Abstract
Nonsense-mediated mRNA decay (NMD) is a surveillance mechanism that detects and degrades transcripts containing premature translation termination codons. Gene expression profiling experiments have shown that inactivation of the NMD pathway leads to the accumulation of both aberrant, nonsense-containing mRNAs, and many apparently wild-type transcripts. Such increases in transcript steady-state levels could arise from direct changes in the respective mRNA half-lives, or indirectly, as a consequence of the stabilization of transcripts encoding specific regulatory proteins. Here, we distinguished direct from indirect substrates by virtue of their association with the Saccharomyces cerevisiae Upf1 protein. Analyses of this dataset, and its comparison to the sets of transcripts that respectively increase or decrease in abundance when NMD is either inactivated or reactivated, indicate that the number of direct NMD substrates is larger than previously thought and that low abundance, alternatively transcribed mRNAs, i.e., mRNAs whose 5′ ends are derived from previously unannotated 5′ flanking sequences, comprise a significant class of direct substrates. Using thiamine metabolism as an example, we also show that apparent NMD-regulated cellular pathways may actually reflect the detection of low-abundance alternative transcripts under conditions where a pathway is repressed.
Keywords: microarray analysis, mRNA half, lives, nonsense-mediated mRNA decay, translation termination, Upf proteins
Eukaryotic cells have evolved quality-control mechanisms to ensure that aberrant mRNAs do not accumulate as substrates for the translational machinery. One such mechanism, nonsense-mediated mRNA decay (NMD), identifies mRNAs containing premature translation termination codons and targets them for rapid degradation (1, 2). The destabilization of nonsense-containing mRNAs requires their translation and a distinct set of trans-acting factors, including the evolutionarily conserved Upf1, Nmd2 (Upf2), and Upf3 proteins (1). Gene expression profiling of yeast, fly, or human cells lacking the activity of one or more of these factors has suggested that the NMD pathway influences the expression of somewhere between 1% and 10% of cellular transcripts (3–10). Of the affected transcripts, the vast majority accumulate, indicating that their degradation, or the degradation of an mRNA encoding a regulator of their abundance, is controlled by NMD (3–5, 7). Further analyses of the transcripts up-regulated when NMD is inactivated suggested that NMD not only controls the expression of aberrant transcripts, but also that of many apparently wild-type mRNAs. This finding, in combination with the observation that the transcripts displayed an overrepresentation in specific cellular processes, has suggested that NMD may have a regulatory function beyond mRNA surveillance (3, 5, 7–9, 11). However, comparisons of NMD-regulated transcripts and processes between different organisms have revealed a poor overlap, suggesting that the regulatory role is not evolutionarily conserved (7, 9).
Approximately one-third of the transcripts that accumulate in NMD-deficient yeast cells could be divided into categories in which their presumed degradation could be accounted for by a premature or atypical context of the termination codon (3). In addition to mRNAs from known nonsense alleles, these categories included inefficiently spliced intron-containing pre-mRNAs, transcripts from pseudo- and bicistronic genes, transcripts subject to leaky scanning, transcripts derived from transposable elements or their LTRs, and transcripts containing upstream ORFs (uORFs) or a programmed frameshifting site. The majority of the transcripts in these groups are likely to be direct NMD substrates because seven of nine mRNAs randomly selected from this set displayed longer half-lives in a strain deleted for NMD2 (3). The transcripts that could not be assigned to the above-mentioned categories may accumulate as an indirect consequence of altered expression of bona fide NMD substrates. This view is supported by the observation that none of nine NMD-regulated transcripts defined in another study showed altered half-lives in a strain lacking NMD (4). More recently, genomewide mRNA half-life determinations indicated that ≈50% of the transcripts that accumulate in NMD-deficient cells are direct NMD substrates (11). However, the intrinsic difficulties in such half-life determinations suggested that it was important to use an independent approach to classify the substrates.
In this study, we addressed the question of direct versus indirect NMD substrates by characterizing the RNAs associated with the Upf1 protein in the budding yeast Saccharomyces cerevisiae. A similar approach has recently been used to analyze a single mRNA in Schizosaccharomyces pombe (10) and five mRNAs in Caenorhabditis elegans (12), but here we use microarray analysis to delineate the entire population of Upf1p-associated transcripts. Our results suggest that the number of direct NMD substrates is larger than previously thought and that a significant fraction of NMD-regulated “wild-type” transcripts are nonproductive alternative forms with an easily explained NMD reactivity.
Results
Identification of Transcripts Associated with the Upf1 Protein.
To identify transcripts that associate with Upf1p, we constructed a yeast strain expressing, from its normal chromosomal location, a functional C-terminally tandem affinity purification (TAP)-tagged version of the protein [supporting information (SI) Fig. 5A]. Ribonucleoprotein complexes were recovered from extracts by affinity selection on IgG beads, followed by cleavage with tobacco etch virus protease. To control for nonspecifically enriched RNAs, the same procedure was performed with extracts from wild-type cells lacking the TAP-tagged allele. The copurifying RNA population from four independent experiments was then analyzed with yeast genome microarrays (Affymetrix YG98S). Transcripts were considered to be enriched if the signal intensities between the Upf1p-TAP and mock samples showed a relative change of at least 2-fold. In addition, this change had to be reproducible in at least three of the four replicate experiments and demonstrate a statistically significant P value (≤0.05). The analyses revealed that 765 transcripts (781 probe sets) were enriched with Upf1p-TAP (SI Table 1).
The first step in the preparation of cRNA target samples for the microarray hybridization involves oligo(dT)-primed cDNA synthesis. Although NMD in yeast has been shown to promote deadenylation-independent decapping of target mRNAs (13), it was nevertheless important to ensure that cRNA synthesis was not biased by an atypical distribution of poly(A) lengths on Upf1p-TAP-associated mRNAs. The analysis revealed that the poly(A) tails present in the mock and Upf1p-TAP-associated fractions have a slight shift to shorter poly(A) lengths compared with the input samples (SI Fig. 5B), which is presumably because the input, but not the affinity-purified, samples were poly(A)-selected before the analysis. Importantly, the mock and Upf1p-TAP-enriched samples showed similar poly(A) tail length distributions (SI Fig. 5B), suggesting that the cRNA population derived from the Upf1p-TAP-associated mRNAs is not biased by mRNA poly(A) tail length, and that Upf1p interaction with mRNA is not confined to transcripts that are either newly synthesized or extensively deadenylated.
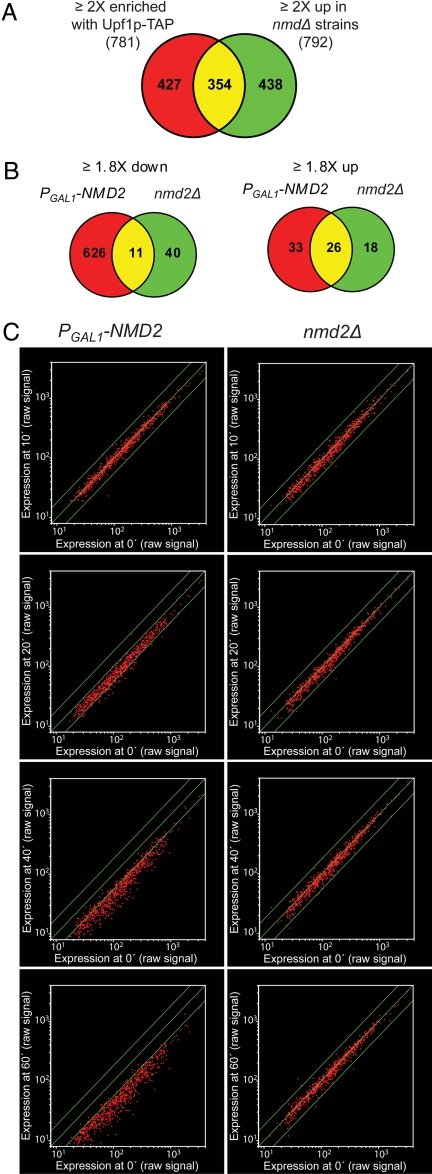
A combined analysis of the transcripts that showed >2-fold increases in upf1Δ, nmd2Δ, or upf3Δ cells (collectively designated nmdΔ cells) defined 746 NMD-regulated transcripts (792 probe sets) (3). A comparison between this dataset and the transcripts enriched with Upf1p-TAP revealed a significant overlap (Fig. 1A). This finding validates the Upf1p-TAP approach and indicates that many of the transcripts that accumulate in NMD-deficient cells represent direct substrates of the NMD pathway. Further, this preferential enrichment of a subset of cellular transcripts is consistent with the notion that Upf1p functions in premature termination, but not in normal termination (2, 14).
Fig. 1.
Transcripts enriched with Upf1p-TAP or down-regulated upon NMD reactivation. (A) Venn diagram showing the overlap of the 781 probe sets representing the transcripts ≥2-fold enriched with Upf1p-TAP and the 792 probe sets that increased ≥2-fold in nmdΔ strains (3). (B) Venn diagrams showing the overlap of the probe sets that exhibited ≥1.8-fold increases or ≥1.8-fold decreases in the PGAL1–NMD2 and nmd2Δ strains during the time-course experiment. (C) Scatter plots of the 626 probe sets uniquely down-regulated in the PGAL1–NMD2 strain upon shift to galactose-containing medium. The average raw signals at 10, 20, 40, and 60 min in the PGAL1–NMD2 (Left) or the nmd2Δ (Right) strain were compared on a logarithmic scale to the average signal at time point 0. The middle lines indicate the line of equivalence. The outer lines indicate a 1.8-fold difference in expression.
Effect of Reactivation of NMD on Global mRNA Levels in nmd2Δ Cells.
The observation that about half of the transcripts enriched with Upf1p-TAP do not seem to accumulate in nmdΔ cells (Fig. 1A) implied that these RNAs either escaped identification in the nmdΔ analysis (3) or that they have a unique requirement for Upf1p. To be able to distinguish between these possibilities, we investigated whether the inducible NMD system (15) can be used as an independent approach to define the set of NMD-regulated transcripts. In this system, the NMD2 (UPF2) gene is under the control of the galactose-inducible GAL1 promoter (PGAL1), and yeast cells harboring this allele are inactive in NMD when grown under noninducing conditions. However, the NMD pathway is fully activated 20 min after PGAL1–NMD2 induction, resulting in rapid degradation of the nonsense-containing mRNAs that accumulated before activation of the pathway (15). Samples of nmd2Δ cells harboring either an empty vector or a plasmid containing the PGAL1–NMD2 allele, hereafter referred to as the nmd2Δ and PGAL1–NMD2 strains, were collected 0, 10, 20, 40, and 60 min after the shift to galactose-containing medium. Transcripts that were differentially expressed at different time points in either strain, or between the two strains, were identified by hybridization to oligonucleotide microarrays. Subsequent data analysis revealed a small number of similar transcripts up-regulated in the PGAL1–NMD2 and nmd2Δ strains, whereas the total number of down-regulated transcripts differed significantly for the two strains (Fig. 1B and SI Fig. 6).
The 597 transcripts (626 probe sets; SI Table 2) uniquely down-regulated upon reactivation of NMD were essentially unchanged at 10 min after induction (Fig. 1C). At 20 min, when the NMD pathway is fully activated, almost all of these transcripts exhibited decreased levels. This decrease was even more pronounced at 40 and 60 min, when the majority of transcripts had decreased by >1.8-fold. Importantly, the levels of these transcripts were not changed in the nmd2Δ strain (Fig. 1C and SI Fig. 6), showing that their down-regulation in the PGAL1–NMD2 strain is not a general consequence of the shift in carbon source. These observations, and the destabilization of seven selected mRNAs whose abundance decreases in NMD2-reactivated cells (SI Fig. 7), lead us to conclude that reactivation of NMD causes rapid down-regulation of a subset of transcripts and that the inducible system provides an independent approach to identify NMD-regulated transcripts. Further, these observations provide additional evidence that, at least in yeast, mRNAs cannot acquire immunity to NMD (2, 15, 16).
A Significant Fraction of NMD-Regulated Transcripts Are Direct Substrates of the NMD Pathway.
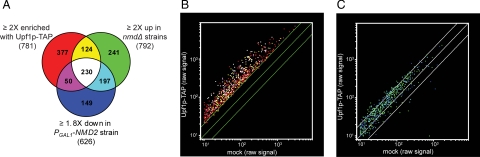
To delineate further the targets of NMD, we examined the overlap between the transcripts enriched with Upf1p-TAP and the transcripts up-regulated in nmdΔ strains or down-regulated upon NMD reactivation (Fig. 2A). This analysis revealed that 395 Upf1p-TAP-enriched transcripts (404 probe sets) were NMD-regulated in the nmdΔ and/or PGAL1–NMD2 experiments (Fig. 2 A and B), suggesting that they represent direct substrates of the NMD pathway. Accordingly, many previously characterized NMD substrates are present in this group, e.g., the transcripts from many transposable elements or their LTRs, the inefficiently spliced REC107 (MER2) and HFM1 (MER3) mRNAs, the uORF-containing CPA1 mRNA, the nonsense-containing ade2-1 and can1-100 mRNAs, and the EST3 mRNA, which contains a programmed translational frameshifting site.
Fig. 2.
A significant fraction of NMD-regulated transcripts are direct substrates for the NMD pathway. (A) Overlap of significantly changed probe sets from the Upf1p-TAP, nmdΔ, and PGAL1–NMD2 experiments. (B and C) Scatter plot of the 781 probe sets that increased >2-fold in the Upf1p-TAP experiment (B) or the 587 probe sets that increased <2-fold in the Upf1p-TAP experiment but that exhibited ≥2-fold increase in nmdΔ strains and/or ≥1.8-fold decrease upon NMD reactivation (C). The average of the raw signal intensity from the four independent affinity selections (Upf1p-TAP or mock) was compared on a logarithmic scale. The middle lines indicate values that represent a Upf1TAP/mock ratio of 1 (no enrichment). The outer lines represent an Upf1p-TAP/mock ratio of 2 and 0.5, respectively. Coloring is according to A.
Transcripts indirectly regulated by NMD were expected to be found within the 554 transcripts (587 probe sets) that are not enriched with Upf1p-TAP, but that were found to be NMD-regulated in the nmdΔ and/or PGAL1–NMD2 experiments (Fig. 2A). However, most of the transcripts in this group were skewed toward enrichment by TAP selection (Fig. 2C), suggesting that a significant fraction of these represent bona fide substrates that have been excluded by our stringent definition of enrichment. This result suggests that a large fraction of the 931 potential NMD-regulated transcripts (represented by 991 probe sets in the nmdΔ and/or PGAL1–NMD2 experiments) are direct substrates of the NMD pathway.
Growth Conditions Influence the NMD Substrate Status of Specific Transcripts.
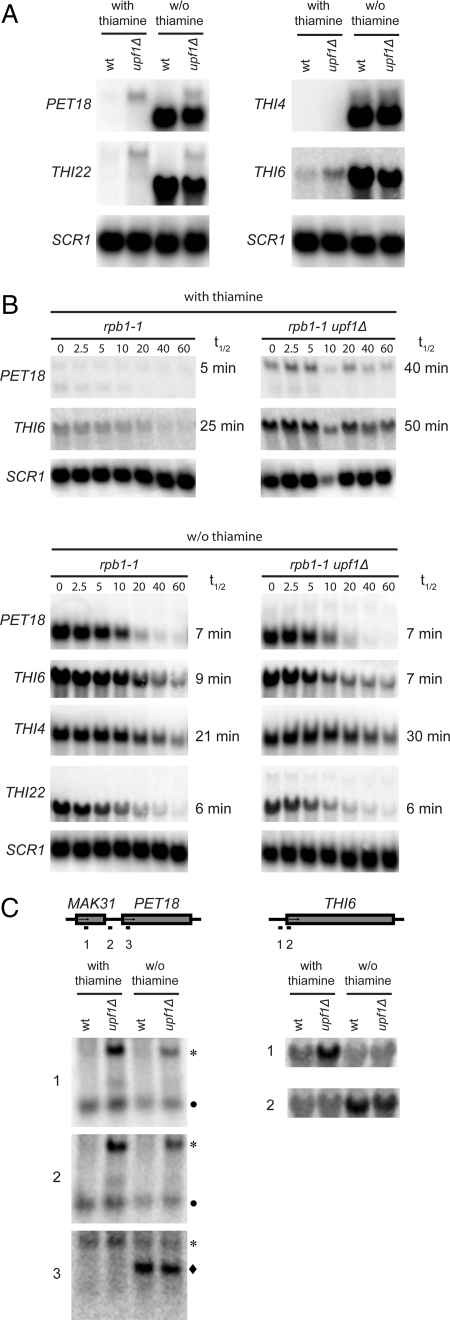
Previous studies have suggested that the NMD pathway may be involved in the regulation of specific cellular processes (3, 5, 7, 11). An examination of the gene ontology classifications associated with the NMD-regulated yeast transcripts revealed that the process with the highest likelihood of being NMD-regulated was thiamine metabolism. However, because the medium used for the relevant microarray experiments provides ample thiamine for growth, the NMD reactivity of individual transcripts may be influenced by culture conditions. To test this possibility, we determined the steady-state levels of the PET18, THI4, THI6, and THI22 mRNAs in wild-type and upf1Δ cells grown in the presence or absence of thiamine. Of these, the PET18, THI4, and THI22 transcripts belong to the group of NMD-regulated transcripts that were significantly enriched with Upf1p-TAP. The blots were also probed for SCR1 RNA, a stable RNA polymerase III transcript, as a loading control. Growth in thiamine-containing medium led to the accumulation of PET18, THI6, and THI22 mRNAs in upf1Δ cells, whereas the THI4 transcript was below the level of detection (Fig. 3A). Interestingly, thiamine starvation induced expression of shorter forms of the PET18, THI6, and THI22 mRNAs, and these transcripts did not show increased steady-state levels in the upf1Δ strain (Fig. 3A). The THI4 transcript was strongly induced under these conditions, but its steady-state level was not affected by inactivation of the NMD pathway (Fig. 3A).
Fig. 3.
The growth condition affects identification of NMD substrates. (A) Northern blot analyses of total RNA isolated from wild-type (HFY113) or upf1Δ (MJY86) cells grown exponentially at 30°C in the presence or absence of thiamine in synthetic minimal medium supplemented with adenine, histidine, leucine, trypthophan, and uracil (SD+Ade+His+Leu+Trp+Ura). The blots were probed for indicated transcripts by using randomly labeled DNA fragments. (B) The rpb1-1 (MJY95) or rpb1-1 upf1Δ (MJY96) strains were grown at 24°C in SD+Ade+His+Leu+Trp+Ura medium with (Upper) or without (Lower) thiamine followed by inhibition of RNA polymerase II transcription by a rapid shift to 37°C. Time points (minutes) after the shift are indicated above the lanes. The signal in each lane was normalized to the corresponding SCR1 RNA signal and the half-life (t1/2) determined from the initial slope of the curve. (C) Northern blot analyses of the total RNA samples described in A by using the indicated oligonucleotide probes. The symbols indicate the PET18 (longer; *), PET18 (shorter; ◆), and MAK31 (●) transcripts.
To determine which of these transcripts are direct NMD substrates, we measured their half-lives in wild-type or upf1Δ cells in the presence or absence of thiamine (Fig. 3B). mRNA decay rates were monitored after inhibition of RNA polymerase II activity in strains harboring a temperature-sensitive rpb1-1 allele (17, 18). Because of their low abundance in wild-type cells, half-lives could not be determined for the THI4 mRNA and the longer THI22 mRNA in thiamine-containing medium. However, the uninduced forms of the PET18 and THI6 transcripts displayed longer half-lives in upf1Δ cells than in wild-type cells, confirming that they are direct NMD substrates (Fig. 3B). In contrast, the half-lives of the shorter, induced forms of the PET18, THI6, and THI22 mRNAs were unaffected by inactivation of NMD (Fig. 3B). The THI4 transcript showed a slightly longer half-life in the upf1Δ strain despite the fact that its steady-state level is unchanged in the same cells (Fig. 3B). This minor change in decay rate might indicate that the induced THI4 mRNA is also subject to feedback regulation under thiamine starvation conditions.
To determine the structural basis of NMD reactivity for the uninduced, longer forms of the PET18 and THI6 transcripts, we used Northern blotting and oligonucleotide probes specific to regions within or upstream of the respective ORFs to localize their 5′ termini (Fig. 3C). These analyses suggested that the longer, NMD-responsive PET18 transcript contains a large part of the upstream MAK31 ORF. Consistent with this observation, a large-scale analysis of cDNAs derived from capped mRNAs identified a PET18 mRNA species with a 362-nt-long 5′ UTR that starts within the first half of the MAK31 ORF (19). The first AUG codon in this mRNA is out-of-frame with the MAK31 ORF, leading to a termination codon after two additional sense codons (19). However, even if translation would initiate at the next AUG codon, which is in-frame with the MAK31 ORF, the stop codon would be in an atypical bicistronic context. The longer THI6 mRNA also contains an extended 5′ UTR (Fig. 3C) that encompasses at least one upstream AUG codon and a short uORF. Thus, the NMD reactivity of the uninduced PET18 and THI6 mRNAs can be explained by their inclusion of premature termination codons. Taken together, these results suggest that the overrepresentation of transcripts encoding proteins involved in thiamine metabolism is a consequence of performing the original microarray experiments under conditions where the respective genes are repressed, thereby making it possible to detect low abundance NMD-responsive transcripts that may arise from transcriptional noise, i.e., inappropriate transcriptional initiation at cryptic promoters and intergenic regions (20).
Low-Abundance Alternative Transcripts May Represent a Significant Fraction of NMD Substrates.
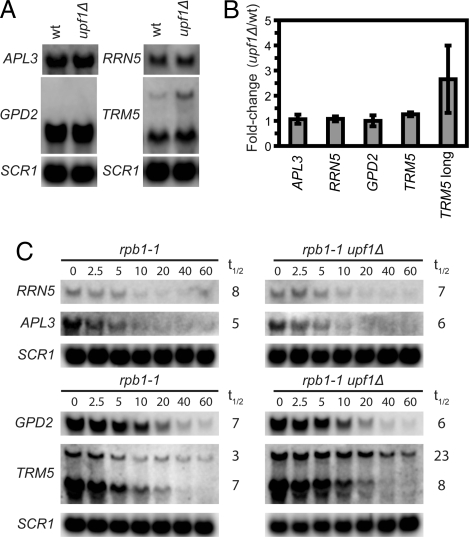
The predominance of transcripts that accumulate in cells lacking NMD activity are of low abundance (3), suggesting that transcripts derived from transcriptional noise (20) may comprise a significant fraction of the total number of naturally occurring NMD substrates. This supposition also implies that the Upf1p-TAP enriched transcripts that showed no apparent accumulation in NMD-deficient cells (Fig. 2 and SI Fig. 8) may represent minor variant forms for which the accumulation is masked by a major form(s), i.e., there may be other genes giving rise to both aberrant and appropriately expressed transcripts similar to those derived from the PET18 and THI22 genes during thiamine starvation (Fig. 3). An alternative explanation is that the half-life but not the steady-state level of such RNAs would be affected by NMD analogous to the THI4 mRNA under thiamine starvation conditions. To investigate the validity of these models, we determined the effect of NMD inactivation on the steady-state levels and half-lives of four randomly selected Upf1p-TAP-enriched transcripts that showed no apparent NMD regulation in the gene expression profiles. This analysis identified two different forms of the TRM5 transcript, and the longer, less abundant form accumulated in upf1Δ cells (Fig. 4 A and B). As expected, the longer, but not the shorter, TRM5 transcript displayed an extended half-life when UPF1 was deleted (Fig. 4C). For the other three transcripts (APL3, GPD2, and RRN5), no obviously longer forms and no apparent effect on either steady-state level or half-life was observed (Fig. 4). Although it is possible that these transcripts are either false positives or regulated by Upf1p at a step different from degradation, we cannot exclude that possibility that the Upf1p-TAP enrichment is caused by the presence of a low abundance species that is below the level of detection or of comparable size to the major mRNA species.
Fig. 4.
Alternative transcripts may represent a significant fraction of NMD substrates. (A) Northern blot analysis of total RNA isolated from wild-type (HFY113) or upf1Δ (MJY86) cells grown exponentially in rich yeast extract/peptone/dextrose (YEPD) medium at 30°C. The blots were probed for indicated transcripts by using randomly labeled DNA fragments. (B) The signal of respective mRNA in A and two additional independent experiments was normalized to the corresponding SCR1 signal, and the value for the upf1Δ strain was expressed relative to that for the wild-type. The standard deviation is indicated. (C) The rpb1-1 (MJY95) or rpb1-1 upf1Δ (MJY96) strains were grown at 24°C in YEPD medium followed by inhibition of RNA polymerase II transcription by a rapid shift to 37°C. Time points (minutes) after the shift are indicated above the lanes. The signal in each lane was normalized to the corresponding SCR1 signal and the half-life (t1/2) was determined from the initial slope of the curve.
Discussion
Direct vs. Indirect NMD Substrates.
In several different eukaryotes, inactivation of the NMD pathway leads to the accumulation of a substantial number of transcripts (3–7). However, the basis for this modulation of mRNA levels has been unclear, i.e., it is unknown whether most NMD-regulated transcripts are directly targeted for accelerated degradation by the NMD pathway, or if their levels change as an indirect consequence of the altered expression of a relatively small number of genuine substrates. Three genomewide approaches have been used to address this problem, namely the determination of mRNA half-lives in NMD-deficient cells (11), the identification of mRNAs whose abundance decreases when NMD is reactivated (Fig. 1), and the characterization of mRNAs that copurify with Upf1p (Figs. 1, 2, and 4). Using the latter approach, we find that 765 transcripts (represented by 781 probe sets) associate with Upf1p. Of these, 395 (404 probe sets) were present among 931 (991 probe sets) potential NMD substrates (Fig. 2), showing that at least 42% of NMD-regulated transcripts are direct substrates. This estimate could be conservative because the vast majority of NMD-regulated transcripts may also be direct substrates (Fig. 2C).
The percentage of direct substrates estimated here is comparable with that obtained in the genomewide mRNA half-life experiment, where 278 of 598 NMD-regulated transcripts were proposed to be direct substrates (11). However, there may be significant difficulty in accurately predicting substrates from global decay rate experiments, a conclusion evident from the lack of correlation between the direct and indirect substrates identified by Guan et al. (11) and either the Upf1p-associated mRNAs or the mRNAs defined herein as direct substrates (SI Fig. 9).
Possible Contribution of Transcriptional Noise to the Pool of Direct NMD Substrates.
Recent studies have suggested that the transcriptomes of eukaryotic cells are much more complex than previously thought (20–22). In yeast, for example, a large number of nonannotated intergenic and antisense RNAs have been identified, and most protein-coding genes appear to have more than one transcriptional start site, a property that leads to both long and short transcripts from the same gene (19, 23). The biological significance of most of these novel transcripts is unclear, but it is likely that a substantial fraction originates from transcriptional noise (20). A role for NMD in degrading such putative “junk” RNA was recently implied by its apparent function in the cytoplasmic degradation of a subset of yeast cryptic unstable transcripts derived from intergenic regions (24). Furthermore, the observation that NMD regulates the expression of the long TRM5 transcript and the uninduced forms of the PET18, THI6, and THI22 mRNAs (Figs. 3 and 4) suggests that atypically transcribed mRNAs may comprise a major class of NMD substrates. Mechanistically, such RNAs are likely to become NMD substrates because the presence of multiple transcriptional start sites often leads to at least one transcript with a uORF in the 5′ UTR (19). This observation, and the expectation that most transcripts generated by transcriptional noise would be of low abundance and thus difficult to detect in the presence of major transcripts, suggests that the number of naturally occurring NMD substrates is probably much larger than previously thought. This conjecture, and NMD's already well documented role in the degradation of cytoplasmic intron-containing pre-mRNAs, transcripts from pseudogenes, and transcripts derived from transposable elements or their LTRs (3, 8) underscores its role as a front-line cellular surveillance mechanism for ensuring that mRNAs that associate with ribosomes have proper ORFs (2).
NMD Regulation of Specific Cellular Processes: A Reassessment.
The overrepresentation of NMD-regulated transcripts in specific cellular processes, e.g., thiamine metabolism, has suggested a regulatory role for NMD beyond its function in mRNA surveillance (3, 5, 7–9, 11). However, many of the NMD-regulated transcripts in thiamine metabolism are low-abundance alternative forms that are not induced upon thiamine starvation and the highly abundant forms that are present during thiamine starvation are generally not NMD substrates (Fig. 3). These observations indicate that thiamine metabolism would not have been identified as an NMD-regulated process if the expression profiling experiments had been performed in medium lacking thiamine. A corollary to this conclusion is that the notion of NMD-regulated processes must be reassessed to take into account the possibility that the data supporting the notion of NMD regulation was obtained under conditions where the specific pathway is repressed or relatively inactive, thereby allowing detection of transcriptional noise (or some other as-yet-uncharacterized sources of aberrant transcripts).
The possible contribution of RNAs derived from transcriptional noise to the set of NMD-regulated transcripts is consistent with the overall low abundance of the latter set in both yeast and flies, the minimal conservation of the apparently affected processes (3, 4, 7, 9), and the Upf1p-TAP enrichment of transcripts that show no apparent accumulation in microarray analyses of nmdΔ cells (3). Likewise, the underrepresentation of transcripts encoding proteins involved in protein synthesis among NMD-regulated yeast transcripts (3, 4) is consistent with their high levels of expression in exponentially growing cells. Although the validity of models suggesting that NMD regulates the expression of several gene products in a single cellular process may be questionable, it is evident that some transcripts are under physiologically relevant NMD regulation. A notable example of this phenomenon is the negative regulation of yeast CPA1 mRNA, where arginine addition to culture media promotes ribosome stalling at a uORF termination codon and consequent NMD destabilization of the transcript (16).
The CPA1 example demonstrates that the significance of NMD regulation should be associated with correlated changes between mRNA and protein levels. Interestingly, the increased level of STN1 mRNA in nmdΔ cells has been shown to correlate with an increased level of the protein (25). Because Stn1p is involved in telomere capping and in regulating recruitment of the telomerase to the ends of chromosomes (26, 27) the increased Stn1p levels have provided a possible explanation for the reduced telomere length and telomeric silencing defect of nmdΔ cells (25, 28, 29). The STN1 mRNA level was originally proposed to be affected indirectly by NMD (29), but our observation that the transcript is ≈3.5-fold enriched with Upf1p-TAP suggests that it may well be a direct substrate. Collectively, these results suggest that the altered expression of specific proteins, and the accumulation of aberrant transcripts, can explain at least some phenotypes of NMD-inactivated cells.
Materials and Methods
Strains, Media, and Genetic Procedures.
The yeast strains used in this study are listed in SI Table 3. Yeast transformations (30), media, and genetic procedures have been described (31). Yeast nitrogen base without thiamine was purchased from MP Biomedicals. For details of strain constructions see SI Text.
RNA and Microarray Analyses.
Procedures for RNA analyses, isolation of Upf1p-TAP-associated transcripts, and identification of transcripts down-regulated upon NMD reactivation are described in SI Text. Microarray procedures, including cDNA and cRNA synthesis, hybridization, and data analysis were performed essentially as described (3). For details see SI Text.
Supplementary Material
ACKNOWLEDGMENTS.
We thank the members of A.J.'s laboratory for helpful discussions and editorial comments. This work was supported by National Institutes of Health Grant R37GM27757 (to A.J.). M.J.O.J. was supported by a postdoctoral fellowship from the Swedish Research Council.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE9482 and GSE9486).
This article contains supporting information online at www.pnas.org/cgi/content/full/0709257105/DC1.
References
- 1.Jacobson A, Izaurralde E. In: Translational Control in Biology and Medicine. Mathews MB, Sonenberg N, Hershey JWB, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2007. pp. 655–688. [Google Scholar]
- 2.Amrani N, Sachs MS, Jacobson A. Nat Rev Mol Cell Biol. 2006;7:415–425. doi: 10.1038/nrm1942. [DOI] [PubMed] [Google Scholar]
- 3.He F, Li X, Spatrick P, Casillo R, Dong S, Jacobson A. Mol Cell. 2003;12:1439–1452. doi: 10.1016/s1097-2765(03)00446-5. [DOI] [PubMed] [Google Scholar]
- 4.Lelivelt MJ, Culbertson MR. Mol Cell Biol. 1999;19:6710–6719. doi: 10.1128/mcb.19.10.6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nat Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 6.Wittmann J, Hol EM, Jack HM. Mol Cell Biol. 2006;26:1272–1287. doi: 10.1128/MCB.26.4.1272-1287.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E. RNA. 2005;11:1530–1544. doi: 10.1261/rna.2160905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He F, Jacobson A. In: Nonsense-Mediated mRNA Decay. Maquat L, editor. Georgetown, TX: Landes Bioscience; 2006. pp. 27–41. [Google Scholar]
- 9.Rehwinkel J, Raes J, Izaurralde E. Trends Biochem Sci. 2006;31:639–646. doi: 10.1016/j.tibs.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 10.Rodriguez-Gabriel MA, Watt S, Bahler J, Russell P. Mol Cell Biol. 2006;26:6347–6356. doi: 10.1128/MCB.00286-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guan Q, Zheng W, Tang S, Liu X, Zinkel RA, Tsui KW, Yandell BS, Culbertson MR. PLoS Genet. 2006;2:e203. doi: 10.1371/journal.pgen.0020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johns L, Grimson A, Kuchma SL, Newman CL, Anderson P. Mol Cell Biol. 2007;27:5630–5638. doi: 10.1128/MCB.00410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muhlrad D, Parker R. Nature. 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- 14.Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A. Nature. 2004;432:112–118. doi: 10.1038/nature03060. [DOI] [PubMed] [Google Scholar]
- 15.Maderazo AB, Belk JP, He F, Jacobson A. Mol Cell Biol. 2003;23:842–851. doi: 10.1128/MCB.23.3.842-851.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaba A, Jacobson A, Sachs MS. Mol Cell. 2005;20:449–460. doi: 10.1016/j.molcel.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 17.Nonet M, Scafe C, Sexton J, Young R. Mol Cell Biol. 1987;7:1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrick D, Parker R, Jacobson A. Mol Cell Biol. 1990;10:2269–2284. doi: 10.1128/mcb.10.5.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miura F, Kawaguchi N, Sese J, Toyoda A, Hattori M, Morishita S, Ito T. Proc Natl Acad Sci USA. 2006;103:17846–17851. doi: 10.1073/pnas.0605645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Struhl K. Nat Struct Mol Biol. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- 21.Johnson JM, Edwards S, Shoemaker D, Schadt EE. Trends Genet. 2005;21:93–102. doi: 10.1016/j.tig.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 22.Willingham AT, Gingeras TR. Cell. 2006;125:1215–1220. doi: 10.1016/j.cell.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 23.David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM. Proc Natl Acad Sci USA. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson DM, Parker R. Mol Cell Biol. 2007;27:92–101. doi: 10.1128/MCB.01023-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Enomoto S, Glowczewski L, Lew-Smith J, Berman JG. Mol Cell Biol. 2004;24:837–845. doi: 10.1128/MCB.24.2.837-845.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grandin N, Reed SI, Charbonneau M. Genes Dev. 1997;11:512–527. doi: 10.1101/gad.11.4.512. [DOI] [PubMed] [Google Scholar]
- 27.Lustig AJ. Nat Struct Biol. 2001;8:297–299. doi: 10.1038/86157. [DOI] [PubMed] [Google Scholar]
- 28.Lew JE, Enomoto S, Berman J. Mol Cell Biol. 1998;18:6121–6130. doi: 10.1128/mcb.18.10.6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dahlseid JN, Lew-Smith J, Lelivelt MJ, Enomoto S, Ford A, Desruisseaux M, McClellan M, Lue N, Culbertson MR, Berman J. Eukaryot Cell. 2003;2:134–142. doi: 10.1128/EC.2.1.134-142.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gietz D, St Jean A, Woods RA, Schiestl RH. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amberg DC, Burke DJ, Strathern JN. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2005. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.