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.