Abstract
Hematopoietic stem cell (HSC) self-renewal and differentiation is regulated by cellular and molecular interactions with the surrounding microenvironment. During ontogeny, the aorta–gonad–mesonephros (AGM) region autonomously generates the first HSCs and serves as the first HSC-supportive microenvironment. Because the molecular identity of the AGM microenvironment is as yet unclear, we examined two closely related AGM stromal clones that differentially support HSCs. Expression analyses identified three putative HSC regulatory factors, β-NGF (a neurotrophic factor), MIP-1γ (a C–C chemokine family member) and Bmp4 (a TGF-β family member). We show here that these three factors, when added to AGM explant cultures, enhance the in vivo repopulating ability of AGM HSCs. The effects of Bmp4 on AGM HSCs were further studied because this factor acts at the mesodermal and primitive erythropoietic stages in the mouse embryo. In this report, we show that enriched E11 AGM HSCs express Bmp receptors and can be inhibited in their activity by gremlin, a Bmp antagonist. Moreover, our results reveal a focal point of Bmp4 expression in the mesenchyme underlying HSC containing aortic clusters at E11. We suggest that Bmp4 plays a relatively late role in the regulation of HSCs as they emerge in the midgestation AGM.
Keywords: aorta–gonad–mesonephros, BMP4, microenvironment, stromal cells
The adult hematopoietic system is sustained by a cohort of hematopoietic stem cells (HSCs) residing in specialized niches within the bone marrow (BM) and liver at the adult and fetal stages of development. These niches provide a supportive microenvironment for the maintenance and growth of HSCs. In the marrow, HSCs are localized near the bone (1), in the endosteal niches (2, 3), and in the vascular niches of the sinusoids (4). Indeed, several signaling pathways play a role in the regulation of adult BM HSC activity. These include hematopoietic cytokines, intracellular regulators, and developmental regulators such as the Notch, Wnt, TGFβ, and Hh signaling pathways (1–3, 5–7). During mouse development, the first adult-type HSCs emerge in the midgestation aorta–gonad–mesonephros (AGM) region (8). These first HSCs are generated de novo, most likely from endothelial cells (possessing hemogenic potential) localized within the ventral wall of the dorsal aorta (9, 10). Despite knowledge concerning the localization of these cells, little is known about the surrounding microenvironment that initiates and supports the growth of these first HSCs.
Attempts to study hematopoietic supportive microenvironments in the adult mouse initially used in vitro culture of primary adherent BM cells (11). Subsequently, a wide variety of stromal cell lines were generated from adult BM and other hematopoietic territories, including the fetal liver (FL), yolk sac, and AGM (12–17). Most stromal cell lines exhibit a vascular smooth muscle phenotype and, under appropriate mesenchymal lineage-differentiation culture conditions, reveal osteo-, adipo-, and chondrogenic, and/or endothelial potential (18–20). Importantly, some of these stromal cell lines support HSC and hematopoietic progenitor growth in coculture systems and, thus, are representative of the in vivo microenvironment. Using previously isolated stromal clones from the aorta and urogenital subregions of the AGM, we found that some clones are potent supporters of HSCs (16, 17). These clones and primary cells from the AGM exhibit mesenchymal differentiation potential, and, hence, the AGM stromal cells appear to be representative of the midgestational HSC-supportive microenvironment (19, 21).
Transcriptional profiling has provided insight into the identity of molecules elaborated by the HSC-supportive microenvironment. The comparative expression signatures of embryonic day (E)14 liver-derived HSC-supportive and -nonsupportive stromal lines (22) show a complex genetic program involving many molecules whose function in hematopoiesis is unknown. The genetic program of the developmentally earlier AGM microenvironment was recently examined within our panel of closely related AGM-derived stromal cell lines (23). In the study presented here, further expression analysis on AGM stromal cell lines identified three additional candidate HSC-regulatory molecules: β-NGF, a neurotrophic factor; MIP-1γ, a chemokine of the C–C family whose activity on HSC potential was previously unexplored; and Bmp4, a known developmental signaling factor. Our in vivo transplantation experiments reveal that all three of these factors affect HSC activity. Most interestingly, the localized expression of Bmp4 in the mesenchymal cells underlying emergent clusters of hematopoietic cells in the ventral midgestation mouse aorta strongly suggests that Bmp4 is involved in the complex regulation of HSC growth.
Results
Identification and Expression of Candidate HSC Regulators in AGM Stromal Cells and Embryonic Hematopoietic Tissues.
Previously, AGM stromal clones derived from E11 urogenital ridges were shown to provide differential support for AGM HSCs in in vitro cocultures: UG26.1B6 provides potent HSC support, whereas UG26.3B5 is less supportive (16, 19). To identify potential AGM HSC regulators, we compared the gene expression profile of these clones using arrays containing >500 genes encoding cytokines, cytokine receptors, and molecules involved in signal transduction pathways and cell migration. The genes highly expressed by both stromal clones include molecules that are known hematopoietic and developmental regulators [supporting information (SI) Table 3]. A few differentially expressed genes were found, including β-NGF (a neurotrophic factor), MIP-1γ (a member of the C–C chemokine family), and Bmp4 (a morphogen). Quantitative real-time PCR (Fig. 1) demonstrated expression of β-NGF and Bmp4 at 2.5- and 4-fold higher levels respectively in UG26.1B6 cells, whereas MIP-1γ was expressed at a 5-fold higher level in UG26.3B5 cells. Thus, β-NGF and Bmp4 may act as positive regulators, and MIP-1γ may act as a negative regulator of AGM HSC growth.
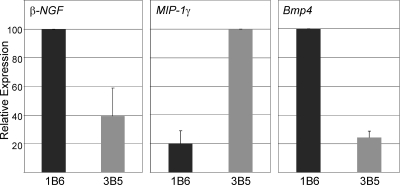
Fig. 1.
Differential expression of candidate HSC regulators by UG26.1B6 and UG26.3B5 stromal cells. Real-time PCR was performed for β-NGF, MIP-1γ, and Bmp4 on three RNA samples from UG26.1B6 (HSC-supportive) and UG26.3B5 (less supportive) stromal clones. Graphs show the relative expression levels of each factor in both cell lines. n = 3. For MIP-1γ and Bmp4, P < 0.001.
To determine whether the candidate regulators are normally expressed within embryonic hematopoietic sites, RT-PCR for β-NGF, MIP-1γ, and Bmp4 and their receptors was performed on aorta mesenchyme, urogenital ridges, liver, yolk sac, and limb bud (positive control) tissues from E11 mouse embryos (Fig. 2). β-NGF is expressed at low levels in both subregions of the AGM but not the liver or yolk sac. Whereas the p75 receptor gene is expressed in aorta, urogenital ridges, and liver, the high-affinity receptor gene TrkA is expressed only in urogenital ridges and liver. MIP-1γ and its receptor Ccr1 are expressed in both subregions of the AGM and the other hematopoietic tissues. Bmp4 and Bmp receptors Alk3, Alk6, and BmprII are expressed in all of the embryonic hematopoietic tissues, as are the intracellular signaling molecules Smad1, 4, and 5 (the latter only at low levels). These results suggest that the three regulatory pathways may be active in vivo in the AGM region and could affect HSC development.
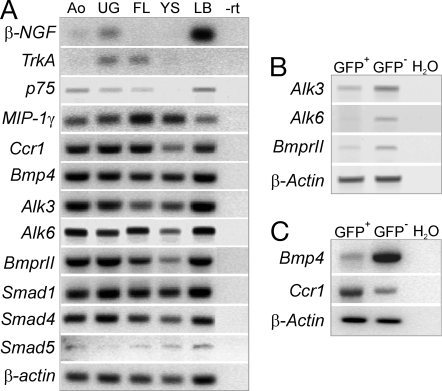
Fig. 2.
Expression of candidate regulators and receptors in E11 embryonic tissues and enriched AGM HSCs. RT-PCR analysis of E11 embryonic tissues for expression of β-NGF and NGF receptors (p75 and TrkA), MIP-1γ and receptor Ccr1, Bmp4, and Bmp4 receptors (Alk3, Alk6, and Bmp-RII), and downstream signaling molecules Smad1, 4, and 5 (A); GFP+ and GFP− sorted AGM cells from E11 Ly-6A GFP embryos for expression of Alk3, Alk6 and BmprII (B); and for expression of Bmp4 and Ccr1 (C). β-actin expression was the normalization control. Ao, aorta and mesenchyme; UG, urogenital ridges; FL, fetal liver; YS, yolk sac; LB, limb bud.−rt, no reverse transcriptase.
Effects of Recombinant β-NGF, MIP-1γ and Bmp4 on AGM HSCs.
Previously, others have suggested that β-NGF and its receptor TrkA participate in the control of hematopoiesis (24, 25). A MIP-1γ family member MIP-1α, which shares 24% sequence homology, is a known inhibitor of BM HSC proliferation (26, 27). Bmp4 is known to affect the commitment of mesoderm to hematopoietic fate in embryos (28, 29) and ES cells (30, 31), primitive erythropoiesis (32), and the expansion of human hematopoietic progenitors (33). Thus, to study the effects of β-NGF, MIP-1γ, and Bmp4 on AGM hematopoiesis and HSCs, we performed in vivo transplantation experiments of cells from E11 AGMs after 3 days of explant culture in the presence of these exogenous factors. This organ culture system has been shown to maintain and promote the autonomous expansion of AGM HSCs and CFU-S (8).
As shown in Table 1, HSC activity is increased almost 2-fold in the presence of 250 ng/ml β-NGF (60% of recipients repopulated) compared with AGM explants cultured in its absence (36% of recipients repopulated) or with 25 ng/ml β-NGF. A 1.3-fold increase in HSC activity was found when AGM explants were cultured with 20 ng/ml Bmp4 (77% of recipients repopulated compared with control 58%). No increase in HSC activity was found in the presence of 2 ng/ml Bmp4 (43% of recipients repopulated). These factor-induced increases in HSC activity suggest that the higher expression of β-NGF and Bmp4 by UG26.1B6 (compared with UG26.3B5) may be responsible, in part, for its in vitro support of HSCs.
Table 1.
Effects of recombinant factors on AGM HSC repopulating activity
| Recombinant factor | Cells injected | Repopulated/transplanted (mice repopulated, %) | Donor chimerism range (mean, %) |
|---|---|---|---|
| None | 0.3 ee | 8/22 (36) | 27–100 (72) |
| β-NGF (25 ng/ml) | 0.3 ee | 3/8 (38) | 29–76 (58) |
| β-NGF (250 ng/ml) | 0.3 ee | 6/10 (60) | 25–100 (75) |
| MIP-1γ (20 ng/ml) | 0.3 ee | 8/13 (62) | 16–100 (75) |
| MIP-1γ (200 ng/ml) | 0.3 ee | 8/15 (53) | 22–100 (74) |
| None | 1 ee | 7/12 (58) | 37–77 (57) |
| Bmp4 (2 ng/ml) | 1 ee | 6/14 (43) | 23–72 (46) |
| Bmp4 (20 ng/ml) | 1 ee | 10/13 (77) | 21–69 (48) |
E11 AGM explants were cultured in the presence of recombinant factors. After 3 days of culture, AGM cells were collected, and 0.3 or 1 embryo equivalents (ee) of cells were injected into irradiated adult recipient mice. Four months after transplantation, mice were analyzed (>10% of donor chimerism is considered positive engraftment). n = 3 for MIP-1γ and Bmp4 and n = 1 for β-NGF.
In the presence of both 20 ng/ml and 200 ng/ml MIP-1γ, AGM HSC activity was also increased (1.5- to 2-fold). This is in contrast to the finding that the UG26.3B5 cells, which are less supportive for BM HSCs, express much higher levels of MIP-1γ than UG26.1B6 cells. Because others have found that MIP-1α inhibits adult BM HSC proliferation, we tested whether MIP-1γ would increase or decrease adult BM HSC activity. When MIP-1γ was added in cocultures of sorted c-kit+Ly-6A GFP+ BM cells and UG26.1B6, HSC activity was diminished (Table 2). Thus, MIP-1γ acts similarly to family member MIP-1α as a negative regulator of adult BM HSCs. The observation that MIP-1γ acts as a positive regulator of AGM HSCs suggests that this factor acts differentially in the embryo and adult.
Table 2.
Effects of recombinant MIP-1γ on enriched BM HSC repopulating activity
| Recombinant protein | Cell dose injected | Repopulated/transplanted (mice repopulated, %) |
|---|---|---|
| None | 5.0 × 103 | 3/6 (50) |
| MIP-1γ (20 ng/ml) | 5.0 × 103 | 1/6 (15) |
| MIP-1γ (200 ng/ml) | 5.0 × 103 | 0/6 (0) |
| None | 2.5 × 103 | 2/6 (30) |
| MIP-1γ (20 ng/ml) | 2.5 × 103 | 2/6 (30) |
| MIP-1γ (200 ng/ml) | 2.5 × 103 | 0/6 (0) |
Adult BM c-kit+Ly-6A GFP+ cells were sorted and cocultured with UG26.1B6 stromal cells in the presence of MIP-1γ. After 10–11 days, all cells were harvested, and the equivalent of 2.5 or 5.0 × 103 input c-kit+Ly-6A GFP+ cells were injected into irradiated adult mouse recipients. Four months after transplantation, mice were analyzed (>10% of donor chimerism is considered positive engraftment). Chimerism ranged from 10% to 100%. n = 2.
Bmp4 Regulates AGM HSCs.
Considering that β-NGF is not highly expressed in the aorta subregion and that MIP1-γ effects on AGM HSCs are complex, we focused further studies on Bmp4. To date, no studies have addressed the functional role of Bmp4 in the regulation of the first HSCs in the AGM microenvironment. If Bmp4 does play a role, it should be possible to modulate HSC activity in AGM explants by inhibiting the Bmp4 pathway with gremlin, an antagonist known to bind Bmps and block Bmp signaling (34).
To examine the role of Bmp4 produced endogenously within the AGM, we added recombinant gremlin to explant cultures. Gremlin activity (250 ng/ml) was verified by its inhibition of Bmp4-mediated differentiation of an osteogenic cell line (SI Fig. 5). When gremlin was added to AGM explants (250 ng/ml), HSC activity was abolished (Fig. 3A). Noggin (250 ng/ml), another Bmp antagonist, also reduced AGM HSC activity, although to a lesser extent (data not shown). The effect of Bmp4 on immature hematopoietic progenitors was also tested by the short-term in vivo Colony Forming Unit–Spleen (CFU-S) and in vitro Colony Forming Unit-Culture (CFU-C) assays. E11 AGM explants cultured in the presence of gremlin yielded a statistically significant decrease (3-fold) in the number of CFU-S11 (Fig. 3B) and CFU-Mix (Fig. 3C; 2-fold decrease) per AGM (P < 0.01). In contrast, gremlin did not significantly affect the number of more mature progenitors (CFU-G, CFU-M, and CFU-GM). Taken together, Bmp4 signaling appears to be involved in positively regulating the number of the most immature AGM hematopoietic progenitors and HSCs.
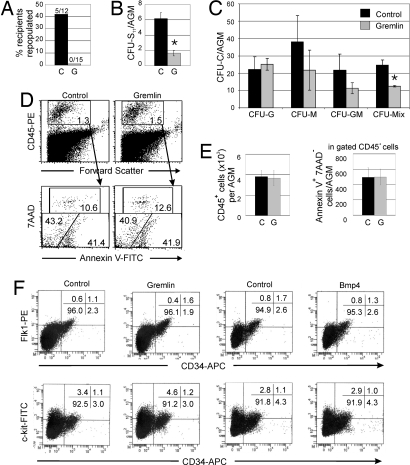
Fig. 3.
Effect of gremlin on the activity and survival of AGM hematopoietic cells. (A–C) E11 AGMs cultured as explants in the presence or absence of gremlin (250 ng/ml) were analyzed after long term HSC repopulation assay (A), CFU-S11 assay (B), and CFU-C assay (C). For HSC repopulation, 0.3 AGM cell equivalents were injected per recipient. For each CFU-S11 condition, 14 mice were injected with 1 AGM equivalent (n = 3, *, P = 0.002). For CFU-C, n = 3. P < 0.01 for CFU-Mix. Data represent the mean ± SD. (D) FACS plots showing 7AAD and Annexin V staining of CD45+ cells from AGMs cultured in the absence or presence of gremlin (250 ng/ml). Percentages of cells shown in gated regions. (E) Graphs show absolute numbers of CD45+ cells and Annexin V+ 7AAD+CD45+ cells per AGM (n = 3). (F) Four-color staining dot plots showing percentages of viable cells from AGM explants cultured in the presence or absence of gremlin or Bmp4. (Upper) Cells gated within the CD45− population. (Lower) Cells gated within the total population. C, control; G, gremlin.
To investigate how Bmp4 mediates its effect on AGM hematopoiesis, the viability of AGM hematopoietic cells cultured in the presence of gremlin was tested. Flow cytometric analysis showed no difference in the percentage and absolute number of CD45+ cells in E11 AGM explants cultured with or without gremlin (Fig. 3 D and E). When these cells were analyzed for 7AAD uptake and expression of the apoptotic marker Annexin V, no differences in the percentages and absolute numbers of CD45+ cells in the preapoptotic or apoptotic stages were observed (Fig. 3 D and E). Therefore, high-dose gremlin is not toxic and does not compromise AGM hematopoietic cell viability or predispose these cells to the apoptotic pathway.
We next examined the effects of gremlin and Bmp4 on endothelial (CD45−Flk-1+CD34+) and hematopoietic progenitor/stem cell (CD34+c-kit+) populations in AGM explants (Fig. 3F). Gremlin affected a small increase and Bmp4 a decrease in the percentages of CD45−Flk-1+CD34+ cells in AGM explants. However, no obvious differences were found for the CD34+c-kit+ population cultured with either gremlin or Bmp4. These data indicate that Bmp4 signaling is not involved in the survival of preexisting HSCs or hematopoietic progenitors and, together with the in vivo functional results, suggest that Bmp signaling plays a role in maintaining HSC and hematopoietic progenitor potential.
Localized Expression of Bmp4 in the E11 AGM Region.
Although Marshall et al. (35) showed a polarized gradient of Bmp4 protein underlying the ventral wall of the human aorta at week 5 of gestation, no comparable study has been performed in the mouse E11 AGM, at the time when HSC activity is highest. Bmp4 immunostaining analysis of an E11 mouse trunk section (Fig. 4A) shows strong localized Bmp4 expression ventral to the dorsal aorta. A patchy signal is seen in areas surrounding the notochord, mesonephros, the region between the dorsal aorta and gut, and dorsal to the cardinal veins. Ventral to the aorta, Bmp4 is found mainly in mesenchymal cells and appears throughout the cell cytoplasm (Fig. 4B). A control stained section (Fig. 4C) and adjacent anti-Bmp4 stained section (Fig. 4D) verified the specificity of staining. To determine whether these cells are actively transcribing Bmp4, in situ analysis was performed. Bmp4 expression was found in cells in the same mesenchymal region underlying the ventral aspect of the dorsal aorta (Fig. 4 E and G).
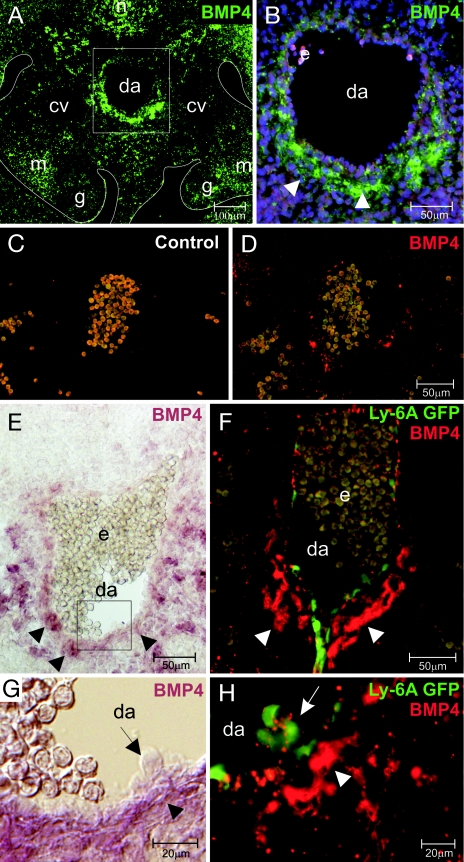
Fig. 4.
Expression of Bmp4 in the midgestation AGM. Transverse sections through E11 AGM (dorsal at top; ventral at bottom) stained (A) with anti-Bmp4 antibody. (B) Framed region in A showing Bmp4 staining (green) in the mesenchyme underlying the ventral wall of the dorsal aorta (arrowheads). Nuclear DAPI staining is blue. (C) Immunostaining control with fluorescent (red) secondary antibody used in D in combination with anti-Bmp4 antibody (late E10/early E11). (E) In situ hybridization analysis showing Bmp4 mRNA (purple). Expression is restricted to mesenchyme underlying the ventral aspect of the dorsal aorta (arrowheads). (F) Transverse section through E11 Ly-6A GFP aorta stained with anti-Bmp4 antibody. GFP+ cells (green) are located in the ventral endothelium of the aorta with Bmp4 protein (red) detected in the mesenchyme underlying GFP+ cells (arrowheads). (G) Framed region in E showing Bmp4-expressing cells (arrowheads) underlying a hematopoietic cluster (arrow). (H) Ventral aspect of aorta in F showing Bmp4 in mesenchymal cells (arrowhead) underlying GFP+ hematopoietic cells (arrow). Some GFP+ cells show punctate Bmp4 signal. cv, cardinal vein; da, dorsal aorta; e, erythrocytes; g, gonad; m, mesonephros; n, notochord.
Immunostaining for Bmp4 was also performed on sections from E11 transgenic Ly-6A GFP embryos to test whether Bmp4-expressing cells are associated with emerging HSCs in the cell clusters along the ventral luminal wall of the dorsal aorta. It has been shown that AGM HSCs are exclusively found in the GFP+ fraction (9). A concentrated localization of Bmp4 expression was found in mesenchymal cells underlying GFP+ cells in the ventral endothelial wall (Fig. 4F). Moreover, whereas the mesenchymal cells are positive for Bmp4 throughout the cytoplasm, some cells, particularly the GFP+ cells in the hematopoietic clusters (Fig. 4H), appear to have foci of Bmp4 on their surface suggestive of an interaction with surface receptors.
Because Bmp4 receptors are known to be expressed by adult BM HSCs (33), we tested whether they are also expressed on AGM HSCs. A enriched HSC population (GFP+) was sorted from Ly-6A GFP transgenic E11 aortas and analyzed by RT-PCR for expression of Bmp receptors Alk3, Alk6, and BmprII. GFP+ cells express Alk3 and BmprII but not Alk6 (Fig. 2B), whereas all three receptors were expressed by GFP− cells. RT-PCR analysis was also performed for Bmp4. As expected, Bmp4 is highly expressed in GFP− cells. GFP+ cells express only low levels of Bmp4 (Fig. 2C). Taken together, these data suggest that Bmp4 produced by the mesenchymal cells underlying the aortic hematopoietic clusters directly influences AGM HSC development.
Discussion
Identification of Regulatory Molecules Expressed by the AGM Hematopoietic Supportive Microenvironment.
Gene expression profiling of HSC-supportive and -nonsupportive cell lines as first reported by Moore and colleagues (22) has provided insight into the molecular players in the fetal/adult hematopoietic microenvironment. Our studies focused on the AGM microenvironment (23) with the UG26.1B6 clone, the most highly HSC-supportive AGM stromal line (16). It was derived from E11 urogenital (UG) tissue along with a closely related stromal line that does not efficiently support HSCs (UG26.3B5). Although it was initially surprising to find HSC-supportive cells in the UG subregion of the AGM, HSCs have been found in E11 UGs after explant culture and in vivo in E12 UGs (9). Similar to bone marrow-derived stromal cells, UG26.1B6 express the Sca-1 marker (17). Sca-1 is expressed at high levels in the UG region of E11 embryos (36), strongly suggesting that UG26.1B6 represents a component of the in vivo AGM HSC niche.
β-NGF, MIP-1γ, and Bmp4, identified through their differential expression in the UG stromal clones, are able to affect AGM HSC growth in vitro. Both β-NGF and Bmp4 were more highly expressed in supportive UG26.1B6 cells than in nonsupportive UG26.3B5 cells. As expected for positive regulators, addition of exogenous β-NGF or Bmp4 to AGM explants resulted in approximately a 2-fold increase in HSCs (as determined by in vivo transplantation). In contrast, MIP-1γ was expressed at higher levels by the nonsupportive cell line UG26.3B5. When MIP-1γ was exogenously added to AGM explants, it did not inhibit HSCs but, instead, showed a positive effect. Hence, expression studies must be complemented by functional assays.
The stimulatory effect of MIP-1γ on AGM HSCs was surprising, given that this factor inhibited adult BM HSCs. We have found by RT-PCR that BM c-kit+Ly-6A GFP+ cells express the MIP-1γ receptor Ccr1 (data not shown), suggesting that BM HSCs can directly respond to MIP-1γ. The effect of MIP-1γ on AGM HSCs appears to be more complex, because both Ly-6A GFP+ and GFP− AGM cells express Ccr1 (Fig. 2C). It is possible that MIP-1γ directly affects HSCs. Alternatively or additionally it could act through nonhematopoietic cells in the microenvironment and they, in turn affect HSCs. UG26.1B6 cells would then represent only a part of the AGM microenvironment. Because we have found that Ccr1 is expressed by both the aortic and UG regions of the AGM, identification of the responding cells will be of great interest. Notwithstanding, MIP-1γ is one of the few factors identified to date that differentially regulates adult and embryonic HSCs.
It is intriguing that the well studied neuronal growth factor β-NGF is expressed by AGM stromal lines and increases AGM HSC repopulating activity. Parallels between neuronal and hematopoietic development have been suggested by several studies (37), particularly in the formation of the enteric nervous system and the lymphoid Peyer's patches (38). Both require signaling through the same receptor system, the RET receptor tyrosine kinase and Gfrα-3, a member of the GDNF family of neuronal receptors. Moreover, many neuronal factors were found to be expressed by fetal liver stromal lines, including neurotactin, neuromodulin, neuroligin2, neuregulin, and pro-NDF-β1 (22). Thus, factors traditionally associated with one lineage may also regulate the development of a different lineage. Further studies with neuronal factors will be particularly interesting because, simultaneous to the generation of HSCs, neural crest cells are migrating through the AGM region of the mouse embryo (39).
Bmp4 and Its Role in the Growth of AGM HSCs.
Several laboratories have shown that the Bmp signaling pathway, and more specifically Bmp4, is closely involved in the commitment of mesodermal cells to the hematopoietic lineage in both Xenopus and mammals (29), plays a role in the hematopoietic differentiation of mouse ES cells (40) and controls the expansion of human BM immature progenitors (33). Our findings of high-level Bmp4 expression by an HSC-supportive AGM stromal cell line, the localized expression of Bmp4 in mouse AGMs, and the results of transplantation studies implicate Bmp4 as a positive regulator of AGM HSCs in vivo.
The response of AGM HSCs to Bmp4 appears to be dose-dependent. Addition of exogenous Bmp4 to AGM cultures at 20 ng/ml resulted in an increase in HSC activity. Addition of 200 ng/ml Bmp4 did not further increase the HSC activity (data not shown), whereas 2 ng/ml had no (or a slightly negative effect) on AGM HSCs compared with the control. Bhatia and colleagues (33) found that human cord blood CD34+CD38−Lin− cells cultured with a similar medium dose of BMP4 (25 ng/ml) increased CFU-C numbers and the frequency of engraftment in NOD/SCID recipients. A lower dose of Bmp4 (5 ng/ml) resulted in a slight decrease in CFC numbers and a much lower frequency of engraftment. Interestingly, the addition of Bmp antagonist gremlin had a more dramatic effect on AGM HSCs. Gremlin (250 ng/ml) abolished AGM HSC activity and decreased CFU-S and CFU-Mix numbers. This was not due to a toxic effect, because high-dose gremlin did not decrease the absolute numbers of hematopoietic cells in AGM explants nor did it induce apoptosis. Preliminary flow-cytometric analytical data have revealed that the numbers of CD45+CD34intermediatec-kit+ cells are increased and the numbers of CD45−CD34+Flk1+ cells are decreased in AGM explants cultured in the presence of Bmp4, suggesting that Bmp4 may induce hematopoietic fate, particularly to immature hematopoietic progenitors and HSCs in the aortic clusters. Because Bmp4 is locally produced in AGM mesenchymal cells adjacent to aortic endothelium and hematopoietic clusters and Bmp4 receptors Alk3 (Type I) and BmpRII (Type II) are expressed by AGM HSCs, we suggest that endogenous Bmp4 regulates HSC development in the AGM in vivo. Future studies will focus on demonstrating the in situ activation of the Bmp4 signaling pathway and the downstream events in specific subpopulations of AGM cells and possible in vivo roles of antagonists, e.g., gremlin, noggin, and follistatin and other Bmps, such as Bmp2 and Bmp7 (29) in regulating HSCs.
In conclusion, the molecular analysis of two closely related AGM stromal clones has allowed the identification and functional characterization of three regulators (MIP-1γ, β-NGF, and Bmp4) and one known regulator (Bmp4) of hematopoietic cells. Our studies suggest that the signaling pathways stimulated by these factors are functional in the midgestation AGM and provide positive signals to increase AGM HSC activity. Moreover, Bmp4 does not act as a survival factor but may promote the differentiation and/or expansion of AGM HSCs. Future studies identifying the specific target populations of Bmp4 signaling (as well as those of MIP-1γ and β-NGF), such as AGM HSCs, hemangioblasts, or hemogenic endothelial cells, could provide insights into how HSCs are generated and/or expanded in embryonic and perhaps adult vascular and mesenchymal microenvironments.
Materials and Methods
Animals.
Embryos were generated from human β-globin (41), Ly-6A GFP transgenic (9), and C57BL/6 or (B10xCBA)F1 mice. The day of vaginal plugging is day 0. BM was from adult Ly-6A GFP mice. Mice were bred at Erasmus Medical Center according to institutional guidelines and procedures carried out in compliance with the Standards for Humane Care and Use of Laboratory Animals.
Expression Analyses.
RNA was prepared from TRIzol-lysed cells (Life Technologies), RNase-free DNaseI (Promega) treated, extracted in phenol/chloroform/isoamyl alcohol, ethanol precipitated, and quantified by spectrophotometry. RNA integrity was confirmed by gel electrophoresis.
Total RNA (2 μg) from UG26.1B6 and UG26.3B5 cells was used for mouse cytokine array screening (R & D Systems). Gene expression was quantitated by PhosphorImaging and autoradiography (X-O Mat, Kodak) by using ImageQuant 5.2 (Molecular Dynamics). Confirmation of differential expression was by real-time PCR. cDNA samples were amplified on an Icycler thermal cycler (Bio-Rad) by using SYBR green fluorescence and analyzed with Icycler IQ detection system software. GAPDH was used to normalize cDNA dilutions. After amplification of serial dilutions of cDNAs, cycle threshold was plotted against cDNA amount, and the slope was used to calculate PCR efficiency. Quantitation of samples was by comparison of the number of cycles required to reach reference and target threshold values.
Tissue-specific gene expression was analyzed by RT-PCR on total RNA from mouse embryonic tissues and sorted AGM cells. OligodT primers (Promega) and reverse transcriptase (SuperscriptII; Stratagene) were used for the cDNA synthesis. Specific primers are listed in SI Table 4.
Hematopoietic Assays.
Recombinant factors Bmp4, gremlin, MIP-1γ, and NGF were from R & D Systems.
In Vitro Cultures.
E11 AGMs were dissected and cultured as explants on 0.65-μm filters at the liquid–air interface in long-term medium (M5300; Stem Cell Tech) with 1 μM hydrocortisone (42). After 3 days at 37°C, explants were dissociated in collagenase. Adult BM cells were sorted for c-kit and Ly-6A GFP expression. Sorted cells (15× 103 to 25 × 103 cells per well) were cocultured with confluent irradiated (30 Gy) stromal cells in M5300 medium with 1 μM hydrocortisone at 33°C, 5% CO2. Medium (half) was changed after 1 week of culture. After 10–11 days, adherent and nonadherent cells were assayed.
In Vivo Transplantation Assay.
Cells harvested from collagenase-treated explants or pooled cells from BM cocultures (2,500 and 5,000 input cells) were injected intravenously into irradiated recipients (9 Gy). Syngeneic spleen cells (2 × 105) were coinjected to confer short-term survival of recipient. Four months after transplantation, peripheral blood DNA was analyzed by semiquantitative PCR for the donor marker (male Ymt, hβ-globin, or GFP). Recipients were considered positive only if >10% of peripheral blood DNA was of donor genotype.
CFU-S11 Assay.
AGM cells were injected intravenously into irradiated recipients (10 Gy) (43) and analyzed 11 days after transplantation. Spleens were fixed in Tellesnicky's solution and CFU-S counted under a dissection microscope.
CFU-C Assay.
Cells were seeded in methylcellulose medium (StemCell Tech) supplemented with SCF, IL-3, IL-6, and Epo, incubated at 37°C in 5% CO2. CFU-G (granulocyte), M (macrophage), GM (granulocyte-macrophage), Mix (granulocyte, erythroid, macrophage, and megakaryocyte) were scored with an inverted microscope at day 10.
Immunostaining and in Situ Hybridization.
E11 Ly-6A GFP and wild-type embryos were fixed in 4% PFA, embedded in PBS/gelatin/sucrose, quick-frozen, and cut at 10 or 20 μm. For Bmp4 protein detection, frozen sections were permeabilized in PBS/0.1% Triton X-100. Immunostainings were as described (44) by using goat anti-human Bmp4 primary antibody (1:50; Santa Cruz Biotechnology) and HRP- (DAKO) or biotin-(Vector Laboratories) conjugated rabbit anti-goat secondary antibodies with streptavidin-FITC (PharMingen) or -Cy3 (Jackson ImmunoResearch).
In situ hybridization used an anti-sense riboprobe to full-length murine Bmp4 cDNA (kind gift of B. Hogan, Duke University, Durham, NC) labeled by using in vitro transcription kit (Promega) and Digoxigenin-11-UTP (Roche). Frozen sections were incubated overnight at 65.5°C in hybridization mix (1× salt, 50% formamide, 10% dextran sulfate, 1 mg/ml yeast RNA, 1× Denhardt's) with Bmp4 probe. Slides were washed at 65.5°C in 1× SSC/50%formamide/0.1%Tween-20 and then in PBS at room temperature (RT). Sections were blocked in PBS/10% heat-inactivated FCS/0.1% Triton X-100 at RT and incubated overnight at 4°C with alkaline phosphatase-conjugated anti-digoxigenin antibody (1:2000, Roche). Slides were washed in PBS/0.1% Triton X-100 and incubated in 2% NBT/BCIP (Roche) in 0.1 M Tris, pH 9.5/0.1 M NaCl several hours at 37°C.
Flow-Cytometric Analysis.
Antibodies (BD Pharmingen) included: FITC-anti-c-kit, PE-anti-Flk1, APC-anti-CD34, FITC-anti-AnnexinV, phycoerythrin (PE), and PerCP-Cy5-anti-CD45. After 20 min of antibody incubation, cells were washed in PBS/10%FCS/PS and stained with 7AAD or Hoechst 33258 (1 μg/ml, Molecular Probes). Analysis was on a FACScan or FACS ARIA (Becton Dickinson). For AnnexinV staining, cells were incubated 15 min with FITC-anti-AnnexinV and 7AAD and with PE-anti-CD45.
Statistical Analysis.
Student's t test was used to determine statistical significance. P values <0.05 were considered significant.
Supplementary Material
ACKNOWLEDGMENTS.
We thank all laboratory members for constructive discussions; K. van der Horn, F. Wallberg, and R. van der Linden for cell sorting; F. Cerisoli for RNA isolations; and Dr. S. Philipsen for critical reading of the manuscript. M.H.B. thanks D. Mohn for technical assistance. This work was supported by l'ARC (C.D.), National Institutes of Health Grants R37DK51077 (to E.D.) and R01 DK52191 (to M.H.B.), Netherlands Besluit Subsidies Investeringen Kennisinfrastructuur Stem Cells in Development and Disease Grant 03038 (to E.D.), Nederlandse Organisatie voor Welenschappelijk Onderzock Vici Grant 916.36.601 (to E.D.), Marie Curie Grants HPMF-CT-2000-00871 (to C.R.), and MEIF-CT-2006-025333 (to K.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0706923105/DC1.
References
- 1.Taichman RS, Reilly MJ, Emerson SG. Hematology. 2000;4:421–426. doi: 10.1080/10245332.1999.11746468. [DOI] [PubMed] [Google Scholar]
- 2.Zhang J, Niu C, Ye L, Huang H, He X, Tong WG, Ross J, Haug J, Johnson T, Feng JQ, et al. Nature. 2003;425:836–841. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]
- 3.Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, Knight MC, Martin RP, Schipani E, Divieti P, Bringhurst FR, et al. Nature. 2003;425:841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- 4.Kiel MJ, Yilmaz OH, Iwashita T, Terhorst C, Morrison SJ. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 5.Duncan AW, Rattis FM, DiMascio LN, Congdon KL, Pazianos G, Zhao C, Yoon K, Cook JM, Willert K, Gaiano N, Reya T. Nat Immunol. 2005;6:314–322. doi: 10.1038/ni1164. [DOI] [PubMed] [Google Scholar]
- 6.Reya T, Duncan AW, Ailles L, Domen J, Scherer DC, Willert K, Hintz L, Nusse R, Weissman IL. Nature. 2003;423:409–414. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- 7.Bhardwaj G, Murdoch B, Wu D, Baker DP, Williams KP, Chadwick K, Ling LE, Karanu FN, Bhatia M. Nat Immunol. 2001;2:172–180. doi: 10.1038/84282. [DOI] [PubMed] [Google Scholar]
- 8.Medvinsky A, Dzierzak E. Cell. 1996;86:897–906. doi: 10.1016/s0092-8674(00)80165-8. [DOI] [PubMed] [Google Scholar]
- 9.de Bruijn MF, Ma X, Robin C, Ottersbach K, Sanchez MJ, Dzierzak E. Immunity. 2002;16:673–683. doi: 10.1016/s1074-7613(02)00313-8. [DOI] [PubMed] [Google Scholar]
- 10.North T, de Bruijn M, Stacy T, Talebian L, Lind E, Robin C, Binder M, Dzierzak E, Speck N. Immunity. 2002;16:661–672. doi: 10.1016/s1074-7613(02)00296-0. [DOI] [PubMed] [Google Scholar]
- 11.Dexter TM. Acta Haematol. 1979;62:299–305. doi: 10.1159/000207593. [DOI] [PubMed] [Google Scholar]
- 12.Moore KA, Ema H, Lemischka IR. Blood. 1997;89:4337–4347. [PubMed] [Google Scholar]
- 13.Yoder MC, Papaioannou VE, Breitfeld PP, Williams DA. Blood. 1994;83:2436–2443. [PubMed] [Google Scholar]
- 14.Ohneda O, Fennie C, Zheng Z, Donahue C, La H, Villacorta R, Cairns B, Lasky LA. Blood. 1998;92:908–919. [PubMed] [Google Scholar]
- 15.Xu MJ, Tsuji K, Ueda T, Mukouyama YS, Hara T, Yang FC, Ebihara Y, Matsuoka S, Manabe A, Kikuchi A, et al. Blood. 1998;92:2032–2040. [PubMed] [Google Scholar]
- 16.Oostendorp RA, Harvey KN, Kusadasi N, de Bruijn MF, Saris C, Ploemacher RE, Medvinsky AL, Dzierzak EA. Blood. 2002;99:1183–1189. doi: 10.1182/blood.v99.4.1183. [DOI] [PubMed] [Google Scholar]
- 17.Oostendorp RA, Medvinsky AJ, Kusadasi N, Nakayama N, Harvey K, Orelio C, Ottersbach K, Covey T, Ploemacher RE, Saris C, Dzierzak E. J Cell Sci. 2002;115:2099–2108. doi: 10.1242/jcs.115.10.2099. [DOI] [PubMed] [Google Scholar]
- 18.Charbord P, Oostendorp R, Pang W, Herault O, Noel F, Tsuji T, Dzierzak E, Peault B. Exp Hematol. 2002;30:1202. doi: 10.1016/s0301-472x(02)00895-0. [DOI] [PubMed] [Google Scholar]
- 19.Durand C, Robin C, Dzierzak E. Haematologica. 2006;91:1172–1179. [PubMed] [Google Scholar]
- 20.Chateauvieux S, Ichante JL, Delorme B, Frouin V, Pietu G, Langonne A, Gallay N, Sensebe L, Martin MT, Moore KA, Charbord P. Physiol Genomics. 2007;29:128–138. doi: 10.1152/physiolgenomics.00197.2006. [DOI] [PubMed] [Google Scholar]
- 21.Mendes SC, Robin C, Dzierzak E. Development. 2005;132:1127–1136. doi: 10.1242/dev.01615. [DOI] [PubMed] [Google Scholar]
- 22.Hackney JA, Charbord P, Brunk BP, Stoeckert CJ, Lemischka IR, Moore KA. Proc Natl Acad Sci USA. 2002;99:13061–13066. doi: 10.1073/pnas.192124499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oostendorp RA, Robin C, Steinhoff C, Marz S, Brauer R, Nuber UA, Dzierzak EA, Peschel C. Stem Cells. 2005;23:842–851. doi: 10.1634/stemcells.2004-0120. [DOI] [PubMed] [Google Scholar]
- 24.Chevalier S, Praloran V, Smith C, MacGrogan D, Ip NY, Yancopoulos GD, Brachet P, Pouplard A, Gascan H. Blood. 1994;83:1479–1485. [PubMed] [Google Scholar]
- 25.Auffray I, Chevalier S, Froger J, Izac B, Vainchenker W, Gascan H, Coulombel L. Blood. 1996;88:1608–1618. [PubMed] [Google Scholar]
- 26.Graham GJ, Wright EG, Hewick R, Wolpe SD, Wilkie NM, Donaldson D, Lorimore S, Pragnell IB. Nature. 1990;344:442–444. doi: 10.1038/344442a0. [DOI] [PubMed] [Google Scholar]
- 27.Dunlop DJ, Wright EG, Lorimore S, Graham GJ, Holyoake T, Kerr DJ, Wolpe SD, Pragnell IB. Blood. 1992;79:2221–2225. [PubMed] [Google Scholar]
- 28.Winnier G, Blessing M, Labosky PA, Hogan BL. Genes Dev. 1995;9:2105–2116. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- 29.Snyder A, Fraser ST, Baron MH. J Cell Biochem. 2004;93:224–232. doi: 10.1002/jcb.20191. [DOI] [PubMed] [Google Scholar]
- 30.Choi K, Kennedy M, Kazarov A, Papadimitriou JC, Keller G. Development. 1998;125:725–732. doi: 10.1242/dev.125.4.725. [DOI] [PubMed] [Google Scholar]
- 31.Zafonte BT, Liu S, Lynch-Kattman M, Torregroza I, Benvenuto L, Kennedy M, Keller G, Evans T. Blood. 2007;109:516–523. doi: 10.1182/blood-2006-02-004564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmerer M, Evans T. Blood. 2003;102:3196–3205. doi: 10.1182/blood-2003-04-1094. [DOI] [PubMed] [Google Scholar]
- 33.Bhatia M, Bonnet D, Wu D, Murdoch B, Wrana J, Gallacher L, Dick JE. J Exp Med. 1999;189:1139–1148. doi: 10.1084/jem.189.7.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balemans W, Van Hul W. Dev Biol. 2002;250:231–250. [PubMed] [Google Scholar]
- 35.Marshall CJ, Kinnon C, Thrasher AJ. Blood. 2000;96:1591–1593. [PubMed] [Google Scholar]
- 36.Miles C, Sanchez MJ, Sinclair A, Dzierzak E. Development. 1997;124:537–547. doi: 10.1242/dev.124.2.537. [DOI] [PubMed] [Google Scholar]
- 37.Terskikh AV, Easterday MC, Li L, Hood L, Kornblum HI, Geschwind DH, Weissman IL. Proc Natl Acad Sci USA. 2001;98:7934–7939. doi: 10.1073/pnas.131200898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Veiga-Fernandes H, Coles MC, Foster KE, Patel A, Williams A, Natarajan D, Barlow A, Pachnis V, Kioussis D. Nature. 2007;446:547–551. doi: 10.1038/nature05597. [DOI] [PubMed] [Google Scholar]
- 39.Gammill LS, Gonzalez C, Gu C, Bronner-Fraser M. Development. 2006;133:99–106. doi: 10.1242/dev.02187. [DOI] [PubMed] [Google Scholar]
- 40.Adelman CA, Chattopadhyay S, Bieker JJ. Development. 2002;129:539–549. doi: 10.1242/dev.129.2.539. [DOI] [PubMed] [Google Scholar]
- 41.Strouboulis J, Dillon N, Grosveld F. Genes Dev. 1992;6:1857–1864. doi: 10.1101/gad.6.10.1857. [DOI] [PubMed] [Google Scholar]
- 42.Robin C, Dzierzak E. In: Developmental Hematopoiesis Methods and Protocols. Baron MH, editor. Vol 105. Totowa, NJ: Humana; 2005. pp. 257–272. [Google Scholar]
- 43.Medvinsky AL, Samoylina NL, Muller AM, Dzierzak EA. Nature. 1993;364:64–67. doi: 10.1038/364064a0. [DOI] [PubMed] [Google Scholar]
- 44.Jaffredo T, Gautier R, Eichmann A, Dieterlen-Lievre F. Development. 1998;125:4575–4583. doi: 10.1242/dev.125.22.4575. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.