Abstract
Multiple sclerosis (MS) is a common inflammatory disease of the central nervous system unsurpassed for variability in disease outcome. A cohort of sporadic MS cases (n = 163), taken from opposite extremes of the distribution of long-term outcome, was used to determine the role of the HLA-DRB1 locus on MS disease severity. Genotyping sets of benign and malignant MS patients showed that HLA-DRB1*01 was significantly underrepresented in malignant compared with benign cases. This allele appears to attenuate the progressive disability that characterizes MS in the long term. The observation was doubly replicated in (i) Sardinian benign and malignant patients and (ii) a cohort of affected sibling pairs discordant for HLA-DRB1*01. Among the latter, mean disability progression indices were significantly lower in those carrying the HLA-DRB1*01 allele compared with their disease-concordant siblings who did not. The findings were additionally supported by similar transmission distortion of HLA-DRB1*04 subtypes closely related to HLA-DRB1*01. The protective effect of HLA-DRB1*01 in sibling pairs may result from a specific epistatic interaction with the susceptibility allele HLA-DRB1*1501. A high-density (>700) SNP examination of the MHC region in the benign and malignant patients could not identify variants differing significantly between the two groups, suggesting that HLA-DRB1 may itself be the disease-modifying locus. We conclude that HLA-DRB1*01, previously implicated in disease resistance, acts as an independent modifier of disease progression. These results closely link susceptibility to long-term outcome in MS, suggesting that shared quantitative MHC-based mechanisms are common to both, emphasizing the central role of this region in pathogenesis.
There can be few diseases with as much variation in outcome as seen in multiple sclerosis (MS). Clinical manifestations, disease course, severity, and underlying pathological processes vary considerably among patients (1–6). For some it is a lifelong occasional nuisance, whereas for others it can be fatal in <1 year. Clinical and demographic factors have been associated with variation in disease severity and course, but efforts to elicit biological factors that determine such heterogeneity have to date not been fruitful.
There is reason to believe that genes may not only play a role in MS susceptibility (7), but also in clinical outcome as evidenced by twin studies. These data indicate that monozygotic twins are much more concordant for disease course than dizygotic twins or siblings (8). Further evidence of common genetic variants influencing disease severity exists in other putative autoimmune disorders, such as rheumatoid arthritis (9, 10).
The major histocompatibility complex (MHC) is associated with many putative autoimmune diseases, including MS (11–13). In the case of MS, susceptibility is unambiguously linked to the HLA class II region with susceptibility associated with the extended haplotype HLA-DQA1*0102-DQB1*0602-DRB1*1501-DRB5*0101 (14). Several studies have investigated the role of HLA alleles on disease outcome; however, the results have been unclear and conflicting (15–24). Among affected individuals, despite the presence of a common susceptibility region, there is marked clinical heterogeneity. Both the quality of clinical manifestations and the degree of disability outcomes vary substantially from patient to patient.
Approaches to this question have typically used unselected populations and cross-sectional measures or less often, rates of disability over time. This strategy is often underpowered because the outcome measures used in studies of this type frequently encompass only a minority of the entire disease spectrum. Effects sought may be effectively diluted by the frequent inclusion of patients yet to reach important outcome milestones. Insight into any role that the HLA class II region might play in determining clinical outcome in MS could come from the comparison of HLA-DRB1 alleles in patients who are extremely discordant for long-term clinical outcome. This approach is demonstrably more powerful (25–27).
The present study investigates the role of HLA-DRB1 alleles in influencing disease outcome in sporadic MS cases lying at opposite extremes of the distribution of long-term outcome (benign vs. malignant disease) identified with stringent clinical criteria. This unique cohort provides >99% power to detect a quantitative trait locus contributing to 2% of the additive phenotypic variance. This calculation assumes a normal distribution of disability in the MS population, with no dominance and an allele frequency of 0.2, tagged by a biallelic marker in complete linkage disequilibrium with the disease locus.
An additional feature of this study is the use of an independent cohort of affected sibling pairs (i.e., both siblings affected with the disease), a type of affected relative pair widely available for many complex traits. This cohort permits the validation of a positive allelic association with disease outcome identified in “extremes” cohorts. Rates of disease progression can be directly compared within sibling pairs discordant for the allele in question. To clarify whether HLA-DRB1 or genes/regulatory regions in linkage disequilibrium with this locus mediate effects on disease progression, we genotyped a high-density SNP panel spanning the genes within the MHC and its flanking regions. A role for HLA-DRB1*01 in protecting against disease progression in MS was supported.
Results
The Influence of HLA-DRB1 Alleles on MS Susceptibility.
The allele phenotype frequencies for each of the HLA-DRB1 alleles were compared in the sporadic MS cases vs. controls (Table 1). As expected, a significantly greater proportion of MS patients was positive for the HLA-DRB1*15 allele compared with the controls (56% vs. 24%; P < 0.0001). The only other allele to differ between MS patients and controls was the HLA-DRB1*11 allele, which was underrepresented in the MS cases (7.9% vs. 15%; P = 0.05). A separate study investigating HLA-DRB1 alleles in a cohort of affected sibling pairs showed a similar frequency profile to our sporadic MS cohort (28). To determine whether or not the strong association with the HLA-DRB1*15 allele created misleading deviations in the frequencies of the other alleles, the relative predispositional effects comparison method was used. It revealed that the HLA-DRB1*17 allele trended toward being overrepresented in MS patients compared with controls (P = 0.06).
Table 1.
HLA-DRB1 allele phenotype frequencies in sporadic MS patients and controls
| Allele | MS, n = 163 | Control, n = 202 | MS vs. control |
|
|---|---|---|---|---|
| P value | OR (95% C.I.) | |||
| 1 | 23 (14%) | 41 (20%) | NS | |
| 4 | 39 (24%) | 59 (29%) | NS | |
| 7 | 25 (15%) | 49 (24%) | NS | |
| 8 | 11 (6.7%) | 9 (4.5%) | NS | |
| 9 | 1 (0.61%) | 7 (3.5%) | NS | |
| 10 | 1 (0.61%) | 2 (0.99%) | NS | |
| 11 | 13 (7.9%) | 31 (15%) | 0.050 | 0.51 (0.26–0.98) |
| 12 | 5 (3.1%) | 10 (5.0%) | NS | |
| 13 | 33 (20%) | 47 (23%) | NS | |
| 14 | 6 (3.7%) | 12 (5.9%) | NS | |
| 15 | 92 (56%) | 49 (24%) | <0.0001 | 3.00 (2.03–4.43) |
| 16 | 3 (1.8%) | 8 (4.0%) | NS | |
| 17 | 40 (25%) | 45 (22%) | NS | |
NS, not significant.
The Influence of HLA-DRB1 Alleles on Disease Outcome.
Upon stratification of the MS cases into benign and malignant subtypes, an underrepresentation of the HLA-DRB1*01 allele in malignant MS (MMS) was observed when compared with benign MS (BMS) (BMS, 19%; MMS, 3.9%, P = 0.027) (Table 2). This association was independent of gender and confirmed by logistic regression. No other alleles differed significantly between MS outcome groups, including the HLA-DRB1*15 allele (BMS, 56%; MMS, 57%), and the overall frequency distribution of the alleles was not statistically significant between them.
Table 2.
HLA-DRB1 allele phenotype frequencies for BMS, MMS, and controls
| Allele | BMS(n = 112) | MMS(n = 51) | Control(n = 202) | BMS vs. MMS |
BMS vs. control |
MMS vs. control |
|||
|---|---|---|---|---|---|---|---|---|---|
| P value | OR (95% C.I.) | P value | OR (95% C.I.) | P value | OR (95% C.I.) | ||||
| 1 | 21 (19%) | 2 (3.9%) | 41 (20%) | 0.027 | 4.85 (1.11–21.16) | NS | 0.016 | 0.19 (0.04–0.79) | |
| 4 | 27 (24%) | 12 (24%) | 59 (29%) | NS | NS | NS | |||
| 7 | 17 (15%) | 8 (16%) | 49 (24%) | NS | NS | NS | |||
| 8 | 9 (8.0%) | 2 (3.9%) | 9 (4.5%) | NS | NS | NS | |||
| 9 | 0 (0%) | 1 (2.0%) | 7 (3.5%) | NS | 0.049 | 0 | NS | ||
| 10 | 1 (0.89%) | 0 (0%) | 2 (0.99%) | NS | NS | NS | |||
| 11 | 6 (5.4%) | 7 (14%) | 31 (15%) | NS | 0.013 | 0.33 (0.13–0.80) | NS | ||
| 12 | 4 (3.6%) | 1 (2.0%) | 10 (5.0%) | NS | NS | NS | |||
| 13 | 20 (18%) | 13 (25%) | 47 (23%) | NS | NS | NS | |||
| 14 | 5 (4.5%) | 1 (2.0%) | 12 (5.9%) | NS | NS | NS | |||
| 15 | 63 (56%) | 29 (57%) | 49 (24%) | NS | <0.0001 | 2.90 (1.90–4.42) | <0.0001 | 3.27 (1.91–5.59) | |
| 16 | 2 (1.8%) | 1 (2.0%) | 8 (4.0%) | NS | NS | NS | |||
| 17 | 30 (27%) | 10 (20%) | 45 (22%) | NS | NS | NS | |||
NS, not significant.
The genotype (1-1, 1-X, X-X) frequencies between BMS and MMS were compared (where 1 is the HLA-DRB1*01 allele and X represents any HLA-DRB1 allele other than HLA-DRB1*01). A significant underrepresentation of the frequency of genotypes containing the HLA-DRB1*01 allele (i.e., 1-1, 1-X) in MMS vs. BMS was observed (Table 3).
Table 3.
HLA-DRB1*01 genotype frequencies in BMS and MMS
| Genotype | BMS, n | MMS, n | Total, n |
|---|---|---|---|
| 1-1 | 2 (2%) | 0 (0%) | 2 |
| 1-X | 19 (17%) | 2 (4%) | 21 |
| X-X | 91 (81%) | 49 (96%) | 140 |
| Total | 112 | 51 | 163 |
The HLA-DRB1*15 allele has been occasionally associated with more severe disease outcome. In the current study, no significant differences in genotype frequencies between outcome groups were found. However, a somewhat greater proportion of malignant cases were homozygous for HLA-DRB1*15 compared with BMS (MMS, 15% vs. BMS, 8%).
HLA-DRB1 Alleles in HLA-DRB1*15 Negative Patients.
To investigate the effect of alleles other than HLA-DRB1*15 on disease outcome, the allele phenotype frequencies of patients and controls negative for the HLA-DRB1*15 allele were compared. MMS patients carried HLA-DRB1*01 significantly less than BMS cases (MMS, 11% vs. BMS, 43%, P = 0.039). In addition, the HLA-DRB1*01 allele was significantly overrepresented in BMS compared with controls (BMS, 43% vs. control, 24%; P = 0.033).
Interaction of HLA-DRB1 Alleles and Its Influence on Disease Outcome.
To determine whether or not interactions between HLA-DRB1 alleles influenced disease outcome, the HLA-DRB1 alleles that differed significantly from controls were tested for interactions by using logistic regression. No significant interactions were found between any of these risk and protective alleles on disease outcome. In addition, the HLA-DRB1 allele frequencies in patients who are positive for HLA-DRB1*15 were compared. The HLA-DRB1*01 allele was underrepresented in both BMS and MMS when compared with unaffected controls.
Transmission Disequilibrium Test (TDT) Analysis.
A TDT analysis was performed to test for transmission distortion of the HLA-DRB1 alleles. The HLA-DRB1*01 allele was significantly undertransmitted in MMS but not in BMS. TDT analysis also showed that HLA-DRB1*15 was preferentially overtransmitted to malignant (5:1) compared with benign cases (2.3:1) (Table 4). To control for the overrepresentation of the HLA-DRB1*15 allele, a TDT analysis using HLA-DRB1*15 negative parents was used. It was found that the HLA-DRB1*01 allele was undertransmitted to affected offspring with MMS. There was no evidence of transmission distortion for any of the other alleles investigated in BMS, MMS, or all MS cases combined. However, the number of families wherein both parents were HLA-DRB1*15 negative was small (n = 13), and these results should be taken with caution.
Table 4.
Transmission of HLA-DRB1 alleles in MS
| Allele | All MS cases |
BMS |
MMS |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TR, n | NT, n | χ2 | P value | TR, n | NT, n | χ2 | P value | TR, n | NT, n | χ2 | P value | |
| 1 | 12 | 20 | 2 | NS | 6 | 7 | 0.08 | NS | 1 | 7 | 4.5 | 0.03 |
| 4 | 21 | 25 | 0.35 | NS | 9 | 7 | 0.25 | NS | 6 | 8 | 0.29 | NS |
| 7 | 13 | 17 | 0.53 | NS | 6 | 6 | 0 | NS | 3 | 4 | 0.14 | NS |
| 8 | 8 | 3 | 2.27 | NS | 4 | 0 | 4 | 0.05 | 1 | 1 | 0 | NS |
| 9 | 1 | 0 | 1 | NS | 0 | 0 | 0 | NS | 1 | 0 | 1 | NS |
| 11 | 6 | 14 | 3.2 | NS | 2 | 7 | 2.78 | NS | 2 | 2 | 0 | NS |
| 12 | 4 | 0 | 4 | 0.05 | 2 | 0 | 2 | NS | 1 | 0 | 1 | NS |
| 13 | 10 | 26 | 7.11 | 0.008 | 1 | 12 | 9.31 | 0.002 | 2 | 6 | 2 | NS |
| 14 | 4 | 4 | 0 | NS | 2 | 3 | 0.2 | NS | 0 | 1 | 1 | NS |
| 15 | 49 | 17 | 15.52 | 0.00008 | 14 | 6 | 3.2 | NS | 15 | 3 | 8 | 0.004 |
| 16 | 2 | 4 | 0.67 | NS | 2 | 1 | 0.33 | NS | 0 | 0 | 0 | NS |
| 17 | 13 | 13 | 0 | NS | 6 | 5 | 0.09 | NS | 4 | 4 | 0 | NS |
TR, transmitted; NT, not transmitted; NS, not significant.
Validation of Protective Effect of HLA-DRB1*01 on Disease Outcome.
Independent affected sibling-pair cohort.
To validate the protective effect of the HLA-DRB1*01 allele on disease outcome, the progression indices (disability scores/time) of a prospective cohort of affected sibling pairs discordant for the HLA-DRB1*01 allele were compared by using a Wilcoxon paired samples test. The results confirm the protective effect conferred by the HLA-DRB1*01 allele as the sibling carrying the HLA-DRB1*01 allele had a progression index (PI) of 0.27 compared with 0.57 for the sibling not carrying the protective allele (P = 0.009). In other words, the affected sibling with the HLA-DRB1*01 allele reached an Expanded Disability Status Scale (EDSS) = 6 in a mean of 22.2 years of disease onset compared with the affected sibling without HLA-DRB1*01, who reached the same level of disability 10.5 years after disease onset. Upon further analysis of the sibling cohort data, the protective effect conferred by the HLA-DRB1*01 allele is only significant when it is part of the HLA-DRB1*15-HLA-DRB1*01 (15-1) genotype (15-1, PI = 0.25; 1-X, PI = 0.49; P = 0.01; where X represents any HLA-DRB1 allele other than HLA-DRB1*01 and HLA-DRB1*15).
Sardinian cohort.
We genotyped benign and malignant patients from Sardinia for the presence of HLA-DRB1*01. There were 19 benign and 0 malignant patients positive for the allele (P = 0.006).
HLA-DRB1 subtypes.
HLA-DRB1*0101, HLA-DRB1*0401, and HLA-DRB1*0404 all share a common stretch of amino acids at positions 67–74 in the peptide binding groove of the DR β chain (29). Twenty-six of 112 (23%) benign patients were positive for either HLA-DRB1*0401 and HLA-DRB1*0404, compared with only 5 of the 51 (10%) malignant patients (P = 0.03).
High-Density SNP Typing of the MHC Region.
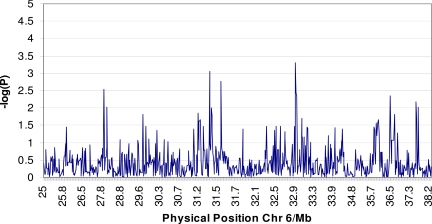
In the Canadian benign and malignant patients we typed the MHC genomic region with a dense panel of SNPs. We accepted markers for analysis if they met the following criteria: (i) a minimum call rate of 90%; (ii) consistency with Hardy–Weinberg equilibrium at P > 0.001; and (iii) a minimum minor allele frequency of >2%. These criteria identified 703 successful assays. Using a previously published definition (30), we identified 104 haplotype blocks spanning the MHC. Analysis of both single markers and haplotypes could not identify any statistically significant differences between benign and malignant patients (Fig. 1). The single SNP marker most significantly associated with outcome was rs2071541 in the HLA class II region (P = 0.005, uncorrected; see www.well.ox.ac.uk/∼sreeramr for a full list of markers and associated P values). No association remained significant after correction for multiple testing using a permutation test (n = 10,000 permutations). Given the number of samples and markers used, no SNP may ever reach significance after correction, so we attempted to replicate all nominally associated SNPs in the additional cohorts. No SNP association replicated.
Fig. 1.
Results of high-density SNP typing of the MHC region in benign vs. malignant patients (negative logarithm P value of difference in SNP frequency between benign and malignant patients plotted against physical position of SNP in MHC).
Discussion
The only genetic association with MS in Northern Europeans has been with extended MHC haplotypes, especially those containing HLA-DRB1*1501 (7). However, IL-7 receptor (IL7RA) and IL-2 receptor (IL2RA) alleles have recently been shown to have slight influence on risk (31, 32). These effects [maximum odds ratios (OR) = 1.3] are much smaller compared with those within the MHC (OR = 5.4). It is now clear that the MHC is the key susceptibility locus in MS with other susceptibility genes contributing relatively little (33). Reports of linkages or associations improbably reported on nearly every chromosomal arm appear to be false positives for the most part (34).
The role of the MHC and, in particular, the HLA-DRB1 locus in determining clinical outcome in MS has been poorly understood. The present study examines the distribution of HLA-DRB1 alleles in two types of large MS cohorts. The first consists of sporadic cases lying at the two extreme ends of long-term clinical outcome, and the second is the common type of affected relative pair in this disease–concordant sibling pairs. In the first cohort of sporadic MS cases, the classic association of the HLA-DRB1*15 allele to disease susceptibility was confirmed in both case-control and family-based (i.e., TDT) analyses. In keeping with other studies, a modest association of allele HLA-DRB1*17 with MS was also observed, after taking into account the relative predispositional effect of the HLA-DRB1*15 allele (22, 28). As an independent control for the HLA-DRB1 allele frequencies noted in this study, the HLA-DRB1 allele frequency profiles of the sporadic cases were compared with those of a large independent cohort of affected sibling pairs (28). The distribution of allele frequencies between cohorts was found to be remarkably similar. This finding implies that sporadic MS may not differ qualitatively from familial MS in terms of genetic susceptibility to the disease and invites direct comparisons.
Several groups investigating HLA class II genotypes in relapsing remitting MS and primary progressive MS have sought evidence for genetic heterogeneity in MS, but there has been little consensus to indicate that different alleles confer susceptibility to different clinical subsets of the disease (1, 15, 35). Comparison of HLA-DRB1 allele profiles between BMS and MMS in the current dataset showed no difference in allele frequency for the main associated allele HLA-DRB1*15. These findings cohere with observations within families and imply that these extreme clinical phenotypes are not separate disease entities. Even though MS is clinically heterogeneous in terms of age of onset, duration of disease, and gender distribution, the frequency of the main Northern European MS susceptibility allele, HLA-DRB1*15, was similar in both BMS (56%) and MMS (57%). Taken together with the observation that few other diseases [chronic sarcoidosis (36), narcolepsy (37), and pernicious anemia (38)] are positively associated with this allele, the notion that BMS and MMS are probably extreme clinical manifestations of the same disease finds support.
With similar HLA-DRB1*15 allele frequencies in the extremes, comparison of the frequency of other alleles conveniently isolates their outcome effect on a common roughly equivalent background of both risk and the major allele associated with its associated susceptibility. The strategies used effectively minimize the problems of population stratification by these inherent features of internal matching.
The analyses here yielded several lines of evidence suggesting that HLA-DRB1*01 protects against adverse clinical outcome in MS. First, genotyping data from sporadic MS cases showed HLA-DRB1*01 to be significantly underrepresented in malignant cases compared with both the benign and control groups, indicating that the absence of the allele may accentuate a progressive course in MS. The fact that the HLA-DRB1*01 allele frequency did not differ between benign and control cases implied this effect diverges from susceptibility itself and determines outcome rather than risk, but higher-order interactions have been demonstrated (28). Second, stratification of cases for HLA-DRB1*15 revealed that HLA-DRB1*01 was significantly underrepresented in MMS compared with BMS and was overrepresented in BMS compared with controls in cases negative for HLA-DRB1*15. These findings show that the phenotype-based allele frequencies of HLA-DRB1*01 observed in BMS and MMS are not misleading deviations caused by the strong association of other alleles, such as HLA-DRB1*15, and that the presence of HLA-DRB1*01 protects against malignant disease. Third, in a TDT analysis using the genotype data from nuclear family members, it was found that the HLA-DRB1*01 allele was significantly undertransmitted to affected offspring with MMS, whereas no transmission distortion for this allele was observed in offspring with benign disease. The results from the TDT analysis provide another dimension of support to the assertion that HLA-DRB1*01 protects against the rapid acquisition of disability in MS, further attenuating the possibility that population substructure could confound the data. Fourth, in a replication cohort of Sardinian benign and malignant patients there was a significant overrepresentation of HLA-DRB1*01 in benign patients. Remarkably, this was found despite the primary MS association in this population being with non-HLA-DRB1*1501 alleles. Fifth, in an independent cohort of affected sibling pairs discordant for HLA-DRB1*01, the mean PI was considerably lower in the sibling carrying the HLA-DRB1*01 allele compared with their affected HLA-DRB1*01-negative sibling. Not only does the affected sibling-pair dataset serve to replicate the genotype findings from the sporadic MS cohort, it serves to validate the claim that the protective effect on clinical outcome comes from the HLA-DRB1*01 allele or a nearby variation. Sibling pairs share a common genetic background and perhaps environmental factors, thereby increasing the likelihood that a discordant locus or allele (i.e., HLA-DRB1*01) when associated with a discordant phenotype, contributes to the clinical picture observed. Finally, although the HLA-DRB1 allele appears to play a significant role in modifying the disease course and severity in MS, it is important to emphasize that the protective effect does not necessarily arise directly from the HLA-DRB1*01 allele itself. It is plausible that a locus or regulatory region in tight linkage disequilibrium with the HLA-DRB1*01 allele, or a more complex protective haplotype, may be disease-modifying. However, the results of dense SNP typing of the MHC region did not identify any variants differing significantly between benign and malignant patients. Although this study argues against any involvement of genes encoding the MHC apart from HLA-DRB1 in determining outcome, it remains possible that the SNP panel used may have missed variants with disease-modifying activity or was underpowered to do so, especially given the difficulty with SNP typing in this region (39).
The greater proportion of benign patients positive for either HLA-DRB1*01 or HLA-DRB1*0401/HLA-DRB1*0404 than of malignant cases is intriguing. Ironically, such analogy is not readily ascertained for DRB1 alleles involved in MS susceptibility, possibly dissociating these two effects. It suggests that the known shared binding characteristics of these molecules could pinpoint the precise site that allows them to participate in underlying severity-related mechanisms. For DRB1*0401/HLA-DRB1*0404, this finding is converse to the role of these alleles in rheumatoid arthritis, where these alleles predispose to severe disease but the genetic pattern seems similar. There are several reported examples of peptides that bind both HLA-DRB1*0401 and HLA-DRB1*0101 (29).
The finding that the protective effect of HLA-DRB1*01 in sibling pairs occurs primarily within the HLA-DRB1*15/01 genotype suggests that interactions between specific HLA-DRB1 alleles/haplotypes influence phenotypic expression of MS. This notion is analogous to what we have previously shown for susceptibility (28) and also applies to animal models where epistasis between HLA-DRB1*15 and HLA-DRB5*0101 has been shown to influence the clinical course of experimental autoimmune encephalomyelitis (28) (40). Because interactions between HLA-DRB1*15 and DRB1*01 have been shown to confer resistance to acquiring the disease, susceptibility and outcome in MS may be intrinsically linked (28, 41). Overlap between these effects may only be partial. Although HLA-DRB1*15 is by far the commonest susceptibility allele, it did not differ in allele frequency between benign and malignant cases in contrast to DRB1*01.
There may well be similar effects from other DRB1 resistance alleles of lower frequency or with smaller effect size on outcome, which might be most efficiently sought by using larger numbers of extremes. Given the striking ethnic differences for class II allele frequency, epistatic mechanisms identified here and previously are plausible candidates for some of the remarkable differences in MS outcome that have been reported among racial groups. Perhaps less obvious is the potential explanation offered by epistatic effect of the type shown here for population-specific differences in outcome and allelic association. Here, we showed that in populations stratified by outcome the primary allelic association did not differ but the epistatic allele had a major influence on disability. This finding has potential relevance to the vexing problems of replicating disease associations, suggesting that some findings attributed to false positivity could be manifestations of inadvertent comparison of populations unmatched for important epistatic effects.
In conclusion, HLA-DRB1*01 segregates both with disease resistance and favorable outcome, is an independent contributor to the risk of disease progression, and operates in both sporadic and familial MS (sibling pairs). Future genetic and functional studies will investigate the manner in which the HLA-DRB1*01 allele exerts its protective effect on outcome. This will be crucial for developing potential therapeutic strategies based on these findings, which might modify the progression of this often devastating disease.
Subjects and Methods
Patients and Controls.
All subjects used in the study were ascertained through the ongoing Canadian Collaborative Project on the Genetic Susceptibility to MS (CCPGSMS), for which the methodology has been described (42, 43). A total of 163 sporadic MS patients along with their nuclear family members (n = 625) were selected for analysis from London, Ontario, and Vancouver. The disability of the sporadic MS patients was carefully assessed and recorded at entry with the EDSS by neurologists involved in the CCPGSMS. The PI for each affected individual was calculated [PI = EDSS score/time from disease onset (years)]. Patients were classified as having either BMS or MMS based on EDSS scores that were maintained over or achieved within designed time intervals (see Table 5 for clinical details). The BMS cases fall under the relapsing–remitting clinical subtype wherein minimal disability only (i.e., EDSS ≤3) was attained over a period >20 years from disease onset. In contrast, MMS cases, a subgroup of MS patients acquiring significant disability (i.e., EDSS >6; requiring the use of a cane or worse) within 5 years of disease onset, had primary or relapsing–progressive forms of the disease.
Table 5.
Clinical and demographic data on BMS and MMS patients
| Feature | Clinical/demographic group |
||||
|---|---|---|---|---|---|
| BMS | MMS | SBMS | SMMS | CCPGSMS | |
| Sample size, n | 112 | 51 | 62 | 19 | 1,816 |
| Sex ratio, female/male | 87:25 (3.48:1) | 31:21 (1.48:1) | 49:13 (3.76:1) | 12:7 (1.7:1) | 1310:506 (2.58:1) |
| Mean age of onset, years | 25.1 | 37.3 | 26.2 | 35.3 | 30.9 |
| Mean duration of disease, years | 26 | 3.6 | 24.7 | 4.1 | |
BMS, Canadian BMS = EDSS ≤ 3 minimum 20 years from disease onset; MMS, Canadian MMS = EDSS > 6 within 5 years of disease onset; SBMS, Sardinian BMS; SMMS, Sardinian MMS; CCPGSMS, a sample taken from the CCPGSMS database to show average values.
Validation of Positive Allelic Associations.
To validate positive allelic associations with disease outcome found in the sporadic cases, a replication cohort from Sardinia comprised of 62 patients fitting the benign criteria and 19 patients fitting the malignant criteria was used. An additional independent cohort of affected Canadian sibling pairs (i.e., both siblings affected with the disease) (n = 104) discordant for the allele in question was examined, and the rates of disease progression between sibling pairs were compared.
HLA-DRB1 Genotyping.
Total genomic DNA, extracted from whole blood as part of the CCPGSMS (42), was used to type HLA-DRB1 alleles via a nonradioactive high-resolution allele-specific PCR amplification method. All genotypes were generated by observers blind to pedigree structure and disease status of the individual. Seventy-two PCRs were carried out to amplify allelotypes corresponding to alleles HLA-DRB1*01, HLA-DRB1*04, HLA-DRB1*07, HLA-DRB1*08, HLA-DRB1*09, HLA-DRB1*10, HLA-DRB1*11, HLA-DRB1*12, HLA-DRB1*13, HLA-DRB1*14, HLA-DRB1*15, HLA-DRB1*16, HLA-DRB1*17, HLA-DRB1*18, and amplicons for the DRB3, DRB4, and DRB5 genes. Each HLA-DRB1 genotype was scored twice by independent observers. Hardy–Weinberg equilibrium was confirmed for all HLA-DRB1 alleles. Allele frequencies were estimated from 202 unrelated and unaffected individuals of Caucasian descent. All patients and controls were stratified by HLA-DRB1 allele frequencies. Additional MS cases (i.e., neither benign nor malignant) derived from the families of the sporadic MS cohort were included in MS susceptibility analyses.
SNP Selection and Genotyping.
SNP selection and genotyping was conducted as described (39).
Statistical Analyses.
For the sporadic MS cases, the frequencies of HLA-DRB1 alleles in patients and controls were compared bt using a χ2 test or Fisher's exact test to determine probability values for contingency tables. OR and 95% C.I. were calculated for each allele. Logistic regression analysis was used to (i) estimate the relationship between HLA status and affectation status, (ii) determine the effect of allele interactions on clinical status, and (iii) check for any relationship between HLA status, affectation, and gender. Where family data were available, PedCheck (44) was used to check for Mendelian inconsistencies, and the TDT was performed by using Genehunter 2.0 (45).
For the validation cohort, the effect of HLA-DRB1 on the PI of affected sibling pairs discordant for a given allele was evaluated by using a Wilcoxon paired samples test. For SNP analysis we used Haploview (46) and logistic regression analyses to test for single-marker and haplotypic associations.
ACKNOWLEDGMENTS.
We thank all patients who generously participated in this study; the neurologists participating in the CCPGSMS; Julian Knight, Jordana Bell, Andrew Morris, William Valdar, Katie Morrison, Samantha Holmes, Holly Armstrong, and Maria Criscuoli for help with genotyping and data analysis; and Lars Fugger for helpful comments. This work was funded by the Multiple Sclerosis Society of Canada Scientific Foundation, the U.K. MS Society, Genome Quebec, and Genome Canada. G.C.E. is the Action Research Professor of Clinical Neurology at the University of Oxford. G.C.D. is funded by a Multiple Sclerosis Society of Canada Studentship and a Clarendon Scholarship (University of Oxford). S.V.R. is funded by the Medical Research Council of the United Kingdom. T.J.H. is the recipient of a Clinician-Scientist Award in Translational Research by the Burroughs Wellcome Fund and an Investigator Award from Canadian Institutes of Health Research. A.D.S. is a Michael Smith Foundation for Health Research Distinguished Scholar.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Olerup O, Hillert J, Fredrikson S, Olsson T, Kam-Hansen S, Moller E, Carlsson B, Wallin J. Proc Natl Acad Sci USA. 1989;86:7113–7117. doi: 10.1073/pnas.86.18.7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thompson PD, Day BL, Rothwell JC, Dick JP, Cowan JM, Asselman P, Griffin GB, Sheehy MP, Marsden CD. J Neurol Sci. 1987;80:91–110. doi: 10.1016/0022-510x(87)90224-3. [DOI] [PubMed] [Google Scholar]
- 3.Hillert J, Gronning M, Nyland H, Link H, Olerup O. J Neurol Neurosurg Psychiatry. 1992;55:887–890. doi: 10.1136/jnnp.55.10.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Revesz T, Kidd D, Thompson AJ, Barnard RO, Mcdonald WI. Brain. 1994;117:759–765. doi: 10.1093/brain/117.4.759. [DOI] [PubMed] [Google Scholar]
- 5.Deluca GC, Ebers GC, Esiri MM. Brain. 2004;127:1009–1018. doi: 10.1093/brain/awh118. [DOI] [PubMed] [Google Scholar]
- 6.Lucchinetti C, Bruck W, Parisi J, Scheithauer B, Rodriguez M, Lassmann H. Ann Neurol. 2000;47:707–717. doi: 10.1002/1531-8249(200006)47:6<707::aid-ana3>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 7.Dyment DA, Ebers GC, Sadovnick AD. Lancet Neurol. 2004;3:104–110. doi: 10.1016/s1474-4422(03)00663-x. [DOI] [PubMed] [Google Scholar]
- 8.Willer CJ, Dyment DA, Risch NJ, Sadovnick AD, Ebers GC. Proc Natl Acad Sci USA. 2003;100:12877–12882. doi: 10.1073/pnas.1932604100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weyand CM, Hicok KC, Conn DL, Goronzy JJ. Ann Intern Med. 1992;117:801–806. doi: 10.7326/0003-4819-117-10-801. [DOI] [PubMed] [Google Scholar]
- 10.Macgregor A, Ollier W, Thomson W, Jawaheer D, Silman A. J Rheumatol. 1995;22:1032–1036. [PubMed] [Google Scholar]
- 11.Naito S, Namerow N, Mickey MR, Terasaki PI. Tissue Antigens. 1972;2:1–4. doi: 10.1111/j.1399-0039.1972.tb00111.x. [DOI] [PubMed] [Google Scholar]
- 12.Jersild C, Svejgaard A, Fog T. Lancet. 1972;1:1240–1241. doi: 10.1016/s0140-6736(72)90962-2. [DOI] [PubMed] [Google Scholar]
- 13.Winchester R, Ebers G, Fu SM, Espinosa L, Zabriskie J, Kunkel HG. Lancet. 1975;2:814. doi: 10.1016/s0140-6736(75)80033-x. [DOI] [PubMed] [Google Scholar]
- 14.Fogdell A, Hillert J, Sachs C, Olerup O. Tissue Antigens. 1995;46:333–336. doi: 10.1111/j.1399-0039.1995.tb02503.x. [DOI] [PubMed] [Google Scholar]
- 15.Engell T, Raun NE, Thomsen M, Platz P. Neurology. 1982;32:1043–1046. doi: 10.1212/wnl.32.9.1043. [DOI] [PubMed] [Google Scholar]
- 16.Stendahl-Brodin L, Link H, Moller E, Norrby E. Acta Neurol Scand. 1979;59:297–308. doi: 10.1111/j.1600-0404.1979.tb02940.x. [DOI] [PubMed] [Google Scholar]
- 17.Madigand M, Oger JJ, Fauchet R, Sabouraud O, Genetet B. J Neurol Sci. 1982;53:519–529. doi: 10.1016/0022-510x(82)90248-9. [DOI] [PubMed] [Google Scholar]
- 18.Runmarker B, Martinsson T, Wahlstrom J, Andersen O. J Neurol. 1994;241:385–390. doi: 10.1007/BF02033356. [DOI] [PubMed] [Google Scholar]
- 19.De La Concha EG, Arroyo R, Crusius JB, Campillo JA, Martin C, Varela De Seijas E, Pena AS, Claveria LE, Fernandez-Arquero M. J Neuroimmunol. 1997;80:172–178. doi: 10.1016/s0165-5728(97)00153-7. [DOI] [PubMed] [Google Scholar]
- 20.Weinshenker BG, Santrach P, Bissonet AS, Mcdonnell SK, Schaid D, Moore SB, Rodriguez M. Neurology. 1998;51:742–747. doi: 10.1212/wnl.51.3.742. [DOI] [PubMed] [Google Scholar]
- 21.Mcdonnell GV, Mawhinney H, Graham CA, Hawkins SA, Middleton D. J Neurol Sci. 1999;165:77–83. doi: 10.1016/s0022-510x(99)00084-2. [DOI] [PubMed] [Google Scholar]
- 22.Masterman T, Ligers A, Olsson T, Andersson M, Olerup O, Hillert J. Ann Neurol. 2000;48:211–219. [PubMed] [Google Scholar]
- 23.Barcellos LF, Oksenberg JR, Begovich AB, Martin ER, Schmidt S, Vittinghoff E, Goodin DS, Pelletier D, Lincoln RR, Bucher P, et al. Am J Hum Genet. 2003;72:710–716. doi: 10.1086/367781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jersild C, Fog T, Hansen GS, Thomsen M, Svejgaard A, Dupont B. Lancet. 1973;2:1221–1225. doi: 10.1016/s0140-6736(73)90970-7. [DOI] [PubMed] [Google Scholar]
- 25.Risch N, Zhang H. Science. 1995;268:1584–1589. doi: 10.1126/science.7777857. [DOI] [PubMed] [Google Scholar]
- 26.Haston CK, Hudson TJ. N Engl J Med. 2005;353:1509–1511. doi: 10.1056/NEJMe058185. [DOI] [PubMed] [Google Scholar]
- 27.Nebert DW. Eur J Pharmacol. 2000;410:107–120. doi: 10.1016/s0014-2999(00)00809-8. [DOI] [PubMed] [Google Scholar]
- 28.Dyment DA, Herrera BM, Cader MZ, Willer CJ, Lincoln MR, Sadovnick AD, Risch N, Ebers GC. Hum Mol Genet. 2005;14:2019–2026. doi: 10.1093/hmg/ddi206. [DOI] [PubMed] [Google Scholar]
- 29.Jones EY, Fugger L, Strominger JL, Siebold C. Nat Rev Immunol. 2006;6:271–282. doi: 10.1038/nri1805. [DOI] [PubMed] [Google Scholar]
- 30.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, Defelice M, Lochner A, Faggart M, et al. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Z, Duvefelt K, Svensson F, Masterman T, Jonasdottir G, Salter H, Emahazion T, Hellgren D, Falk G, Olsson T, et al. Genes Immun. 2005;6:145–152. doi: 10.1038/sj.gene.6364171. [DOI] [PubMed] [Google Scholar]
- 32.Hafler DA, Compston A, Sawcer S, Lander ES, Daly MJ, De Jager PL, De Bakker PI, Gabriel SB, Mirel DB, Ivinson AJ, et al. N Engl J Med. 2007;357:851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 33.Peltonen L. N Engl J Med. 2007;357:927–929. doi: 10.1056/NEJMe078147. [DOI] [PubMed] [Google Scholar]
- 34.Dyment DA, Ebers GC. J Neuroimmunol. 2007;190:5–7. doi: 10.1016/j.jneuroim.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 35.Sotgiu S, Pugliatti M, Solinas G, Castiglia P, Sanna A, Rosati G. Neurol Sci. 2001;22:167–170. doi: 10.1007/s100720170018. [DOI] [PubMed] [Google Scholar]
- 36.Abe S, Yamaguchi E, Makimura S, Okazaki N, Kunikane H, Kawakami Y. Chest. 1987;92:488–490. doi: 10.1378/chest.92.3.488. [DOI] [PubMed] [Google Scholar]
- 37.Kramer RE, Dinner DS, Braun WE, Zachary AA, Teresi GA. Arch Neurol. 1987;44:853–855. doi: 10.1001/archneur.1987.00520200055019. [DOI] [PubMed] [Google Scholar]
- 38.Ungar B, Mathews JD, Tait BD, Cowling DC. Br Med J. 1981;282:768–770. doi: 10.1136/bmj.282.6266.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lincoln MR, Montpetit A, Cader MZ, Saarela J, Dyment DA, Tiislar M, Ferretti V, Tienari PJ, Sadovnick AD, Peltonen L, et al. Nat Genet. 2005;37:1108–1112. doi: 10.1038/ng1647. [DOI] [PubMed] [Google Scholar]
- 40.Gregersen JW, Kranc KR, Ke X, Svendsen P, Madsen LS, Thomsen AR, Cardon LR, Bell JI, Fugger L. Nature. 2006;443:574–577. doi: 10.1038/nature05133. [DOI] [PubMed] [Google Scholar]
- 41.Ramagopalan SV, Morris AP, Dyment DA, Herrera BM, Deluca GC, Lincoln MR, Orton SM, Chao MJ, Sadovnick AD, Ebers GC. Plos Genet. 2007;3:1607–1613. doi: 10.1371/journal.pgen.0030150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sadovnick AD, Risch NJ, Ebers GC. Can J Neurol Sci. 1998;25:216–221. doi: 10.1017/s0317167100034041. [DOI] [PubMed] [Google Scholar]
- 43.Ramagopalan SV, Dyment DA, Valdar W, Herrera BM, Criscuoli M, Yee IM, Sadovnick AD, Ebers GC. Lancet Neurol. 2007;6:604–610. doi: 10.1016/S1474-4422(07)70132-1. [DOI] [PubMed] [Google Scholar]
- 44.O'Connell JR, Weeks DE. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES. Am J Hum Genet. 1996;58:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- 46.Barrett JC, Fry B, Maller J, Daly MJ. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]