Abstract
In bacteria most secretory proteins are transported across the plasma membrane by the interplay of the ATPase SecA with the translocation channel formed by the SecY complex; SecA uses cycles of ATP hydrolysis to “push” consecutive segments of a polypeptide substrate through the channel. Here we have addressed the mechanism of this process by following the fate of stalled translocation intermediates. These were generated by using a polypeptide substrate containing a bulky disulfide-bonded loop, thus preventing the final residues from passing through the channel. Protease protection experiments showed that the intermediates were stable in the presence of ATP and could complete translocation once the block was removed. The translocation intermediate was also stable when SecA associated with ATPγS, a poorly hydrolyzable ATP analog, or ADP plus AlF4, which mimics the transition state during ATP hydrolysis. In contrast, when SecA was in its ADP-bound state, the translocating polypeptide moved back into the cytosol, as indicated by the disappearance of the protected fragment. Backsliding was not significantly altered by deletion of the plug domain, a short helix in the center of the SecY channel, but it was slowed down when changes were introduced into the pore ring, the constriction of the hourglass-shaped channel. In all cases, backsliding was significantly slower than forward translocation. Together, these data suggest that SecA binds the polypeptide chain in its ATP state and releases it in the ADP state. The channel itself does not bind the polypeptide chain but provides “friction” that minimizes backsliding when ADP-bound SecA resets to “grab” the next segment of the substrate.
In bacteria most secretory proteins are transported post-translationally across the plasma membrane (for reviews, see Refs. 1–3). The minimum translocation apparatus comprises the ATPase SecA and the SecY channel. SecA pushes the polypeptide chain through the channel (4–9). It “grabs” a polypeptide segment, moves it into the channel, and resets to grab the next segment. This process is repeated until the polypeptide is all the way through. How exactly the ATPase cycle of SecA is coupled to polypeptide movement is unclear, but several different models may be considered. One possibility is that SecA and the channel bind the polypeptide chain in an alternating manner; when SecA pushes the polypeptide, the channel would allow free movement, whereas when SecA resets, the channel would hold on to the polypeptide and prevent it from sliding back into the cytosol. An alternative possibility is that SecA provides two alternating binding sites by itself; at any given time, one binding domain would be pushing and the other one holding. The two sites could be contained in a single SecA molecule or split between the subunits of a dimer (9). In all these models the polypeptide would be moved by “two hands” that alternately grab and release the polypeptide chain. An alternative would be a “one-hand model,” in which there is only a single peptide binding site in SecA. Here, the resetting of SecA would be fast compared with the spontaneous sliding of the polypeptide in the channel.
SecA is a multidomain protein that contains two nucleotide binding folds (NBF1 and 2) with the nucleotide binding site in between them (10). ATP binding and hydrolysis are linked to conformational changes of the other domains that bind and release the polypeptide chain (5, 11–13). The SecY channel consists of the channel-forming SecY subunit and two small subunits, SecG and SecE (for review, see Ref. 1). The crystal structure of an archaeal channel homolog indicates that the pore has an hourglass shape, with a ring of 6 hydrophobic amino acids at its constriction, referred to as the pore ring (14). The pore is blocked by a central short helix, the plug, which abuts the pore ring. Channel opening moves the plug away (15, 16) and allows the passage of a translocating polypeptide through the pore ring (17). The crystal structure of the closed archaeal channel indicates that even an unfolded polypeptide could not pass through the pore ring. Thus, one has to postulate that the pore widens during translocation. The plug and the pore ring are both in close proximity to translocating polypeptides (8, 17) and could therefore provide either “friction,” reducing the sliding of a polypeptide in the channel, or even bind the polypeptide as SecA resets.
Here we have addressed the mechanism of SecA-mediated translocation by studying the sliding of a polypeptide chain in the channel. To this end, we have generated translocation intermediates in which complete translocation of a polypeptide chain is prevented by a C-terminal disulfide-bridged loop that is too large to move through the channel (18). We show that these polypeptide chains move slowly back into the cytosol when SecA is in its ADP-bound state but not when it is in the ATP state or in the transition state during ATP hydrolysis. Backsliding is not greatly affected by the deletion of the plug domain from SecY, but it is significantly slowed when changes are introduced into the pore ring. These data suggest that the polypeptide is bound and pushed by SecA in its ATP state. The channel does not bind the polypeptide but provides sufficient friction to minimize backsliding when SecA resets in its ADP state.
EXPERIMENTAL PROCEDURES
Production of SecY Mutants—A version of SecY lacking cysteines (17) was used, and mutations were subsequently introduced through PCR-based mutagenesis (QuikChange; Stratagene) of pBAD-EhisYG (19). The plug mutations were described previously (20). Tryptophans and serines were introduced at positions Ile-86, Ile-191, Ile-278, and Ile-408 of SecY. The expression of these constructs in C43 (DE3) cells was induced with arabinose for 4 h at 37 °C. Complexes were purified after solubilization of the membranes in 1% n-dodecyl-β-d-maltopyranoside (Anatrace) by binding to a Ni2+-chelating column (14). Protein concentrations were determined with the Bradford reagent (Bio-Rad Laboratories). Purified SecY derivatives in TNG buffer (10 mm Tris-Cl, pH 8.0, 150 mm NaCl, 10% glycerol, 10 mm DTT,4 0.03% n-dodecyl-β-d-maltopyranoside) were stored at –80 °C.
Preparation of Proteoliposomes and Inverted Membrane Vesicles—Purified SecY mutants were reconstituted with Escherichia coli polar lipids into phospholipid vesicles as described previously (19). Inverted membrane vesicles (IMVs) were prepared as described previously (21).
Generation of proOmpA Mutants—A truncated version of proOmpA (pOA) was used (amino acids 176–297 deleted) with a cysteine introduced at the new position 202. PCR-based mutagenesis was used to introduce a cysteine at position 164 or 175 to allow for disulfide loop formation. DNA templates coding for pOA were used to make mRNA by in vitro transcription with SP6 polymerase. pOA was synthesized in the presence of [35S]methionine by in vitro translation (rabbit reticulocyte lysate; Promega) for 20 min at 30 °C, precipitated with ammonium sulfate for 30 min at 4 °C, pelleted at 14,000 rpm for 10 min at 4 °C, and resuspended in urea buffer (8 m urea, 50 mm Tris, pH 7.5).
Intermediate Formation, Backsliding, and Chase Reactions—To generate an arrested translocation intermediate, proteoliposomes containing SecY complex were mixed with in vitro translated 35S-labeled pOA and 20 μg of cysteine-free SecA (22) in buffer (50 mm KCl, 50 mm HEPES, pH 7.5, 5 mm MgCl2, 0.5 mg/ml bovine serum albumin, 0.1 mm sodium tetrathionate, and 0.2 mm ATP). After 15 min of incubation at 37 °C, a sample was taken for t = 0 and immediately digested with 1 mg/ml proteinase K. The remaining aliquot was depleted of ATP with 0.5 units of hexokinase and 10 mm glucose, and samples were removed at the times indicated and digested immediately with proteinase K. For chase incubations, 10 mm DTT was added instead of hexokinase/glucose. When indicated, 2.5 mm ADP, 1 mm ATPγS, or ADP plus AlF4 (2.5 mm ADP, 5 mm NaF, and 0.3 mm AlCl3) were added to the reactions (either from the start or after intermediate formation). Samples were then incubated in 1 mm 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride for 10 min at 4 °C, precipitated with 10% trichloroacetic acid for 20 min at 4 °C, pelleted at 14,000 rpm for 15 min at 4 °C, and resuspended in sample buffer (50 mm Tris, pH 7.5, 8 m urea, 5% SDS, 10 mm EDTA, 0.25 mg/ml bromphenol blue). Samples were separated on 4–20% Tris-HCl gels (Bio-Rad), visualized by autoradiography, and quantified by phosphorimaging (Fujix BAS 2000). Background subtraction and cropping of three experiments was performed with ImageGauge v4.22.
RESULTS
Backsliding of Translocation Intermediates—We first generated a translocation intermediate of a proOmpA derivative carrying an internal deletion of residues 176–297. The protein was synthesized in a reticulocyte lysate system in the presence of [35S]methionine. It was denatured in urea and treated with tetrathionate to introduce a disulfide bridge between two cysteines close to the C terminus (Cys-175 and Cys-202; the numbers refer to residues in the deletion mutant). This translocation substrate was then diluted into a mixture containing SecA, urea-washed IMVs, and ATP. After incubation for 15 min at 37 °C, a sample was taken and treated with proteinase K to digest all material that was not fully translocated into the vesicles. As expected (18), a protected ∼18k-Da fragment was seen that corresponds to the ∼150 N-terminal amino acids of the substrate (Fig. 1A, lane 2); this corresponds to the segment of the translocation intermediate that is located inside the vesicles when the disulfide loop reaches the channel. In the presence of ATP, SecA would be expected to push the polypeptide chain continuously into the channel, thus maintaining the position of the polypeptide chain and giving rise to a defined proteolytic fragment. The intermediate did not form in the absence of ATP (Fig. 1A, lane 8) and was digested by proteinase K when detergent was added to disrupt the membrane after translocation (lane 9). When DTT was added at the beginning of the incubation to reduce the disulfide-bonded loop, only the full-length polypeptide was seen after proteolysis (Fig. 1A, lane 1; the doublet is caused by cleavage of the signal sequence by signal peptidase in IMVs). Similarly, when DTT was added after the translocation intermediate had formed, the ∼18-kDa fragment disappeared and full-length substrate appeared instead (Fig. 1A, lane 7). These data confirm that the ∼18-kDa fragment corresponds to a productive translocation intermediate that can be chased into the fully translocated polypeptide when the disulfide bridge is reduced and ATP is present.
FIGURE 1.
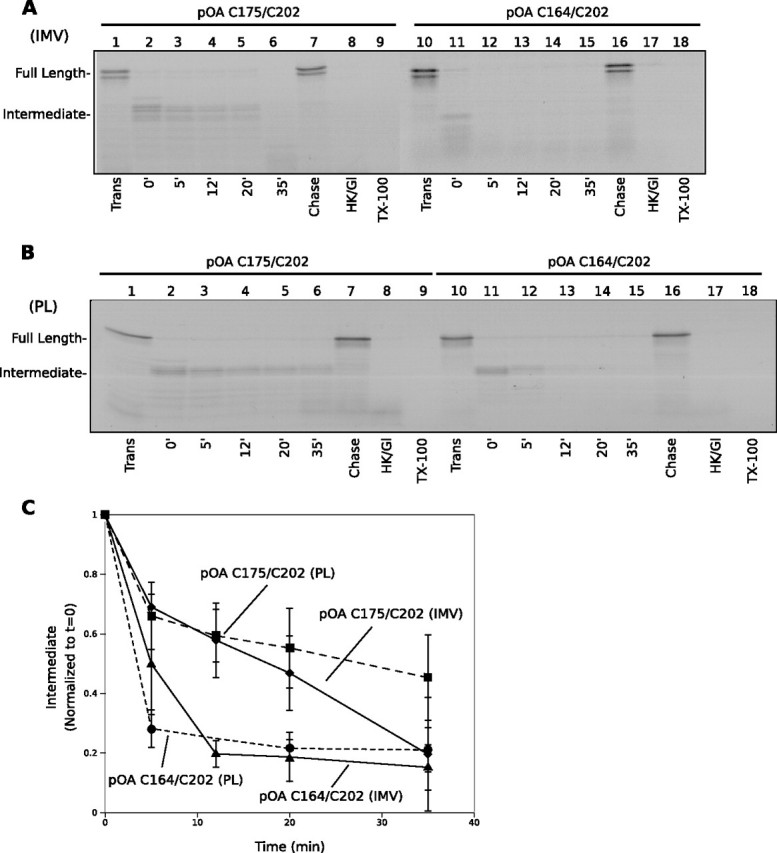
Backsliding of a translocation intermediate after ATP depletion. A, proOmpA (pOA) derivatives with cysteines at positions 175 and 202 or 164 and 202 were translocated into urea-washed inverted membrane vesicles (IMVs) under oxidative conditions for 15 min at 37 °C. After formation of a translocation intermediate (lanes 2 and 11), the samples in lanes 7 and 16 were further incubated with DTT and ATP (Chase). The other samples were depleted of ATP by addition of hexokinase and glucose (HK/Gl), placed at 37 °C, and aliquots were taken at the times indicated (lanes 3–6 and 12–15). The samples in lanes 8 and 17 received HK/Gl at the beginning of the translocation reaction. Lanes 1 and 10 demonstrate full translocation under reducing conditions. Aliquots of samples 2 and 11 were proteolyzed in the presence of Triton X-100 (TX-100)(lanes 9 and 18). All samples were treated with proteinase K, separated by SDS-PAGE, and analyzed by autoradiography. B, as in A, but with proteoliposomes (PL) containing purified E. coli SecY complex. C, quantification of backsliding from A and B. The intensities were normalized to that of the protected fragment at t = 0, and the background was subtracted. The mean and S.D. of three independent experiments are shown.
To test the stability of the translocation intermediate in the absence of ATP, i.e. under conditions where SecA cannot push the polypeptide chain into the channel, we generated the translocation intermediate and then depleted ATP by the addition of hexokinase/glucose. Samples were taken at different time points and analyzed after treatment with proteinase K. The amounts of the ∼18-kDa fragment gradually decreased with time (Fig. 1A, lanes 2–6; quantitation in Fig. 1C). The overall amounts of substrate before addition of protease remained constant (supplemental Fig. S1). We interpret the disappearance of the proteolytic fragment as backsliding of the disulfide-arrested polypeptide into the cytosol; when the disulfide loop moves away from the channel, the protease can cleave at different sites, generating a heterogeneous population of protected chains that can no longer be visualized as a defined band. Our data show that backsliding occurs in the absence of ATP. A similar observation has been made with reconstituted proteoliposomes containing the yeast Sec complex and the luminal ATPase BiP: a stalled prepro-α-factor intermediate was found to slide back into the cytosol in the absence of ATP (23).
Backsliding was significantly faster with a proOmpA substrate in which a cysteine was introduced at position 164 rather than position 175, generating a larger disulfide-bonded loop at the C terminus (Fig. 1A, lanes 11–15; quantitation in Fig. 1C). These data suggest that the rate of sliding depends on the amino acid sequence present in the SecY channel.
To test whether the observed backsliding rate is dependent on proteins other than the SecY channel, we performed similar experiments with proteoliposomes containing the purified E. coli SecY complex. Again, with a disulfide bond at the C terminus between Cys-175 and Cys-202, the proOmpA substrate was only partially translocated (Fig. 1B, lane 2). When DTT was added either at the beginning of the incubation (Fig. 1B, lane 1) or after the intermediate had formed (lane 7), translocation was completed. No protease-protected material was seen if ATP were depleted before translocation (Fig. 1B, lane 8) or when detergent was present during proteolysis (lane 9). When ATP was depleted after formation of the intermediate, the amount of intermediate decreased over time with kinetics similar to that seen with IMVs (Fig. 1B, lanes 2–6; quantitation in Fig. 1C). These results show that the backsliding rate is determined by the SecY channel and not by other proteins present in crude membranes. As before, backsliding was significantly faster with a proOmpA substrate containing a disulfide loop between positions 164 and 202 (Fig. 1B, lanes 11–15; quantitation in Fig. 1C).
Next we tested whether backsliding only occurs in the absence of ATP. To this end, we first generated a translocation intermediate with reconstituted proteoliposomes, using the proOmpA substrate with cysteines at positions 164 and 202. The intermediate was isolated by sedimenting the membranes. The pellet was washed to remove residual nucleotides and resuspended. As expected, a prominent protease-resistant fragment was observed (Fig. 2A, lane 2). In the presence of ATP and DTT, this band disappeared and a fully translocated species appeared instead (Fig. 2A, lane 21). When the disulfide-bonded intermediate was incubated at 37 °C in the absence of added nucleotides, the protease-protected band disappeared as before (Fig. 2A, lanes 3–5; quantitation in Fig. 2B). Backsliding was seen in the presence of ADP (Fig. 2A, lanes 6–8) or if hexokinase/glucose were added (lanes 9–11; the identity of the smaller fragment is unclear). In contrast, in the presence of ATP, when SecA is continuously pushing the polypeptide chain into the channel, the intermediate remained stable (Fig. 2A, lanes 12–14; quantitation in Fig. 2B). When the poorly hydrolyzable ATP analog ATPγS was added, a slightly larger protected fragment was observed (Fig. 2A, lanes 15–17). In the presence of ADP and AlF4, which are thought to generate a transition state of ATP hydrolysis, an additional fragment was seen that was ∼0.5 kDa larger than seen with ATPγS (Fig. 2A, lanes 18–20; the increased intensity of the band is due to the protection of an additional labeled methionine). In both conditions, the fragments were stable over time, indicating that no backsliding occurred. It should be noted that the addition of hexokinase/glucose, ATPγS, or ADP plus AlF4 at the beginning of the reaction prevented translocation, demonstrating that they effectively block SecA function (Fig. 2A, lanes 22–24). Taken together, these data show that for backsliding to occur, the ATP bound to SecA needs to be hydrolyzed.
FIGURE 2.
Nucleotide dependence of backsliding. A, a translocation intermediate was generated with proteoliposomes as in Fig. 1B, using proOmpA (pOA) with cysteines at positions 164 and 202. The intermediate was isolated by centrifugation at 14,000 rpm for 15 min at 4 °C. After washing in nucleotide-free buffer, a sample was taken for t = 0(lane 2) and digested with proteinase K (PK) immediately. The remaining sample was aliquoted, and the indicated components were added, incubated at 37 °C, and digested with proteinase K on ice at the times indicated (lanes 3–20). Lane 1 demonstrates full translocation under reducing conditions, and lanes 22–24 had the indicated components present from the onset of translocation. Lane 21 is an aliquot of sample 2 that was treated with DTT and incubated at 37 °C for 30 min. Lane 25 is an aliquot of sample 2 treated with detergent. B, The mean and S.D. of three independent experiments are shown.
Features of SecY Affecting Backsliding of a Polypeptide—Next we determined features of SecY that affect the sliding of a polypeptide chain in the channel. We first tested the possible role of the plug domain. During translocation the plug is displaced from the center of the channel (15, 16), but it could contact the translocating polypeptide chain and thereby restrict its movement. We used two deletion mutants, lacking either the entire plug domain or half of it (full-plug and half-plug deletions) (20). These mutants regenerate a new plug from neighboring segments, but the new plugs have lost many interactions that are present in the wild type protein (20). Therefore, the mutant channels may open more easily and make less contact with the translocating polypeptide.
Proteoliposomes containing the mutant SecY complexes were active in overall translocation (Fig. 3A, lanes 9 and 17 versus wild type shown in lane 1; see also Ref. 20). When the intermediates were incubated in the absence of ATP, backsliding was slightly slower than with wild type SecY (Fig. 3A, lanes 10–13 and 18–21 versus lanes 2–5; quantitation in Fig. 3B). It thus appears that the plug has a relatively small effect on the rate at which a polypeptide chain slides inside the channel.
FIGURE 3.

Features of SecY influencing backsliding. A, SecY complexes containing SecY lacking plug residues (half plug or full plug deletions, see Ref. 20) were isolated and reconstituted into proteoliposomes. Backsliding was studied as in Fig. 1B, using proOmpA (pOA) with cysteines at positions 175 and 202. The disappearance of the intermediate was followed over time (lanes 2–5, 10–13, and 18–21). Lanes 1, 9, and 17 demonstrate translocation under reducing conditions. Lanes 6, 14, and 22 received DTT after intermediate formation (t = 0) and were incubated at 37 °C for 30 min. Lanes 7, 15, and 23 received hexokinase and glucose (HK/Gl) from the onset of translocation, and lanes 8, 16, and 24 received TX-100 before proteolysis. B, shown are the mean and S.D. of three experiments performed as in A. C, a mutant of SecY with 4 tryptophan residues (SecY-4W) introduced in the pore ring was tested for backsliding as in A. D, shown are the mean and S.D. of three experiments performed as in C. E, as in C, but with a mutant of SecY with 4 serine residues (SecY-4S) introduced in the pore ring. F, shown are the mean and S.D. of three experiments performed as in E.
Another SecY feature that contacts a translocating polypeptide and could therefore determine the rate of its movement is the pore ring (14). To test the role of the pore ring residues, we mutated 4 of the isoleucines to tryptophans (4W). The introduction of these bulky and hydrophobic residues into the pore ring significantly slowed the backsliding of a translocation intermediate (Fig. 3C, lanes 10–13 versus lanes 2–5; quantitation in Fig. 3D); with the 4W mutant, the intermediate was stable for at least 1 h at 37 °C. The forward translocation rate of this mutant was somewhat lower than with wild type SecY complex (data not shown), but after 15 min the amount of translocated material was about the same (Fig. 3C, lane 9 versus lane 1). Apparently, SecA can still efficiently push the polypeptide through these channels.
We also mutated 4 pore ring residues into serines. The overall translocation of this 4S mutant was about the same as that of wild type SecY, but the backsliding rate of an intermediate was reduced (Fig. 3E, lanes 10–13 versus lanes 2–5; quantification in Fig. 3F), although not quite as dramatically as with the 4W mutant. Thus, making the pore ring residues smaller and more hydrophilic also increases the friction of a translocating chain in the channel.
Comparison of the Rates of Backsliding and Translocation—Previous measurements of translocation rates were performed by adding translocation substrate to inverted vesicles or reconstituted proteoliposomes (4, 6, 7, 24). However, these measurements include the time required for the binding of substrate to SecA and SecY and for other steps and therefore do not report the true rate of polypeptide movement through the channel. To determine the actual rate of translocation, we first generated a translocation intermediate with proteoliposomes, using a substrate that contained a disulfide-bonded loop formed between cysteines at positions 175 and 202. A high concentration of DTT was then added to instantly reduce the disulfide bridge and allow the substrate to complete translocation. To prevent new, labeled substrate molecules from initiating translocation, we also added an excess of unlabeled proOmpA, purified in chemical amounts, at the beginning of the chase period. As expected, the unlabeled proOmpA effectively competed with labeled substrate when both were added at the same time (Fig. 4A, lane 9 versus lane 1). During the chase incubation, the translocation intermediate disappeared and the fully translocated substrate appeared with the same kinetics (Fig. 4A, lanes 2–6; quantitation in Fig. 4B). The half-time for forward translocation of the stalled intermediate was ∼1 min, corresponding to ∼50–60 amino acid residues/min. The same rate was measured with even higher DTT concentrations (data not shown), indicating that disulfide bridge reduction is not rate-limiting. With a substrate containing the cysteines at positions 164 and 202, for which a faster backsliding rate had been determined (half-time ∼5 min; Fig. 1C), the forward translocation half-time was 1–2 min (data not shown). Similar experiments, performed with IMVs and a substrate containing cysteines at positions 175 and 202, gave a somewhat higher rate of ∼150 amino acids/min (supplemental Fig. S2). The higher rate was not caused by the proton motive force that can be generated with IMVs, but not with proteoliposomes, because the addition of the uncoupler carbonyl cyanide 3-chloro phenylhydrazone had no effect (supplemental Fig. S2). While our translocation rates are somewhat lower than reported in the literature (270 amino acids/min) (24), the main conclusion is that SecA-mediated forward translocation in the presence of ATP is always faster than backsliding in the absence of ATP.
FIGURE 4.

Forward translocation after reduction of the disulfide-bonded loop. A, a translocation intermediate of radiolabeled proOmpA (pOA) with cysteines at positions 175 and 202 was generated with proteoliposomes as in Fig. 1B, and an aliquot was immediately digested with proteinase K (lane 2). 10 mm DTT and excess unlabeled pOA were added, and samples were taken at the times indicated and digested with proteinase K (lanes 3–6). An aliquot of sample 2 was proteolyzed in the presence of Triton X-100 (TX-100)(lane 8). Lane 1 demonstrates translocation under reducing conditions. Lane 7 had hexokinase and glucose (HK/Gl) present from the onset of translocation. The sample in lane 9 received excess unlabeled pOA at the beginning of the translocation reaction. B, quantification of translocation assays performed as in A. For the disappearance of the intermediate, the intensities were normalized to that observed at t = 0. For the appearance of the full-length species, intensities were normalized with respect to the plateau level at t = 8 min.
DISCUSSION
Our data on backsliding of polypeptides suggest that SecA engaged in translocation binds a substrate in the ATP-bound state and releases it in the ADP-bound state. This is consistent with cross-linking experiments indicating that isolated, nontranslocating SecA interacts with a preprotein in the ATP-bound state (12). ATP binding to SecA might cause a peptide binding domain to close around the polypeptide segment. Simultaneously, the polypeptide would be moved into the channel. After ATP hydrolysis, the peptide binding pocket would open, releasing the polypeptide, allowing SecA to reset and bind the next segment. According to this model, SecA would remain bound to SecY during the ATP hydrolysis cycle, which is consistent with the observation that upon removal of excess SecA by urea washing, forward translocation can be restored by the addition of ATP and DTT.5 The model also predicts that non-hydrolyzable ATP analogs or transition state analogs would lock SecA with a bound polypeptide segment at a position close to the channel, in agreement with our results. In fact, we observed that the protease-protected fragments of the translocation intermediate were slightly larger in the presence of ATPγS or ADP plus AlF4 compared with ATP, indicating that the polypeptide chain can move back a small distance after hydrolysis of an ATP molecule. The polypeptide chain appears to be pushed to the maximum extent into the channel in the presence of ADP plus AlF4, which mimics the transition state of ATP hydrolysis.
Our results show that while SecA is in the ADP-bound state a polypeptide can slide backward in the SecY channel. This shows that SecY does not actually bind the polypeptide chain. Nevertheless, backsliding is relatively slow. In ∼5–20 min, depending on the substrate, only half of the polypeptide population moves back from the channel sufficiently to be cleaved by added protease into a heterogeneous population. The backsliding rate is significantly slower than the forward translocation rates seen in the presence of ATP (50–60 amino acids in 1–2 min with purified SecY in proteoliposomes). The actual difference in the kinetics may be significantly larger because in the backsliding assay even the movement of a few amino acids will result in the loss of a defined band. Taken together, these results suggest that SecY simply provides enough friction so that the polypeptide chain does not slide back to a significant extent while SecA resets in its ADP-bound state to grab the next polypeptide segment. Both a rapid conformational change by SecA and a significant, but not too strong, interaction of SecY with the substrate are required for this mechanism to work.
The backsliding rates determined by us for the fastest substrate are about the same as those seen for prepro-α-factor in the yeast Sec complex (23). Although the driving force for translocation is provided in entirely different ways in bacteria and yeast (SecA pushes polypeptides through the channel, whereas the endoplasmic reticulum luminal ATPase BiP binds to the polypeptide as it emerges in the endoplasmic reticulum lumen) (23), the interaction of the channel with a translocating polypeptide appears to be conserved during evolution.
Disulfide bridge cross-linking had shown that a translocating polypeptide chain contacts residues in both the pore ring and the plug (8, 17). Our present data now suggest that the pore ring in SecY is one source of friction, because the replacement of 4 of the 6 normally occurring pore residues decreased the backsliding rate of a polypeptide in the channel. As might have been expected, introducing large, hydrophobic tryptophan residues at the narrowest point of the channel increased the friction encountered by a translocating polypeptide. Surprisingly, however, we also found that the introduction of small and hydrophilic serine residues caused increased friction. Perhaps this is caused by interactions of the hydroxyl groups of the serines with the carbonyl groups of the polypeptide backbone. Replacing 4 of the pore residues with glycines or aspartates resulted in mutant proteins that did not express well and seemed to be toxic to cells.5 It thus appears that the normally occurring pore residues (isoleucines, leucines, and valines) are optimal with respect to size and polarity. The side chains of these residues would not be expected to make strong contacts with the backbone or the side chains of the translocation substrate. In addition, these residues may have just the right size to not obstruct the movement of a polypeptide through the pore and yet still restrict the permeation of small molecules. It should be noted, however, that amino acid residues outside the actual pore ring may come in contact with the translocating chain once the channel has opened, as suggested by molecular dynamics simulations (25).
Interestingly, the forward translocation of the pore mutants was less affected than their backsliding, perhaps because SecA in its ATP form changes the conformation of SecY or exerts enough force to “push” the polypeptide chain through the altered pore ring. The plug domain is another potential source of friction encountered by a polypeptide chain in the channel. However, we found that backsliding was unaffected in the plug deletion mutants. Although in these mutants new plugs are formed, the amino acids that would contact the polypeptide chain are different from the wild type situation (20) and would not be expected to interact equally strongly with a translocating polypeptide chain.
The friction provided by SecY also appears to depend on the amino acid sequence of the translocating polypeptide located in the channel. We observed differences in backsliding rates with different segments in the channel. Interestingly, forward translocation also occurs at different rates, dependent on the amino acid sequence in the channel; hydrophobic sequences significantly slow translocation (7). Comparing fast and slow backsliding substrates, we have not seen a large difference in hydrophobicity between the polypeptide segments predicted to be inside the channel. It is therefore unclear which features of the amino acid sequence are important for the observed difference.
The comparison between crude inverted vesicles and liposomes containing the purified SecY complex indicates that the backsliding rate of a substrate is mostly determined by the SecY channel. A previous study showed that changes of the SecD/SecF levels in inverted vesicles influence the movement of a polypeptide chain both in the forward and backward direction (26). Although it is possible that SecD/SecF interacts with the translocation substrate and thereby restricts its movement, variation of the SecD/SecF levels might also indirectly affect SecA-mediated translocation. In vivo, translocation at late stages can be driven by a proton motive force. The proton motive force might slow down backsliding or replace SecA in providing the forward driving force.
Our results indicate that a one-hand model, in which a polypeptide chain is bound and released by a single binding site in SecA, is sufficient to mediate translocation. The data exclude a two-hand model in which both polypeptide binding sites are present in a single SecA molecule; in this case, one would have expected that no backsliding occurs in ADP. Our data do not strictly rule out two-hand models in which the two alternating binding sites are present in two different SecA molecules (9). They also do not exclude that the ATPase cycle of SecA could cause conformational changes in SecY that would lead to the alternating tightening and loosening of the “grip” on the translocating polypeptide. However, the grip is never strong enough to prevent backsliding, indicating that this could only be an auxiliary mechanism. The simplest model is therefore that, upon ATP hydrolysis, SecA undergoes a conformational change fast enough to grab the next polypeptide segment before significant backsliding in the channel can occur.
Supplementary Material
Author's Choice—Final version full access.
This work was supported, in whole or in part, by National Institutes of Health Grant GM052586 (to T. A. R.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: DTT, dithiothreitol; IMV, inverted membrane vesicle; pOA, proOmpA.
K. J. Erlandson and T. A. Rapoport, unpublished results.
References
- 1.Rapoport, T. A. (2007) Nature 450 663–669 [DOI] [PubMed] [Google Scholar]
- 2.Osborne, A. R., Rapoport, T. A., and van den Berg, B. (2005) Annu. Rev. Cell Dev. Biol. 21 529–550 [DOI] [PubMed] [Google Scholar]
- 3.Johnson, A. E., and van Waes, M. A. (1999) Annu. Rev. Cell Dev. Biol. 15 799–842 [DOI] [PubMed] [Google Scholar]
- 4.Schiebel, E., Driessen, A. J., Hartl, F. U., and Wickner, W. (1991) Cell 64 927–939 [DOI] [PubMed] [Google Scholar]
- 5.Economou, A., and Wickner, W. (1994) Cell 78 835–843 [DOI] [PubMed] [Google Scholar]
- 6.Uchida, K., Mori, H., and Mizushima, S. (1995) J. Biol. Chem. 270 30862–30868 [DOI] [PubMed] [Google Scholar]
- 7.Sato, K., Mori, H., Yoshida, M., Tagaya, M., and Mizushima, S. (1997) J. Biol. Chem. 272 5880–5886 [DOI] [PubMed] [Google Scholar]
- 8.Osborne, A. R., and Rapoport, T. A. (2007) Cell 129 97–110 [DOI] [PubMed] [Google Scholar]
- 9.Tomkiewicz, D., Nouwen, N., and Driessen, A. J. (2007) FEBS Lett. 581 2820–2828 [DOI] [PubMed] [Google Scholar]
- 10.Hunt, J. F., Weinkauf, S., Henry, L., Fak, J. J., McNicholas, P., Oliver, D. B., and Deisenhofer, J. (2002) Science 297 2018–2026 [DOI] [PubMed] [Google Scholar]
- 11.Karamanou, S., Vrontou, E., Sianidis, G., Baud, C., Roos, T., Kuhn, A., Politou, A. S., and Economou, A. (1999) Mol. Microbiol. 34 1133–1145 [DOI] [PubMed] [Google Scholar]
- 12.van Voorst, F., Vereyken, I. J., and de Kruijff, B. (2000) FEBS Lett. 486 57–62 [DOI] [PubMed] [Google Scholar]
- 13.Papanikou, E., Karamanou, S., and Economou, A. (2007) Nat. Rev. Microbiol. 5 839–851 [DOI] [PubMed] [Google Scholar]
- 14.Van den Berg, B., Clemons, W. M., Jr., Collinson, I., Modis, Y., Hartmann, E., Harrison, S. C., and Rapoport, T. A. (2004) Nature 427 36–44 [DOI] [PubMed] [Google Scholar]
- 15.Harris, C. R., and Silhavy, T. J. (1999) J. Bacteriol. 181 3438–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tam, P. C., Maillard, A. P., Chan, K. K., and Duong, F. (2005) EMBO J. 24 3380–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cannon, K. S., Or, E., Clemons, W. M., Jr., Shibata, Y., and Rapoport, T. A. (2005) J. Cell Biol. 169 219–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tani, K., Tokuda, H., and Mizushima, S. (1990) J. Biol. Chem. 265 17341–17347 [PubMed] [Google Scholar]
- 19.Collinson, I., Breyton, C., Duong, F., Tziatzios, C., Schubert, D., Or, E., Rapoport, T., and Kuhlbrandt, W. (2001) EMBO J. 20 2462–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, W., Schulman, S., Boyd, D., Erlandson, K., Beckwith, J., and Rapoport, T. A. (2007) Mol. Cell 26 511–521 [DOI] [PubMed] [Google Scholar]
- 21.Douville, K., Price, A., Eichler, J., Economou, A., and Wickner, W. (1995) J. Biol. Chem. 270 20106–20111 [DOI] [PubMed] [Google Scholar]
- 22.Osborne, A. R., Clemons, W. M., Jr., and Rapoport, T. A. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 10937–10942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matlack, K. E., Misselwitz, B., Plath, K., and Rapoport, T. A. (1999) Cell 97 553–564 [DOI] [PubMed] [Google Scholar]
- 24.Tomkiewicz, D., Nouwen, N., van Leeuwen, R., Tans, S., and Driessen, A. J. (2006) J. Biol. Chem. 281 15709–15713 [DOI] [PubMed] [Google Scholar]
- 25.Gumbart, J., and Schulten, K. (2006) Biophys. J. 90 2356–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duong, F., and Wickner, W. (1997) EMBO J. 16 4871–4879 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



