Abstract
RNAi is a powerful tool for interrogating gene function in ES cells. Combining the high penetrance of a microRNA-embedded shRNA (shRNA-mir) cassette with a locus-defined, inducible expression strategy, we developed a system for RNAi in mouse ES cells. An shRNA-mir cassette is targeted near the constitutively active HPRT locus under a tetracycline (tet)-regulatable promoter through Cre-mediated site-specific recombination. The major advantage of this system is that the shRNA-mir cassette can be targeted to a precise locus, allowing for control of shRNA-mir expression in an inducible fashion. Induction of an shRNA-mir directed against the pluripotency factor, Nanog, resulted in the loss of self-renewal and differentiation to parietal endoderm-like cells, which can be rescued by the introduction of an RNAi-immune version of Nanog cDNA. Knockdown efficiency can be enhanced by using multiple shRNA-mir hairpins against the target gene, which was further validated by knocking down two additional ES cell factors. This site-directed, virus-free, and tet-inducible RNAi system, designated as SDVFi RNAi in our study, presents an efficient option for controlled gene silencing in ES cells.
Keywords: microRNAs, Nanog, RNA interference, tetracycline
Since the initial discovery in Caenorhabditis elegans that dsRNA can silence the expression of homologous genes (1), RNAi has evolved into an essential tool for genetic experiments. In mammalian cells, where dsRNA elicits a nonspecific IFN response, different approaches have been taken to induce RNAi, including the expression of artificial ≈21-nt siRNA duplexes (2), ≈70-nt shRNA transcripts (3), and longer, ≈150-nt microRNA precursors, in which the target-specific shRNA is flanked by sequences of the naturally occurring miR30 gene (4). The resultant microRNA-embedded shRNA (shRNA-mir) stably suppresses gene expression even when present in the genome at single copy (5) and yields a higher level of siRNA and more efficient knockdown than a simple shRNA expression vector (6). The potent RNAi response and the ability to be regulated by Pol II promoters have made shRNA-mir vectors the basis for second-generation shRNA libraries in the mouse and human genomes (7).
In this study, we sought to take advantage of the high penetrance of shRNA-mir vectors to establish an efficient system for inducible RNAi in murine ES cells. RNAi has provided valuable insights into the pathways regulating ES cell self-renewal, pluripotency, and lineage determination, especially in combination with microarray expression profiling (8, 9). Inducible RNAi raises the prospect of temporal and dosage-sensitive control over the induction of gene knockdown. Previously reported strategies for inducible RNAi in ES cells or transgenic mice include the use of tamoxifen-inducible Cre-recombinase to activate vector-based shRNAs and shRNAs integrated in a defined locus (10, 11), as well as the use of a doxycycline-inducible shRNA cassette integrated in a defined locus (12, 13). Here we describe an alternative drug-inducible system based on the Ainv15 ES cell line, in which an shRNA-mir cassette is directed to the constitutively active hypoxanthine-guanine phosphoribosyltransferase (HPRT) locus upon Cre-mediated site-specific recombination.
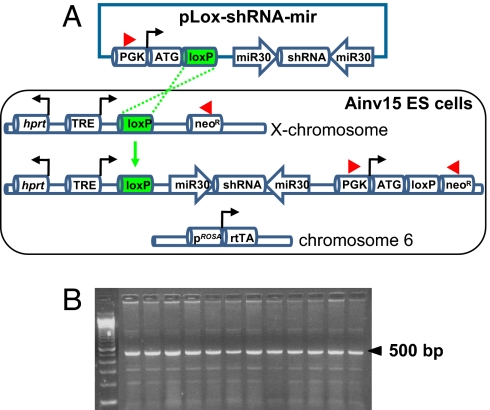
The Ainv15 line was derived from the E14 ES cell line (14), which was modified previously (15) by inserting the reverse tetracycline transactivator (rtTA) for the tet-ON system into the ubiquitously expressed Rosa26 locus on chromosome 6. In addition, the HPRT locus on the X chromosome was engineered to contain a tet response element (TRE), a loxP site, and a neomycin-resistance gene lacking an ATG initiation codon (see Fig. 1A) (15). This system allows for Cre-mediated targeting and selection of a loxP-containing vector (referred to as the pLox construct) (15) and the simultaneous introduction of tet-regulatable transgene expression (see Fig. 1A). The advantages of this inducible system are: (i) the rtTA is controlled by an endogenous gene regulatory system; (ii) the transgene or shRNA-mir expression cassette is directed to a location that has a lower propensity for silencing; and (c) the protocol is virus-free, which prevents complications associated with insertional mutagenesis and silencing because of methylation of the provirus genome or binding of transacting repressive factors to the long terminal repeats of the viral promoter (16).
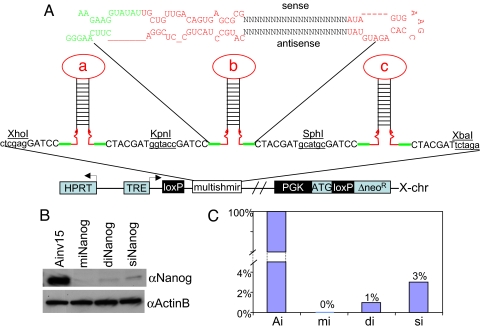
Fig. 1.
Site-directed, virus-free, and inducible (SDVFi) RNAi in Ainv15 ES cells. (A) Schematic depiction of Ainv15 cells harboring genetic modifications of chromosome 6 and X chromosome (modified from ref. 15). An shRNA-mir cassette is integrated into the region 5′ to the HPRT locus by loxP-mediated, site-specific recombination indicated by green dashed lines. Reconstitution of neomycin expression upon recombination enables efficient selection of correctly targeted clones. Red arrowheads denote primers used for PCR screening for positive clones. (B) PCR screening for recombinants by using primers indicated in A. A 500-bp band indicates the correct integration of shRNA-mir cassette into the targeted locus. ATG, methionine translation initiation codon; neoR, neomycin-resistance cassette; PGK, phosphoglycerokinase promoter; rtTA, reverse tetracycline transactivator; TRE, tetracycline response element.
We predicted that the integration of shRNA-mir technology within the Ainv15/pLox system would result in a potent and reliable method for inducible RNAi in ES cells. As a proof of principle, we tested whether this site-directed, virus-free, and inducible system (dubbed SDVFi RNAi) could be used for targeting an shRNA-mir cassette against the homeoprotein Nanog, a transcription factor that plays a fundamental role in ES cell self-renewal and pluripotency (17, 18). We determined whether the generated phenotype was a specific effect of the Nanog shRNA-mir and whether penetrance could be further enhanced by multiplying the number of shRNA-mir cassettes. Finally, we demonstrated applicability of the SDVFi RNAi to study other critical factors involved in stem cell pluripotency.
Results
Establishment of the SDVFi RNAi System in ES Cells.
To establish a site-directed, tet-regulatable RNAi system in Ainv15 ES cells, we used an shRNA directed against the 3′ untranslated region (UTR) of Nanog mRNA (RNAi codex) that was tested for efficient knockdown of endogenous Nanog during retrovirus-mediated delivery in ES cells (data not shown). The shRNA was cloned into a retroviral vector containing miR30 sequences flanking the shRNA to enhance the processing of miRNA into siRNA (4). This shRNA-mir cassette was then amplified by PCR and subcloned into the pLox-targeting vector (15). The pLox construct contains a phosphoglycerokinase (PGK) promoter and methionine translation initiation codon (ATG) to restore resistance to G418 after recombination into the inducible locus. It also has a loxP site placed between the promoter-ATG and the shRNA-mir-Nanog cassette. As shown in Fig. 1A, transfection of the pLox-shRNA-mir construct along with a Cre-expression vector results in site-specific recombination by loxP sites and incorporation of the shRNA-mir cassette into a locus next to the HPRT locus on the X chromosome. The tet-regulatable promoter unit consists of seven tet operator (tetO) sites flanked by minimal promoters derived from the CMV, which allows for multiple rtTA binding in the presence of doxycycline (19, 20). Upon recombination of the pLox-shRNA-mir construct into the inducible locus, reconstitution of neomycin expression (resistance to G418) enables efficient selection of correctly targeted clones. PCR analysis confirmed that 12 of 12 G418-resistant clones underwent site-specific recombination (Fig. 1B).
Kinetics and Dose-Responsiveness of Nanog Regulation in Ainv15 shRNA-mir-Nanog Cells.
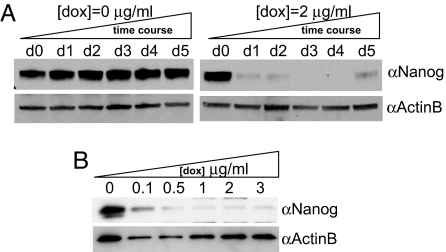
To study the kinetics of Nanog knockdown in Ainv15 shRNA-mir-Nanog cells, whole-cell lysates were collected from doxycycline-treated and untreated samples over a period of 5 days and subjected to Western blot analyses by using anti-Nanog antibody (Fig. 2A). In contrast to the nontreated samples, which maintained Nanog expression over the time course (Fig. 2A Left), Nanog protein expression in 2 μg/ml doxycycline-treated cells was greatly down-regulated during the first 2 days, followed by further reduction to a nondetectable level from day 3 to day 4. However, slight reexpression of Nanog was detected at day 5 (Fig. 2A Right) presumably because of the competitive outgrowth of residual low knockdown cells (see Discussion).
Fig. 2.
Kinetics and dose-responsiveness of Nanog regulation in Ainv15 shRNA-mir-Nanog cells. (A) Detection of Nanog expression over a 5-day period in the total cell extracts of untreated and treated samples with the indicated concentration. (B) Knockdown of Nanog expression after treatment for 5 days with increasing concentrations of doxycycline as indicated. Twenty micrograms of total lysates per data point was subjected to Western blotting with indicated antibodies. β-actin serves as loading control.
To address the concern that doxycycline administration may be toxic to ES cells and to titrate the minimum requirement for the drug to be effective, parental as well as shRNA-mir-Nanog-expressing Ainv15 cells were subjected to treatment for 5 days with a series of doxycycline concentrations up to 3 μg/ml (Fig. 2B). No obvious toxic effect or morphological changes were observed in Ainv15 parental cells that were not transfected with pLox-shRNA-mir-Nanog (data not shown). In contrast, Nanog protein expression was reduced in transfected cells tested with 0.1 μg/ml doxycycline. Moreover, knockdown of Nanog was further enhanced with increasing concentrations of doxycycline tested (Fig. 2B). This finding is consistent with the notion that the doxycycline–rtTA complex increases transgene expression through a cooperative mechanism: The seven tetO sites allow for multiple rtTA binding to promote transcription of a single inducible transgene (20). Considering this cooperative effect and our finding that no toxicity was observed in Ainv15 shRNA-mir-Nanog cells up to 3 μg/ml, we chose 2 μg/ml as the optimal concentration to ensure efficient shRNA-mir expression while avoiding any potential toxic effect.
Functional Consequences and Rescue of Nanog Knockdown in Ainv15 shRNA-mir-Nanog Cells.
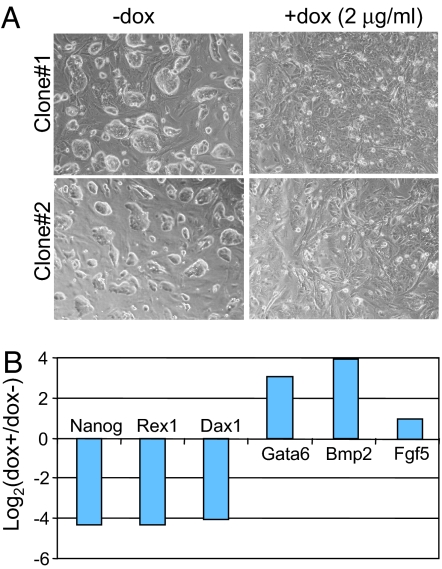
Targeted disruption of one of the two Nanog alleles in mouse ES cells (50% of wild-type expression level) results in multilineage differentiation (21). KO of both Nanog alleles results in more specific differentiation to parietal endoderm-like cells and derepression of endoderm transcription factor Gata6 (18). In Ainv15 shRNA-mir-Nanog cells, 2 μg/ml doxycycline treatment for 4 days resulted in differentiation into dispersed cells (Fig. 3A). This finding resembled the parietal endoderm phenotype observed in Nanog KO ES cells (18) and that after knockdown of Nanog by chemically modified RNAi compounds (22). Quantitative real-time PCR (qRT-PCR) analysis of total RNA collected from induced, feeder-free Ainv15 shRNA-mir-Nanog cells indicated a reduction in stem cell-specific (Nanog, Rex1, and Dax1) gene expression, but a derepression of endoderm-specific (Gata6 and Bmp2) genes (Fig. 3B). As in heterozygous Nanog ES cells (21), but unlike full KO ES cells (18), the ectoderm-specific marker, Fgf5, was slightly induced. This result shows that inducible expression of shRNA-mir-Nanog results in a cellular response similar to that observed in nullizygous Nanog ES cells, but includes some overlap with heterozygous Nanog ES cells. Thus, the functional consequences of inducible RNAi by using an shRNA-mir-Nanog cassette are consistent with previous reports of Nanog depletion.
Fig. 3.
Knockdown of Nanog results in ES cell differentiation and changes of stem cell as well as lineage-specific gene expression. (A) Morphology of Ainv15 shRNA-mir-Nanog cells grown without (Left) and with (Right) doxycycline. Pictures were taken from two independent clones grown at day 4 with indicated treatment. (B) qRT-PCR analysis of gene expression in Ainv15 shRNA-mir-Nanog (clone 2) cells. Gene expression levels in treated cells relative to untreated cells at day 4 were measured after normalizing to β-actin levels in each sample.
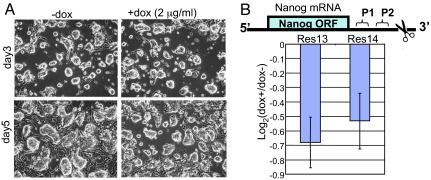
To test whether the observed consequences of doxycycline treatment were a specific effect of Nanog knockdown and not because of off-target effects, we determined whether the undifferentiated phenotype could be rescued by coexpression of RNAi-resistant Nanog cDNA encoding the ORF only. Ainv15 shRNA-mir-Nanog cells were cotransfected with a constitutive, CAG-regulated Nanog cDNA (ORF) expression vector, pPyCAGIZ (kindly provided by Ian Chambers, University of Edinburgh, Edinburgh, U.K.), and stable clones were established with 50 μg/ml zeocin selection. Because the shRNA was directed against the 3′ UTR of endogenous Nanog mRNA, the constitutively expressed Nanog cDNA (ORF) transcript should not be silenced by the RNAi machinery. Upon doxycycline treatment, no changes were observed for rescued cells in morphology (Fig. 4A) or in marker gene analysis by qRT-PCR (data not shown). In contrast, qRT-PCR analysis by using primers directed against sites in the 3′ UTR of the endogenous transcript demonstrated that endogenous Nanog was still down-regulated, albeit with smaller fold change, compared with that in nonrescued cells (Fig. 3B) in two doxycycline-induced rescue clones (Fig. 4B). Fold change was small presumably because of partial reexpression of the endogenous Nanog and/or the autoregulatory loop of Nanog (see Discussion). Because coexpression of an RNAi-resistant Nanog cDNA rescued the undifferentiated phenotype in the presence of doxycycline, the effects of our inducible shRNA-mir-Nanog cassette could be specifically attributed to Nanog gene depletion.
Fig. 4.
Rescue study of Ainv15 shRNA-mir-Nanog cells by using a cotransfected Nanog cDNA (ORF) expression vector. (A) Representative morphology of rescued cells grown with and without doxycycline treatment for 3 and 5 days. (B) qRT-PCR analysis of endogenous mRNA levels of Nanog upon doxycycline treatment for 5 days. (Upper) Schematic depiction of endogenous Nanog mRNA. P1 and P2 stand for the region amplified by qRT-PCR. The scissors indicate the target sequence of Nanog shRNA. (Lower) Measurement of the endogenous Nanog RNA levels in day 5 samples by qRT-PCR. Normalized data from both P1 and P2 regions in two independent rescued clones (Res13 and Res14) were presented.
Enhanced Knockdown Efficiency Using Multiple miR30-Based shRNA Hairpins.
The incorporation of multiple miRNA hairpins in a single vector reportedly improves the efficiency of target gene knockdown (23). To test whether knockdown efficiency in our system could be enhanced, Ainv15 ES cells were transfected with shRNA-mir vectors containing a single (siNanog), double (diNanog), or multiple (miNanog) shRNAs directed against Nanog. In the diNanog and miNanog constructs, the shRNAs were linked in a series and separated by short linker sequences containing minimal miR sequences required for shRNA processing and restriction sites for cloning (Fig. 5A). After Cre-mediated recombination and induction with 2 μg/ml doxycycline for 5 days, whole-cell lysates from the three samples and an uninfected Ainv15 control line were subjected to Western blot analysis with anti-Nanog antibody (Fig. 5B). Consistent with the results in Fig. 2, a single microRNA-embedded shRNA hairpin (siNanog) already significantly diminished Nanog protein expression (3% of the control level) (Fig. 5C). Furthermore, the use of double (diNanog) and triple (miNanog) shRNA-mir cassettes diminished Nanog protein expression progressively further to a nondetectable level (Fig. 5 B and C).
Fig. 5.
Artificial, multi-shRNA-mir hairpin strategy for enhanced gene knockdown. (A) Schematic depiction of the structure of a single shRNA-mir hairpin (Upper) and site-directed integration of multi-shRNA-mir expression cassette into X chromosome (Lower). Restriction sites used for pLox cloning are indicated. (B) Western blot analysis of Nanog protein expression in knockdown samples by using a single hairpin (siNanog), double hairpins (diNanog), and multiple (triple) hairpins (miNanog). Whole-cell lysates were prepared from cells upon induction with 2 μg/ml doxycycline for 5 days. β-Actin served as loading control. (C) Quantitative presentation of the data presented in B. Ai, Ainv15; mi, miNanog; di, diNanog; si, siNanog. The signal of control Ainv15 sample was arbitrarily set to 100%.
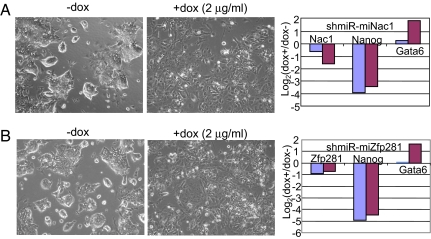
To test whether the SDVFi system is applicable to silence stem cell pluripotency factors other than Nanog, we used the same multi-shRNA-mir strategy to confirm knockdown effects of two newly identified factors in ES cell pluripotency. Nac1, a BTB-domain-containing protein, and Zfp281, the mouse homologue of human zinc-finger protein ZBP99, were identified as Nanog-associated proteins after affinity purification of Nanog under native conditions (24). Retrovirus-mediated delivery of shRNAs against Nac1 or Zfp281 led to compromised self-renewal of cells upon passage to gelatin in the presence of LIF and a striking derepression of Gata6 (24). We established Ainv15 ES cell lines carrying three shRNA-mir cassettes directed against either Nac1 or Zfp281 (designated as shmiR-miNac1 and shmiR-miZfp281 in Fig. 6). Doxycycline induction for 3 days resulted in loss of self-renewal and differentiation to endoderm-like cells in the case of both Nac1 (Fig. 6A) and Zfp281 (Fig. 6B). qRT-PCR analysis of total RNA showed that a reduction in Nac1 mRNA increased from day 1 to day 3 (Fig. 6A), whereas the reduction in Zfp281 mRNA was slightly weakened (Fig. 6B). In both cases, however, Nanog was strongly repressed, whereas Gata6 was derepressed during prolonged induction. These results are consistent with the previously reported phenotype of these Nanog-associated factors and confirm that inducible RNAi can be accomplished by using multiple hairpins directed against a single gene target.
Fig. 6.
Inducible knockdown of the ES cell pluripotency factors Nac1 (A) and Zfp281 (B) by using multi-shRNA-mir hairpin strategy. (Left) Morphology of ES cells before and after treatment with doxycycline to induce the knockdown. Pictures were taken 3 days after doxycycline treatment. (Right) qRT-PCR analysis of gene knockdown levels as well as changes of stem cell marker (Nanog) and primitive endoderm marker Gata6 after 1 day (blue bars) and 3 days (red bars) of doxycycline treatment.
Discussion
This study describes a system for site-directed, virus-free, and tet-inducible (SDVFi) RNAi to controllably diminish Nanog expression in mouse ES cells. Our system provides a number of advantages over conventional approaches for RNAi in mouse ES cells: (i) the integration of the RNAi cassette is specifically directed to the constitutively active HPRT locus on the X chromosome, avoiding the insertional mutagenesis and silencing sometimes associated with viral integration events; (ii) the system avoids the possibility of an immune response to virus-associated proteins; (iii) the activation of the RNAi cassette is inducible by doxycycline, providing greater control over the amount and timing of gene knockdown; (iv) the shRNA against the target gene of interest is embedded in a microRNA structure, resulting in efficient shRNA processing and high penetrance knockdown; and (v) by incorporating multiple shRNA-mir hairpins within a single targeting construct, the efficiency of gene knockdown can be improved and multigene knockdown may be achieved from a single vector.
We tested the ability of SDVFi RNAi to reduce the expression of the pluripotency factor, Nanog. Expression kinetics in the Ainv15-inducible ES cell line were previously characterized by using GFP as an inducible gene and FACS analysis to examine levels of GFP expression. At a doxycycline concentration of 1 μg/ml, the maximum number of cells expressing GFP in ES cell culture was reached by 8 h, and a steady-state mean level of GFP was reached after ≈24 h (20). Although the same inducible system was used here, we felt it was important to recharacterize the Ainv15 line with regard to induction kinetics for Nanog knockdown. The time required to synthesize sufficient GFP to be detectable by FACS may differ from the time required for the shRNA-mir-Nanog transcript to be processed into the RNA-induced silencing complex and deplete levels of Nanog. In addition, the characterization using GFP only examined induction kinetics to 50 h, whereas we were interested in designing an inducible system that could reduce target gene expression for a longer period. When Ainv15 cells expressing shRNA-mir-Nanog were exposed to 2.0 μg/ml doxycycline, Nanog protein expression was greatly down-regulated for the 5-day test period and reduced to nondetectable levels between days 3 and 4 (Fig. 2A). Replicate experiments using different Ainv15 shRNA-mir-Nanog clones confirmed that our system provides a time window of ≈4 days, in which target gene expression is diminished. After 5 days of doxycycline induction, however, we noticed that some Nanog protein expression returned (Fig. 2A Right). Consequently, colonies resembling undifferentiated ES cells reappeared after 7 days in culture (data not shown). During a prolonged induction period, imbalances in drug distribution may confer a competitive growth advantage to colonies that experienced a diminished knockdown effect. The time window is sufficient, however, to perform functional studies of genes (such as Nanog) implicated in ES cell regulation and for most other in vitro applications.
Progressive knockdown of Nanog expression was achieved with increasing concentrations of doxycycline (Fig. 2B) and without any obvious toxicity (data not shown). Others reported that 1 μg/ml doxycycline was sufficient to achieve a stable level of transgene expression by 24 h in the Ainv15 system (20), but we decided that 2 μg/ml doxycycline would provide most effective shRNA-mir induction for experiments lasting several days while avoiding any potential toxic effects. Therefore, this doxycycline concentration was used for all subsequent experiments.
Changes in morphology in response to Nanog knockdown within 5 days (Fig. 3A) were consistent with a loss of self-renewal and differentiation to endoderm, as described previously (18, 22). qRT-PCR analysis confirmed the up-regulation of endoderm-specific genes (Gata6 and Bmp2) and the down-regulation of stem cell-specific genes (Nanog, Rex1, and Dax1). The slight induction of ectoderm marker, Fgf5, however, indicates that induced shRNA-mir-Nanog ES cells, unlike nullizygous Nanog KO ES cells, were not fully committed to the endoderm lineage. Rather, induced shRNA-mir-Nanog ES cells seem to have assumed an intermediate identity between full KO (18) and heterozygous Nanog ES cells (21) because the latter show multilineage differentiation, including derepression of Fgf5. Considering the dosage-dependent effects of Nanog (21), these data indicate that the phenotype generated by our inducible RNAi strategy closely approximates the cellular response to the targeted KO of one or both Nanog alleles. Ainv15 shRNA-mir-Nanog cells that were cotransfected with a Nanog cDNA expression vector maintained self-renewal and ES cell morphology under exposure to doxycycline (Fig. 4A). This rescue experiment confirms that the phenotypic changes reported in Fig. 3 were not because of off-target effects, but represented the specific effect of Nanog down-regulation. The smaller fold change reduction of Nanog mRNA in the rescue experiment (Fig. 4B), compared with that in the knockdown study (Fig. 3B), is likely because of the partial reexpression of endogenous Nanog at day 5 samples, as discussed earlier (see also Fig. 2A). Additionally, it may be explained by a positive autoregulatory loop, in which transgenic Nanog up-regulates expression of the endogenous gene. ChIP analysis in mouse ES cells indicated that both Oct4 and Nanog are involved in autoregulating their own promoter sites (9).
Sun et al. (23) demonstrated that lentiviral delivery of multiple shRNA-mir hairpins against the same or distinct target genes improves gene knockdown efficiency and provides linked multigene knockdown. The SDVFi system in this study seems to be more efficient in that a single shRNA-mir already achieves high efficiency knockdown (see Figs. 2 and 5). This finding could be attributable to the high penetrance of a single-copy shRNA-mir in a defined locus, rather than random integration. However, we do observe, by multiplying the number of shRNA-mir hairpins against Nanog in the pLox-shRNA-mir construct, that the efficiency of Nanog knockdown was further enhanced (from 3% to a nondetectable level) (Fig. 5C). Inducible knockdown of Nac1 and Zfp281 was also achieved by using triple-hairpin shRNA-mir vectors (Fig. 6), which generated a phenotype consistent with the reported response to simple shRNA treatment (24). More attractively, the multi-shRNA-mir strategy may be adapted in our system to simultaneously reduce the expression of multiple, distinct target genes in ES cells. This approach would enable epistatic analysis of functional interactions among genes implicated in ES cells and their derivatives. Finally, the SDVFi system reported here may potentially be adapted to enable inducible RNAi in vivo upon blastocyst injection of Ainv15 shRNA-mir ES cells. After administration of doxycycline to mice, target gene expression could be controllably reduced.
Materials and Methods
ES Cell Culture.
The Ainv15 ES line was maintained in ES medium (DMEM) supplemented with 15% FCS, 10−4 M 2-mercaptoethanol, 2 mM l-glutamine, 0.1 mM each nonessential amino acid, 1% nucleoside mix (100× stock; Sigma–Aldrich), 1,000 units/ml recombinant leukemia inhibitory factor [(LIF) Chemicon], and 50 units/ml penicillin/streptomycin. ES cells were cultured on a feeder layer for photography or gelatin-adapted to deplete the feeder cells for RNA extraction.
Plasmid Construction.
The Nanog1523 shRNA sequence was as follows: 5′- TGCTGTTGACAGTGAGCGCGGAGACAGTGAGGTGCATATATAGTGAAGCCACAGATGTATATATGCACCTCACTGTCTCCATGCCTACTGCCTCGGA-3′ (the target sequence is underlined and located in the 3′-UTR of Nanog cDNA). It was designed by using RNAi Central (RNAi Codex) as previously described (25) and cloned into LMP vector containing the miR30 sequence flanking the shRNA (4). The full shRNA-mir cassette (≈357 bp) was then PCR-amplified by using the forward primer miR5′(KpnI)-F (5′-AGGTACCCAGGGTAATTGTTTGAATGAGGC-3′) and the reverse primer miR3′(XbaI)-R (5′-GTCTAGAGTCTTCCAATTGAAAAAAGTGA-3′) and cloned into KpnI and XbaI sites in the pLox vector. For multiple shRNA-mir hairpin strategy, the minimal miR sequence that was required for shRNA processing was amplified (≈150 bp) for individual shRNA-mir hairpin to minimize the cloning size. Primers were designed as previously described (23), except that XhoI, KpnI, SphI, and XbaI restriction sites were incorporated and used for cloning the multi-shRNA-mir cassette into the pBluescript vector. The final XhoI–XbaI fragments were released from the pBluescript vector and cloned into a pLox vector previously digested with XhoI and XbaI. The Nac1 shRNA-mir and Zfp281 shRNA-mir sequences described previously (24) were cloned into a pLox vector with the strategy described above. For Nanog cDNA expression plasmid, the ORF of Nanog cDNA was amplified by using Nanog(XhoI)-F (5′-TCTCGAGCCACCATGAGTGTGGGTCTTCCTGGT-3′) and Nanog(NotI)-R (5′-CGCGGCCGCTCATATTTCACCTGGTGGA-3′) as PCR primers and cloned into pPyCAGIZ vector (kindly provided by Ian Chambers) previously digested with XhoI and NotI. All plasmids were sequence-verified.
Establishment of Stable Cell Lines Expressing shRNA-mir.
Twenty micrograms each of pLox derivatives and pSALK-CRE was added to 8 × 106 Ainv15 cells in 800 μl of PBS at room temperature. Electroporation was performed by using the Bio-Rad Gene Pulser with the capacitance extender set to 500 μF and voltage set to 250 V. Electroporated cells were plated onto 10-cm dishes with a dense layer of neomycin-resistant MEFs. Selection was begun the next day by using 350 μg/ml G418 in complete growth media. Medium was changed daily until colonies appeared around days 10–14. Integration was detected by using the following primers, which amplify across the loxP site to give a 500-bp band: LoxinF (5′-CTAGATCTCGAAGGATCTGGAG-3′) and LoxinR (5′-ATACTTTCTCGGCAGGAGCA-3′).
Western Blot Analysis.
Aliquots of ≈20–40 μg of total lysates were fractionated on SDS/PAGE and electroblotted onto PVDF membrane. Antibody incubation and chemiluminescence detection were performed according to the manufacturer's instructions (Amersham Biosciences). Anti-Nanog and anti-ActinB antibodies were purchased from Chemicon International and Sigma–Aldrich, respectively.
RNA Extraction and qRT-PCR Analysis.
Total RNA was isolated by using an RNAeasy kit (Qiagen). First-strand cDNA was synthesized by using iScript cDNA synthesis kit (Bio-Rad). qPCR was performed on an iCycler iQ Thermal Cycler (Bio-Rad) with SyBr Green PCR master mix (Bio-Rad). The average threshold cycle for each gene was determined from triplicate reactions, and the levels of gene expression were normalized to β-actin. Primers for stem cell as well as lineage-specific markers were the same as previously described (24). The primers for qRT-PCR of P1 region in Nanog 3′-UTR were: 5′-CCAGGTTCCTTCCTTCTTCC-3′ (forward) and 5′-TTCCGAAGGTCAGGAGTTCA-3′ (reverse), and the primers for qRT-PCR of P2 region in Nanog 3′-UTR were: 5′-ATCCACTGAGCCATCTCACC-3′ (forward) and 5′-GTGGTATGCCACCTTTGGTC-3′ (reverse).
ACKNOWLEDGMENTS.
We thank Dr. Ian Chambers for pPyCAGIZ vector, Dr. George Daley (Children's Hospital) for Ainv15 cells, Dr. Steve Elledge (Harvard Medical School) for LMP vector, and Dr. Yuko Fujiwara for critical reading and discussion of the manuscript. This work was supported by the Howard Hughes Medical Institute (S.H.O.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 3.Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stegmeier F, Hu G, Rickles RJ, Hannon GJ, Elledge SJ. Proc Natl Acad Sci USA. 2005;102:13212–13217. doi: 10.1073/pnas.0506306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, Lowe SW. Nat Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- 6.Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, et al. Nat Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 7.Chang K, Elledge SJ, Hannon GJ. Nat Methods. 2006;3:707–714. doi: 10.1038/nmeth923. [DOI] [PubMed] [Google Scholar]
- 8.Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- 9.Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 10.Coumoul X, Li W, Wang RH, Deng C. Nucleic Acids Res. 2004;32:e85. doi: 10.1093/nar/gnh083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu J, McMahon AP. Genesis. 2006;44:252–261. doi: 10.1002/dvg.20213. [DOI] [PubMed] [Google Scholar]
- 12.Seibler J, Kleinridders A, Kuter-Luks B, Niehaves S, Bruning JC, Schwenk F. Nucleic Acids Res. 2007;35:e54. doi: 10.1093/nar/gkm122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wegmuller D, Raineri I, Gross B, Oakeley EJ, Moroni C. Stem Cells. 2007;25:1178–1185. doi: 10.1634/stemcells.2006-0106. [DOI] [PubMed] [Google Scholar]
- 14.Hooper M, Hardy K, Handyside A, Hunter S, Monk M. Nature. 1987;326:292–295. doi: 10.1038/326292a0. [DOI] [PubMed] [Google Scholar]
- 15.Kyba M, Perlingeiro RC, Daley GQ. Cell. 2002;109:29–37. doi: 10.1016/s0092-8674(02)00680-3. [DOI] [PubMed] [Google Scholar]
- 16.Doerfler W. Curr Top Microbiol Immunol. 2006;301:125–175. doi: 10.1007/3-540-31390-7_5. [DOI] [PubMed] [Google Scholar]
- 17.Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 18.Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 19.Baron U, Freundlieb S, Gossen M, Bujard H. Nucleic Acids Res. 1995;23:3605–3606. doi: 10.1093/nar/23.17.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ting DT, Kyba M, Daley GQ. Methods Mol Med. 2005;105:23–46. doi: 10.1385/1-59259-826-9:023. [DOI] [PubMed] [Google Scholar]
- 21.Hatano SY, Tada M, Kimura H, Yamaguchi S, Kono T, Nakano T, Suemori H, Nakatsuji N, Tada T. Mech Dev. 2005;122:67–79. doi: 10.1016/j.mod.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 22.Hough SR, Clements I, Welch PJ, Wiederholt KA. Stem Cells. 2006;24:1467–1475. doi: 10.1634/stemcells.2005-0475. [DOI] [PubMed] [Google Scholar]
- 23.Sun D, Melegari M, Sridhar S, Rogler CE, Zhu L. BioTechniques. 2006;41:59–63. doi: 10.2144/000112203. [DOI] [PubMed] [Google Scholar]
- 24.Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 25.Olson A, Sheth N, Lee JS, Hannon G, Sachidanandam R. Nucleic Acids Res. 2006;34:D153–D157. doi: 10.1093/nar/gkj051. [DOI] [PMC free article] [PubMed] [Google Scholar]