Abstract
The recombinant HIV-1 Tat protein contains a small region corresponding to residues 47YGRKKRRQRR57R, which is capable of translocating cargoes of different molecular sizes, such as proteins, DNA, RNA, or drugs, across the cell membrane in an apparently energy-independent manner. The pathway that these peptides follow for entry into the cell has been the subject of strong controversy for the last decade. This peptide is highly basic and hydrophilic. Therefore, a central question that any candidate mechanism has to answer is how this highly hydrophilic peptide is able to cross the hydrophobic barrier imposed by the cell membrane. We propose a mechanism for the spontaneous translocation of the Tat peptides across a lipid membrane. This mechanism involves strong interactions between the Tat peptides and the phosphate groups on both sides of the lipid bilayer, the insertion of charged side chains that nucleate the formation of a transient pore, followed by the translocation of the Tat peptides by diffusing on the pore surface. This mechanism explains how key ingredients, such as the cooperativity among the peptides, the large positive charge, and specifically the arginine amino acids, contribute to the uptake. The proposed mechanism also illustrates the importance of membrane fluctuations. Indeed, mechanisms that involve large fluctuations of the membrane structure, such as transient pores and the insertion of charged amino acid side chains, may be common and perhaps central to the functions of many membrane protein functions.
Keywords: cell-penetrating peptide, antimicrobial peptide, drug delivery, membrane proteins, pore formation
The HIV-1 Tat protein indicated that some proteins might have short sequence segments responsible for their translocation across cell membranes in a seemingly energy-independent manner (1–8). The sequence responsible for the cellular uptake of the HIV-1 Tat corresponds to residues 47YGRKKRRQRR57R. This peptide is called the Tat peptide, and it has been categorized as a member of a family of peptides called cell-penetrating peptides. These peptides consist of short sequences (10–30 aa) that are highly basic and enter the cells in a seemingly energy-independent manner (1). This property makes them extraordinarily good candidates as transporters for drug delivery (7, 9–11). Therefore, significant effort is currently being invested to understand this phenomenon at a fundamental molecular level, as well as to employ these peptides as drug carriers (2, 12–16).
Much debate exists around what makes it possible for these peptides to translocate across biological membranes. A diverse set of experiments indicates that there may be more than one mechanism contributing to the uptake of these peptides (1–7, 11, 17–20). Among those mechanisms is the possibility that these peptides may be able to spontaneously translocate across the cell membrane, as suggested by several experiments that indicate a nonendocytotic or energy-independent pathway for the uptake. Because these peptides are highly charged and therefore highly hydrophilic, their spontaneous translocation across the hydrophobic core of the membrane is difficult to understand. The most common mechanisms suggested for spontaneous translocation across cell membranes, such as the inverse micelle model (20), the carpet model (21), or the pore-formation model (22) formed by amphipathic α-helical peptides, do not provide a satisfactory explanation of how these peptides are able to translocate.
Experimental studies show that the L and D amino acid sequences, as well as the reverse Tat sequence, can translocate across membranes, suggesting that the secondary structure of the peptide does not affect its ability to translocate (18, 23). Another important finding has been that translocation requires a relatively large concentration of peptides. Crystallographic studies conducted by Wong and coworkers (A. Mishra, V. G. L. Yang, R. Coridan, and G. C. L. Wong, personal communication) showed that aqueous solutions of simple phospholipids and Tat peptides undergo a phase transition from a lipid bilayer to a Pn3m double-diamond phase, where the lipid molecules surround large, 6-nm water pores. This phase transition, which occurs at a relatively high concentration of peptides per lipid (P/L) (one peptide per eight lipids), suggests that the ability of the Tat peptides to induce pores in lipid bilayers may be related to their translocation mechanism in cells. Molecular dynamics simulations have proven to be a valuable approach to study protein- or peptide-membrane complexes by providing new insights into the microscopic details of these interactions (25–30). Those details usually complement the experimental observations and help guide the design of new experiments.
In this manuscript, we present results from molecular dynamics simulations of systems composed of a lipid bilayer, Tat peptides, and water molecules to investigate the mechanism of membrane translocation. We perform extensive molecular dynamics simulations of Tat peptides solvated in water and close to a dioleoyl phosphatidyl choline (DOPC) lipid bilayer membrane. We study several systems that vary in number and concentration of peptides, phospholipids, and water molecules. Our simulations reveal an atomistic mechanism for translocation. At a large concentration of Tat peptides on one leaflet of the bilayer, the arginine side chains of the peptides translocate across the membrane attracted by the phosphate groups on the opposite leaflet. This process initiates the formation of a pore, followed by the translocation of the whole peptide.
The penetration of arginine amino acids into the hydrophobic core of the membrane has been postulated to be involved in important protein membrane processes. For instance, the arginine side chains have been implicated in the mechanism of opening and closing voltage-sensitive ion channels in response to transmembrane potentials (31, 32). Furthermore, it has been postulated that the S4 transmembrane helix in the KvAP K+ channel, which contains four arginine side chains, moves within the membrane in response to a transmembrane potential (33, 34). Molecular dynamics studies (35, 36) have shown that arginine insertion into a bilayer is facilitated by a network of hydrogen bonds with the lipid phosphate groups and water molecules. A common feature of these mechanisms is that the lipid bilayer structure can undergo large perturbations in response to interactions with proteins.
The mechanism of Tat translocation revealed by molecular dynamics involves four steps. First, the arginine and lysine side chains on the Tat peptides bind to the phosphate and carbonyl groups of the zwitterionic phospholipids and occupy a region beneath the phosphate groups at the interface with the carbon chains. Second, at a higher concentration, the Tat peptides sequester phosphate groups from neighboring phospholipids and create regions on the bilayer surface that are crowded with Tat peptides and phosphate groups, as well as regions that are depleted of charged groups. This crowding of charged groups is because of the high concentration of charges in the Tat peptide (8 of 11 amino acids) and results in the attraction between the Tat peptide and phosphate groups on the proximal layer and the phosphate groups on the distal layer. As a result, the bilayer becomes thinner with increasing Tat peptide concentration. Third, arginine and lysine side chains on the Tat peptide and the phosphate groups on the distal bilayer attract each other and start penetrating the lipid bilayer. As these charged groups enter the hydrophobic lipid bilayer, water also penetrates the bilayer and solvates the charged groups. Fourth, the water and phosphate penetration nucleates a water pore in the membrane. Once the pore is formed, the Tat peptides translocate across the membrane by diffusing on the pore walls. The pore closes after a few peptides translocate, in contrast to the persistent pores formed by antimicrobial peptides. This mechanism is supported by multiple molecular dynamics simulations that span times on the order of 100 ns.
Results and Discussion
Here we describe the mechanism of attachment and translocation of the HIV-1 Tat peptide that is suggested by extensive molecular dynamics simulations (see Table 1) of a model lipid bilayer. In each simulation, we initially place the Tat peptides near one layer of the lipid bilayer and explore the translocation of the peptides across the bilayer as a function of the number of Tat peptides P/L. We call the layer where the Tat peptides are placed in the simulations the proximal layer and the other layer the distal layer. The following subsections present the details of each of the four steps involved in the Tat peptide translocation.
Table 1.
Systems studied by molecular dynamics simulations
| Simulation | Labels used to identify the simulations |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | J | K | L | |
| Tat | 1 | 2 | 4 | 4 | 4 | 4 | 4 | 4 | 5 | 6 | 7 | 10 |
| Water | 3,552 | 7,104 | 14,208 | 3,552 | 3,552 | 3,552 | 8,795 | 18,597 | 18,597 | 18,597 | 18,597 | 14,208 |
| Lipid | 72 | 144 | 288 | 72 | 72 | 72 | 92 | 195 | 195 | 195 | 195 | 288 |
| Temperature, K | 323 | 323 | 323 | 323 | 323 | 343 | 363 | 323 | 323 | 323 | 323 | 323 |
| Time, ns | 200 | 80 | 80 | 200 | 200 | 200 | 400 | 80 | 80 | 180 | 80 | 50 |
| Near | 1 | 2 | 3 | 3 | 3 | 3 | 2 | 4 | 5 | 5 | 6 | 9 |
| Distal | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
The table includes the following rows under the heading “Simulation” (in order from top to bottom): the number of Tat peptides, the number of water molecules, the number of lipids, the temperature, and the simulation time. The last two rows list the number of peptides in the proximal and distal layers, respectively. The number of translocating peptides is not listed in either of the last two rows.
The Tat Peptides Bind to the Bilayer Surface.
Several experiments reported strong ionic interactions between Tat peptides and the membrane surface (4, 37). Time-lapse confocal microscopy experiments indicate that dense aggregates of Tat peptides are formed on the surface of the cells that exhibit uptake. To investigate the relative tendency of the peptide to bind to the surface versus stay in bulk water according to its hydrophilic nature, we perform a simulation containing a single peptide close to the surface (simulation A in Table 1). Figs. 1 and 2a show a snapshot after 200 ns of simulation where we can see the peptide bounded to the surface of the membrane. The peptide is partially exposed to water and surrounded by the phosphate and carbonyl groups of the phospholipids. The peptide locates itself underneath the positively charged choline groups and the negatively charged phosphate groups, with the exception of the peptide C-terminal carbonyl group of the peptide. The Tyr residue also is buried in the bilayer. The arginine and lysine amino acid-charged side chains bind to the phosphate and carbonyl groups of multiple lipids. In this configuration, there are 14 phosphate groups in the vicinity of the Tat peptide.
Fig. 1.
Binding of a Tat peptide to a zwitterionic lipid bilayer. (Upper) Snapshot obtained at the end of the 200-ns simulation (simulation A in Table 1). The peptide is partially exposed to water and surrounded by phosphates and carbonyl groups of the phospholipids. (Lower) Positively charged groups of the peptide bind to the phosphate and carbonyl groups of the phospholipid molecules. In this configuration, the Tat peptide binds to 14 phosphate groups.
Fig. 2.
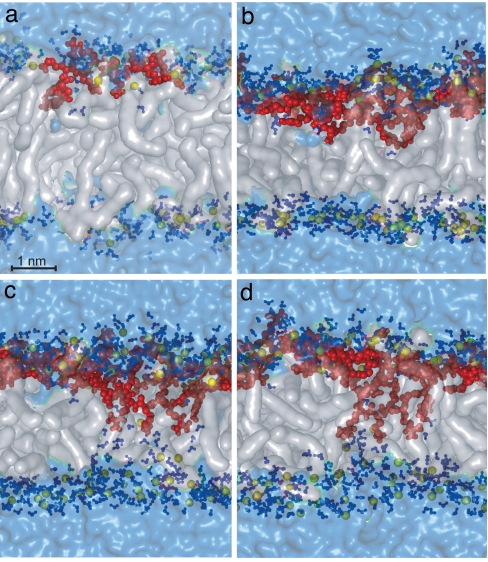
Binding and translocation of Tat peptides (simulations A and D in Table 1). (a) Snapshot of the system comprising 72 DOPC lipids, 3,552 water molecules, and one Tat peptide after a 200-ns simulation. The phospholipid molecules are represented with transparent white surfaces, the phosphate atoms are in yellow spheres, the peptide molecules are in red, any water molecule at a distance of <3.5 Å from any phospholipid or amino acid atom is deep blue, and the rest of the water molecules appear as a pale blue transparent surface. (b–d) Snapshots of the system with four Tat peptides and the same number of DOPC lipids and water molecules as in a after 70, 140, and 200 ns, respectively.
The Effect of Peptide Concentration on Translocation.
Confocal microscopy experiments showed that crowding of peptides on the membrane surface occurs before translocation (4, 37). This finding suggests a mechanism where multiple Tat peptides act cooperatively to translocate, as is the case for antimicrobial peptides (1, 38, 39). Our simulations show that peptide uptake across the bilayer depends on the concentration of peptides. We perform simulations of systems that sample the P/L ratio from 1/72 to 1/18. The system with the lowest peptide concentration consists of one Tat peptide and 72 phospholipids. The system with the highest concentration has four Tat peptides and 72 lipids. At this high concentration, we observe the insertion of arginine side chains of one of the Tat peptides into the hydrophobic region of the bilayer. Fig. 2 b–d shows a time progression of the insertion. We see in Fig. 1 that a Tat peptide binds to 14 phosphate groups in the lipid. As the Tat peptide concentration is increased, not all charged side chains bind to phosphate groups on the proximal layer. Instead, these side chains are attracted by the distal layer phosphate groups, pulling them into the membrane. The charged side chains bring solvation water molecules into the hydrophobic region, thus lowering their free energy of insertion. As the charged side chains reach toward the distal phosphates, some of the distal phosphates and solvating water molecules also reach toward the proximal side (Fig. 2b). Fig. 2d shows the last frame of the 200-ns simulation, where the arginine side chains are bound to phosphates on the distal layer and a chain of water molecules reaches across the bilayer.
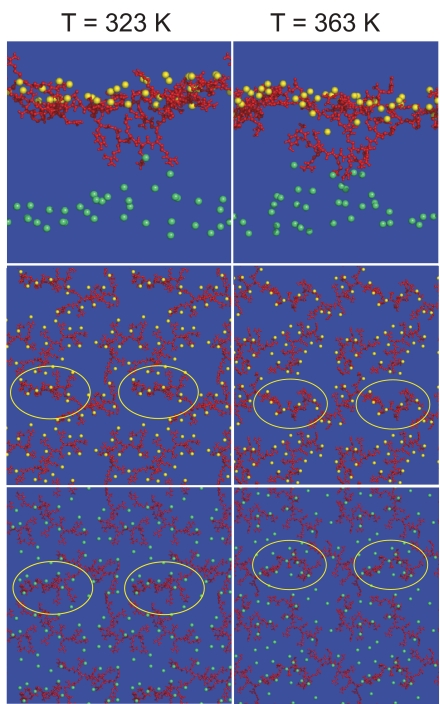
The translocation occurs as a combination of several factors, such as the thinning of the bilayer and the permeation of water and phosphates from the distal layer. Fig. 3 shows a correlation between the positions of the charged side chains of the Tat peptide and the phosphate groups, resulting from the attraction between the phosphate groups and the Tat peptide. Interestingly, the peptide that starts translocating (enclosed in the yellow circles) is attached to fewer phosphate groups on the proximal side of the bilayer, compared with the nontranslocating peptides. The left column corresponds to a simulation at a temperature of 323 K (simulation D in Table 1), and the right column shows similar views, but at a temperature of 363 K (simulation F in Table 1). We also perform a simulation at 342 K (simulation E in Table 1). The strong attraction felt by the phosphate groups toward the peptides becomes clear at higher temperature. Their attraction creates regions over the surface of the membrane that are crowded with phosphates near the peptides and regions that are depleted of phosphates away from the peptides. When an additional peptide attaches to a region with a small number of phosphates, it is attracted by phosphates on the distal layer and eventually translocates to the opposite side.
Fig. 3.
Binding of Tat peptides to phosphate groups. The yellow spheres show phosphates of the proximal layer on which the Tat peptides (in red) are initially bound, and the phosphates of the distal layer are shown by green spheres. For clarity, other molecules are not shown. (Left) Simulation at 323 K (simulation A in Table 1). (Right) Simulation at 363 K (simulation F in Table 1). (Top) Side view of the membrane in which a translocating peptide is visible. (Middle) View of the plane of the membrane and four periodic replicas that only includes the peptides and phosphates of the proximal layer. (Bottom) View of the distal layer of the membrane. These pictures illustrate that the phosphates of both the proximal and distal layers are attracted to the peptides. Heterogeneity induced in the proximal layer with high and low densities of phosphates is visible. Several of the periodic replicas of the translocating peptide are indicated with a yellow ellipse. This peptide is bound to a smaller number of phosphate groups, compared with the rest of the peptides.
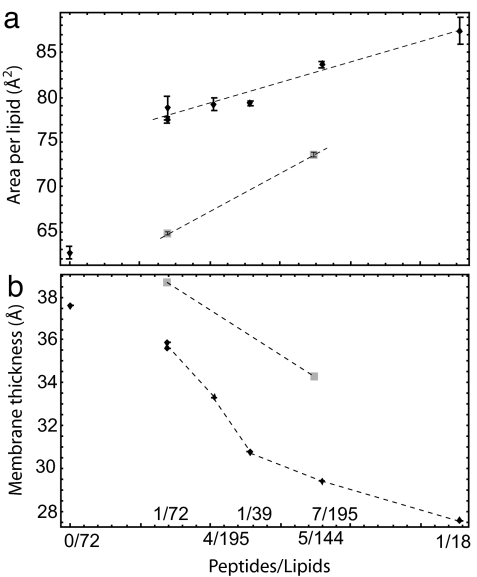
The increase in the number of peptides on one side of the bilayer has several consequences on the bilayer structure that might facilitate the uptake of a peptide. Three main effects are reorientation of the phospholipids near the peptides, the thinning of the bilayer, and the increase of the area per lipid. Fig. 4 shows the area per lipid and the bilayer thickness as a function of the peptide concentration (P/L). The area per lipid increases proportionally with P/L for systems of various sizes. To determine the system size dependence (40) of the results, we simulate two systems with the same concentration (P/L = 1/72), but with a different total number of molecules. One system is composed of one Tat and 72 lipids (simulation A in Table 1), and the other is composed of two Tat peptides and 144 lipids (simulation B in Table 1). We see that the area per lipid and layer thickness for both systems is similar, and that the difference between them is much smaller than the differences obtained by changing the concentrations.
Fig. 4.
Area per lipid and thickness of the bilayer as a function of the number of peptides over the number of phospholipids (simulations A–D and H–L in Table 1). (a) Area per lipid. (b) Thickness of the lipid bilayer. Diamonds indicate results for the cases in which all of the Tat peptides are on the proximal layer. Squares indicate cases in which one Tat peptide is bound to the distal layer and all other peptides are on the proximal layer.
Pore Formation and Translocation Across the Bilayer.
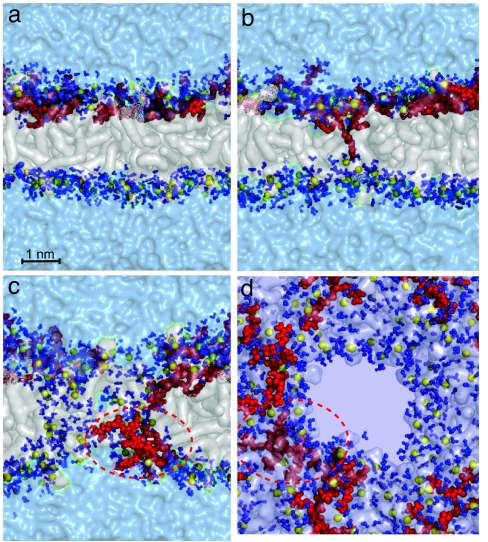
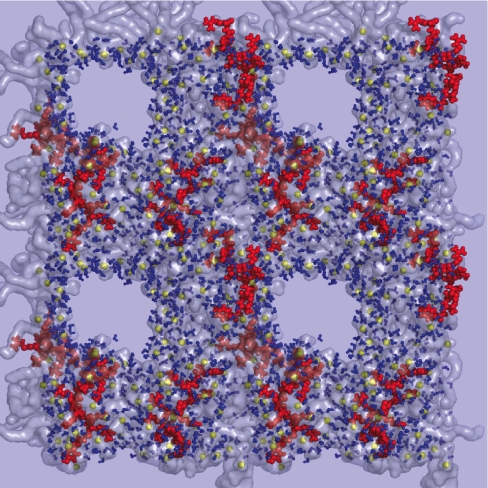
In the simulation described in Fig. 2, we do not see a complete translocation of the Tat peptide. To reduce the effect of periodic boundary conditions on the Tat translocation, we simulate a system with slightly more lipids (92 vs. 72) and significantly more water (8,795 vs. 3,552) (simulation G in Table 1). In this simulation, we observe side chain insertion as in Fig. 2, but we also observe the opening of a 3-nm pore across the bilayer and the diffusion of Tat peptides on the surface of this pore to the other side of the bilayer. Fig. 5 shows the sequence of events that lead to side chain binding (Fig. 5a), insertion (Fig. 5b), the opening of a pore across the bilayer, Tat peptide translocation by diffusion on the pore surface, and the translocated Tat peptide [Fig. 5c; see also supporting information (SI) Movie 1 and SI Figs. 7–11]. We observed this process in two independent simulations. Fig. 5d shows a top view of the water-filled pore across the membrane. Fig. 6 shows the same snapshot as in Fig. 5d, but includes four periodic copies. Pore formation also is observed in a less dense system composed by six Tat peptides, 195 lipids, and 18,597 water molecules (simulation H in Table 1).
Fig. 5.
Snapshots at different times along the simulation of a system composed of 92 DOPC lipids, 8,795 water molecules, and four Tat peptides (simulation G in Table 1). Colors and representations are the same as in Fig. 2. (a) Position of the peptides before translocation. (b and c) Translocation of an arginine amino acid (b) toward the distal layer that nucleates the formation of a water-filled pore (c). (d) Snapshot at the same instant as c but from a direction perpendicular to the membrane, showing another perspective of the pore and translocating peptide.
Fig. 6.
Alternative view of the pore (Fig. 5d), including four periodic images of the system. Phosphate-depletion zones over the surface of the membrane and high concentration of phosphates around the Tat peptides can be clearly seen.
Conclusions
Molecular dynamics simulations provide a mechanism that describes how cell-penetrating peptides rich in arginine amino acids are able to translocate across the membrane in an energy-independent manner. These simulations reveal an atomistic mechanism for translocation, where, at a large concentration of Tat peptides on one leaflet of the bilayer, the Tat peptides are attracted by the phosphate groups on the other layer. This mechanism is composed of four steps. At low Tat concentration, the arginine and lysine side chains on the Tat peptides bind to the zwitterionic phospholipid phosphate and carbonyl groups and occupy a region beneath the phosphate groups, at the interface with the carbon chains of the phospholipids. The Tat peptides act cooperatively to facilitate translocation. At high Tat concentration, the Tat peptides sequester phosphate groups from neighboring phospholipids and create regions on the bilayer surface (proximal layer) that are crowded with Tat peptides and phosphate groups, as well as regions that are depleted of charged groups. This crowding of charged groups is because of the high concentration of charges in the Tat peptide and results in the attraction between the Tat peptide and phosphate groups on one bilayer and the phosphate groups in the distal bilayer. As a result, the bilayer becomes thinner as the Tat peptide concentration increases. The arginine and lysine side chains on the Tat peptide and the phosphate groups on the distal bilayer attract each other and start penetrating the lipid bilayer. As these charged groups enter the hydrophobic lipid bilayer, water also penetrates the bilayer and solvates the charged groups. At longer time scales, we see that water penetration nucleates the formation of a transient water pore in the membrane. The fast transient nature (less than a microsecond) of the pore may explain why cell death because of leakage was not observed with Tat peptides.
The Tat peptides translocate across the membrane by diffusing onto the pore walls while carrying with them the attached phospholipids. Labeling of the phospholipids, according to their initial partitioning between the distal and proximal layers, clearly shows that the phospholipids positions are reshuffled during translocation.
The mechanism revealed by our simulations can explain some of the key elements required by Tat to penetrate the cell observed experimentally. The positively charged arginine and lysine amino acids are important to facilitate the strong interaction with the phosphates groups on both sides of the membrane. When the peptides are bound to the surface, they produce a thinning of the membrane that reduces the hydrophobic free-energy barrier, increasing the probability that the peptides will reach the other side of the membrane. The positive charge also contributes in positioning the peptides between the phosphate groups and the carbon chains and away from the choline groups, which also are positively charged. This location helps enhance the interactions with the phosphate groups on the distal layer. The length of the lysine and arginine side chains facilitates the insertion of the amino acid chains. For shorter amino acid side chains, like histidine, the translocation of one side chain toward phosphates on the distal layer would require either a much thinner membrane or the translocation of more side chains at the same time, making this statistically much less favorable. Experimental reports also show that peptides rich in arginines are more favorably translocated than peptides rich in lysines, and that by increasing the length of the guanidinium side chain the translocation of peptides increases significantly (23). We observe that, in fact, the arginine amino acids insert more easily than lysines. This finding could be partially because of their length and partially because of the way they interact with the phosphate groups. The fundamental role played by the interaction between arginine amino acids and phosphate groups is not exclusive to the present mechanism; it also is a central element in several biological processes. For example, in potassium channels, this interaction is a key ingredient of the voltage gating, and it has been suggested that the usage of arginine in voltage sensors might be an adaptation to the phospholipid composition of cell membranes (32). Previous studies using molecular dynamics simulations found that the arginines also produce a strong local thinning of the membrane around the potassium ion channel up to 1 nm (36). There are other biological examples where the arginine–phosphate interaction does not involve lipid membranes. Such is the case of the arginines of the Tat peptide region of the HIV-1 TAT, which form noncovalent bonds with the phosphate groups in the RNA. This binding of the HIV-1 TAT protein to RNA is critical for transcription (41). This diversity of biological phenomena indicates that the special binding of the arginines to phosphate groups is not only related to its charge and length, but also to the specific structure of the guanidinium group.
We choose conditions that increase the probability of translocation of Tat peptides while including a minimal number of components to maintain the simplicity of our model. Cell membranes are more complex, and there may be other factors that enhance or reduce the uptake through the pathway we propose. For example, the cell membranes are composed of lipid mixtures, and there may be a net electrostatic bias that facilitates the peptide translocation into the cell. The length of the phospholipid carbon chains, the surface tension of the membrane, and the ionic composition of the solution on both sides of the cell are factors that could have a considerable effect. The time scale and system sizes for the pore opening, and the translocation of this peptide across the cell membrane, are at the upper boundary of what is currently reached with molecular dynamic simulations. The present study gives an estimate of the time and size scales that need to be explored to detect and analyze peptide translocation. The addition of salt and counter ions to the solution could potentially slow down the relaxation times, as well as the translocation. However, a high density of peptides enhances the probability of peptide translocation, which is one reason we chose to study relatively high concentrations of peptides (as measured by P/L), but lower than the concentration at which the phospholipids undergo a phase transition (P/L = 1/8) (A. Mishra, V. G. L. Yang, R. Coridan, and G. C. L. Wong, personal communication).
The mechanism proposed here highlights molecular details that have implications on phenomena beyond these of cell-penetrating peptides. The cell membrane is far more complex than an inert hydrophobic barrier and acts as an active participant in membrane protein function. In the translocation of the Tat peptide, we find that the membrane transiently changes its local topology by forming a pore that facilitates the diffusion of the peptide on its surface, allowing it to reach the distal side. This level of membrane flexibility and the ubiquitous nature of the phosphate–guanidinium interactions may be essential in several other biological mechanisms, such as pore formation by antimicrobial peptides (39, 42), voltage-gated ion channels, and flippase regulation of lipid distribution on different faces of a bilayer (43, 44).
Methods
Molecular dynamics simulations were performed to study the translocation mechanism of the protein transduction domain of the HIV-1 Tat across model membranes. The Tat peptides were placed in a periodically repeating box containing a preequilibrated lipid membrane composed of DOPC lipids and water molecules (45, 46). Several simulations performed with different numbers of peptides, lipids, and water molecules are listed in Table 1. The peptides were placed near one side of the bilayer, such that the Tat peptides bound mostly to one layer of the bilayer. These configurations were away from equilibrium. Here we explored how the systems relaxed from these configurations. Because of periodic boundary conditions, there were two paths by which a Tat peptide on one layer moved to bind to the other layer: one that required translocation, and another that required diffusion of the Tat peptide from the initial configuration near the water–bilayer boundary on one layer to the other layer. Our calculations included a large number of water molecules, such that binding to the proximal layer, given the initial conditions we selected, was favored. Two of the systems we generated chose to bind one Tat peptide on the distal layer of the bilayer.
The simulations were performed by using the GROMACS package (47) on a cluster of dual Opteron processors. The overall temperature of the water, lipids, and peptides was kept constant, coupling independently each group of molecules at 323 K with a Berendsen thermostat (48). The pressure was coupled to a Berendsen barostat at 1 atm separately in every dimension (48). The temperature and pressure time constants of the coupling were 0.2 and 2 ps, respectively. The integration of the equations of motion was performed by using a leap frog algorithm with a time step of 2 fs. Periodic boundary conditions were implemented in all systems. A cutoff of 1 nm was implemented for the Lennard–Jones and the direct space part of the Ewald sum for Coulombic interactions. The Fourier space part of the Ewald splitting was computed by using the particle-mesh Ewald method (49), with a grid length of 0.11 nm on the side and a cubic spline interpolation. Periodic boundary conditions and Ewald summations that do not include the surface term ensured system electroneutrality (50). We used the SPC/E (24), the lipid parameters were from Berger et al. (45), and the peptide parameters were from the GROMACS force field (47).
We simulated various systems with varying number of lipids, peptides, and water molecules. These simulations were used to explore the Tat peptide binding to the lipid bilayer, the P/L ratio, and the system size dependence of the binding mode. To observe the initial mechanism of attachment of the peptides to the surface of the lipid bilayer, two systems were built: a system composed of one peptide, 36 lipids per layer, and 3,552 water molecules (simulation A in Table 1); and another system with twice the number of molecules as the first system and the same P/L (simulation B in Table 1). These systems were simulated for 200 and 80 ns, respectively. The area per lipid and the interaction of the peptide with the lipid was similar for both systems, indicating that there was no significant system size dependence in the observed lipid–peptide interaction.
To study the dependence of the area per lipid as a function of the P/L ratio, and to explore the translocation mechanism under other lipid–peptide concentrations, we simulated systems composed of more lipids and water molecules. We increased the P/L ratio by subsequently adding an extra peptide to the system composed of 36 lipids per layer (72 lipids). The peptides were added to the water–bilayer interface. After adding each peptide, the system was energy minimized, equilibrated for 1 ns, and simulated for 30 ns before another peptide was added. This procedure was continued until we had a system with four peptides and 72 lipids. The systems with one (simulation A in Table 1) and four (simulation D in Table 1) Tat peptides were simulated for 200 ns. Two additional 200-ns simulations were performed at higher temperatures. The configuration of the system with four peptides and 72 lipids was used as the initial configuration for two 200-ns simulations at 343 K (simulation E in Table 1) and 363 K (simulation F in Table 1). The features of the simulations at higher temperature were similar to those at 323 K, but the system reached a steady state in shorter simulation time. To cover intermediate values of P/L (between 1/72 and 1/18), we simulated systems with other compositions. In particular, we simulated a system composed of 46 lipids per layer, 8,795 water molecules, and four peptides (simulation G in Table 1; P/L = 1/23). This system was simulated for 60 ns. To cover a broad region of the P/L ratio, we simulated systems that contain 195 lipids (95 in the proximal layer and 100 in the distal layer), 18,597 water molecules, and four (simulation H in Table 1), five (simulation I in Table 1), six (simulation J in Table 1), and seven (simulation K in Table 1) Tat peptides. The systems with five and six Tat peptides were started from the 10-ns configuration of the system with four peptides. The system with seven Tat peptides was started from the 10-ns configuration of the system with five Tat peptides. The system with four Tat peptides was simulated for 80 ns, whereas the other systems were simulated for 40 ns. These systems had P/L values of 4/195, 1/39, 2/65, and 7/195. The two systems where a peptide bound on the distal layer of the bilayer had P/L values of 1/72 and 10/288 (simulations C and L in Table 1). Interestingly, the two systems had a smaller area per lipid than the systems with all Tat peptides on one side, thus giving us a glance at what the final state would be after a peptide translocated through the bilayer. The total simulation time exceeded 1.2 μs.
Supplementary Material
ACKNOWLEDGMENTS.
The authors thank G. Wong for valuable discussions and S. Garde for careful proofreading of the manuscript. This work was supported by National Science Foundation Grant DMR-0117792 and by the Rensselaer Polytechnic Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0706574105/DC1.
References
- 1.Lundberg P, Langel U. J Mol Recognit. 2003;16:227–233. doi: 10.1002/jmr.630. [DOI] [PubMed] [Google Scholar]
- 2.Torchilin VP, Levchenko TS. Curr Protein Pept Sci. 2003;4:133–140. doi: 10.2174/1389203033487298. [DOI] [PubMed] [Google Scholar]
- 3.Torchilin VP, Rammohan R, Weissig V, Levchenko TS. Proc Natl Acad Sci USA. 2001;98:8786–8791. doi: 10.1073/pnas.151247498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ziegler A, Nervi P, Durrenberger M, Seelig J. Biochemistry. 2005;44:138–148. doi: 10.1021/bi0491604. [DOI] [PubMed] [Google Scholar]
- 5.Jarver P, Langel U. Drug Discov Today. 2004;9:395–402. doi: 10.1016/S1359-6446(04)03042-9. [DOI] [PubMed] [Google Scholar]
- 6.Jarver P, Langel U. Biochim Biophys Acta. 2006;1758:260–263. doi: 10.1016/j.bbamem.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 7.Futaki S. Int J Pharm. 2002;245:1–7. doi: 10.1016/s0378-5173(02)00337-x. [DOI] [PubMed] [Google Scholar]
- 8.Chugh A, Eudes F. Biochim Biophys Acta. 2007;1768:419–426. doi: 10.1016/j.bbamem.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 9.Brooks H, Lebleu B, Vives E. Adv Drug Deliv Rev. 2005;57:559–577. doi: 10.1016/j.addr.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 10.El-Andaloussi S, Holm T, Langel U. Curr Pharm Des. 2005;11:3597–3611. doi: 10.2174/138161205774580796. [DOI] [PubMed] [Google Scholar]
- 11.Vives E. J Control Release. 2005;109:77–85. doi: 10.1016/j.jconrel.2005.09.032. [DOI] [PubMed] [Google Scholar]
- 12.Chauhan A, Tikoo A, Kapur AK, Singh M. J Control Release. 2007;117:148–162. doi: 10.1016/j.jconrel.2006.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henriques ST, Melo MN, Castanho MA. Mol Membr Biol. 2007;24:173–184. doi: 10.1080/09687860601102476. [DOI] [PubMed] [Google Scholar]
- 14.Pooga M, Langel U. Methods Mol Biol. 2005;298:77–89. doi: 10.1385/1-59259-877-3:077. [DOI] [PubMed] [Google Scholar]
- 15.Wadia JS, Dowdy SF. Adv Drug Deliv Rev. 2005;57:579–596. doi: 10.1016/j.addr.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 16.Al-Taei S, Penning NA, Simpson JC, Futaki S, Takeuchi T, Nakase I, Jones AT. Bioconjug Chem. 2006;17:90–100. doi: 10.1021/bc050274h. [DOI] [PubMed] [Google Scholar]
- 17.Vives E, Richard JP, Rispal C, Lebleu B. Curr Protein Pept Sci. 2003;4:125–132. doi: 10.2174/1389203033487306. [DOI] [PubMed] [Google Scholar]
- 18.Futaki S, Suzuki T, Ohashi W, Yagami T, Tanaka S, Ueda K, Sugiura Y. J Biol Chem. 2001;276:5836–5840. doi: 10.1074/jbc.M007540200. [DOI] [PubMed] [Google Scholar]
- 19.Vives E. J Mol Recognit. 2003;16:265–271. doi: 10.1002/jmr.636. [DOI] [PubMed] [Google Scholar]
- 20.Derossi D, Calvet S, Trembleau A, Brunissen A, Chassaing G, Prochiantz A. J Biol Chem. 1996;271:18188–18193. doi: 10.1074/jbc.271.30.18188. [DOI] [PubMed] [Google Scholar]
- 21.Pouny Y, Rapaport D, Mor A, Nicolas P, Shai Y. Biochemistry. 1992;31:12416–12423. doi: 10.1021/bi00164a017. [DOI] [PubMed] [Google Scholar]
- 22.Gazit E, Lee WJ, Brey PT, Shai Y. Biochemistry. 1994;33:10681–10692. doi: 10.1021/bi00201a016. [DOI] [PubMed] [Google Scholar]
- 23.Wender PA, Mitchell DJ, Pattabiraman K, Pelkey ET, Steinman L, Rothbard JB. Proc Natl Acad Sci USA. 2000;97:13003–13008. doi: 10.1073/pnas.97.24.13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berendsen HJC, Grigera JR, Straatsma TP. J Phys Chem. 1987;91:6269–6271. [Google Scholar]
- 25.Tieleman DP, Berendsen HJ, Sansom MS. Biophys J. 2001;80:331–346. doi: 10.1016/S0006-3495(01)76018-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tieleman DP, Marrink SJ. J Am Chem Soc. 2006;128:12462–12467. doi: 10.1021/ja0624321. [DOI] [PubMed] [Google Scholar]
- 27.Pastor RW, Venable RM, Feller SE. Acc Chem Res. 2002;35:438–446. doi: 10.1021/ar0100529. [DOI] [PubMed] [Google Scholar]
- 28.Ulmschneider MB, Tieleman DP, Sansom MS. Protein Eng Des Sel. 2005;18:563–570. doi: 10.1093/protein/gzi059. [DOI] [PubMed] [Google Scholar]
- 29.Klauda JB, Kucerka N, Brooks BR, Pastor RW, Nagle JF. Biophys J. 2006;90:2796–2807. doi: 10.1529/biophysj.105.075697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roux B, Schulten K. Structure (London) 2004;12:1343–1351. doi: 10.1016/j.str.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 31.Jiang Y, Ruta V, Chen J, Lee A, MacKinnon R. Nature. 2003;423:42–48. doi: 10.1038/nature01581. [DOI] [PubMed] [Google Scholar]
- 32.Schmidt D, Jiang QX, MacKinnon R. Nature. 2006;444:775–779. doi: 10.1038/nature05416. [DOI] [PubMed] [Google Scholar]
- 33.Hessa T, White SH, von Heijne G. Science. 2005;307:1427. doi: 10.1126/science.1109176. [DOI] [PubMed] [Google Scholar]
- 34.Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, Andersson H, Nilsson I, White SH, von Heijne G. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- 35.Sands ZA, Sansom MS. Structure (London) 2007;15:235–244. doi: 10.1016/j.str.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Freites JA, Tobias DJ, von Heijne G, White SH. Proc Natl Acad Sci USA. 2005;102:15059–15064. doi: 10.1073/pnas.0507618102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ziegler A, Blatter XL, Seelig A, Seelig J. Biochemistry. 2003;42:9185–9194. doi: 10.1021/bi0346805. [DOI] [PubMed] [Google Scholar]
- 38.Huang HW. Biochemistry. 2000;39:8347–8352. doi: 10.1021/bi000946l. [DOI] [PubMed] [Google Scholar]
- 39.Huang HW, Chen FY, Lee MT. Phys Rev Lett. 2004;92:198304. doi: 10.1103/PhysRevLett.92.198304. [DOI] [PubMed] [Google Scholar]
- 40.Herce HD, Garcia AE. J Chem Phys. 2006;125:224711. doi: 10.1063/1.2378893. [DOI] [PubMed] [Google Scholar]
- 41.Calnan B, Tidor B, Biancalana S, Hudson D, Frankel A. Science. 1991;252:1167–1171. doi: 10.1126/science.252.5009.1167. [DOI] [PubMed] [Google Scholar]
- 42.Tang M, Waring AJ, Hong M. J Am Chem Soc. 2007;129:11438–11446. doi: 10.1021/ja072511s. [DOI] [PubMed] [Google Scholar]
- 43.Bretscher MS. Nat New Biol. 1972;236:11–12. doi: 10.1038/newbio236011a0. [DOI] [PubMed] [Google Scholar]
- 44.Rothman JE, Lenard J. Science. 1977;195:743–753. doi: 10.1126/science.402030. [DOI] [PubMed] [Google Scholar]
- 45.Berger O, Edholm O, Jahnig F. Biophys J. 1997;72:2002–2013. doi: 10.1016/S0006-3495(97)78845-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feller SE, Yin D, Pastor RW, MacKerell AD., Jr Biophys J. 1997;73:2269–2279. doi: 10.1016/S0006-3495(97)78259-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 48.Berendsen HJC, Postma JP, van Gunsteren WF, DiNola A, Haak JR. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 49.Darden T, York D, Pedersen L. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 50.Herce HD, Garcia AE, Darden T. J Chem Phys. 2007;126:124106. doi: 10.1063/1.2714527. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.