Abstract
Polymerization of recombinant prion protein (recPrP), which was produced in bacteria, into amyloid fibers was accompanied by the acquisition of prion infectivity. We report here that partially purified preparations of prions seed the polymerization of recPrP into amyloid as detected by a fluorescence shift in the dye Thioflavin T. Our amyloid seeding assay (ASA) detected PrPSc, the sole component of the prion, in brain samples from humans with sporadic Creutzfeldt–Jakob disease, as well as in rodents with experimental prion disease. The ASA detected a variety of prion strains passaged in both mice and hamsters. The sensitivity of the ASA varied with strain type; for hamster Sc237 prions, the limit of detection was ≈1 fg. Some prion strains consist largely of protease-sensitive PrPSc (sPrPSc), and these strains were readily detected by ASA. Our studies show that the ASA provides an alternative methodology for detecting both sPrPSc and protease-resistant PrPSc that does not rely on protease digestion or immunodetection.
Keywords: prion protein, PrPSc, Thioflavin T, protease-sensitive, femtogram
Alternative folding of proteins features in many neurodegenerative diseases, including Parkinson's, Huntington's, and Alzheimer's, as well as the prion diseases (1–3). In several of these diseases, the alternatively folded proteins can adopt β-sheet-rich conformations that facilitate polymerization into amyloid fibers. In prion diseases, amyloid deposition has been observed in both animals and humans (4–6) but is a nonobligatory feature of the disease (7, 8). Recombinant prion proteins (recPrP) with sequences derived from various species and composed of full-length and truncated segments of the protein have been shown to form amyloid fibers (9–11).
In vitro, the kinetics of amyloid formation usually exhibits an initial lag phase, in which no detectable amyloid forms, whereas monomers nucleate to form fibers (12). During this initial phase, monomeric protein molecules adopt partially denatured conformations, which assemble into multimeric species in a process with slow kinetics (13). Once the solution is nucleated, there is a period of amyloid fiber growth that eventually plateaus when the soluble protein pool has been depleted. When added to a fresh pool of soluble protein, fragmented amyloid fibrils can shorten the lag phase and initiate rapid amyloid formation, a phenomenon known as seeding. Therefore, amyloid fiber generation can be initiated through nucleation, which is a slow process, or by seeding the reaction, which leads to amyloid formation after a brief incubation.
Amyloid formation can be monitored in solution by using the dye Thioflavin T (ThT) (14). This dye undergoes a fluorescence shift upon binding to amyloid fibers, from 342/430 to 442/482 nm excitation/emission maxima. Because ThT does not fluoresce significantly at excitation/emission maxima of 442/482 nm in the absence of amyloid fibers, the background signal tends to be quite low, so the dye is a highly sensitive reporter. When used in conjunction with multiwell plates and automated plate readers that record fluorescence over time, ThT offers a facile means of detecting conformational changes of proteins in solution. Using the ThT assay, we find that many prion strains are capable of seeding the polymerization of recPrP into amyloid, and we demonstrate that this seeding property can be used as an assay for the detection of prions in biological samples.
Results
Seeding Accelerates the Formation of Amyloid Fibrils.
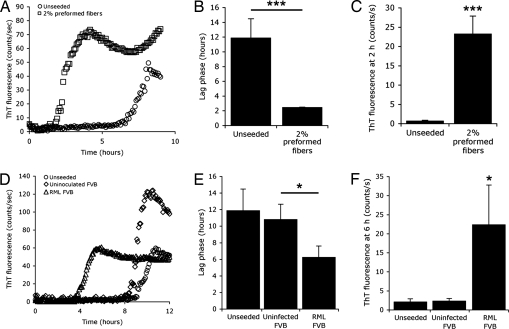
Purified recPrP consisting of mouse residues 89–230 [recMoPrP(89–230)] was diluted from a 5 mg/ml stock solution to 0.05 mg/ml in the presence of low concentrations of guanidine and PBS. Amyloid fibers readily formed as measured by ThT fluorescence (Fig. 1). When preformed fibers of recMoPrP(89–230) were added to the reaction, amyloid formed much more rapidly (Fig. 1A) as previously reported (9, 15). The mean lag phase for seeded reactions was 2 h, compared with 12 h for unseeded reactions (Fig. 1B). At 2 h, before unseeded amyloid-formation reactions have begun to polymerize, a substantial increase in the ThT fluorescence signal was observed in reactions seeded by fragments of fibers compared with control reactions (Fig. 1C). Under the conditions of these experiments, we observed high variability in both the lag phase and the maximum ThT fluorescence attained during fiber formation in the negative control samples. This variability required the testing of four to six replicates to obtain statistically significant results [see supporting information (SI) Fig. 6].
Fig. 1.
Preformed amyloid fibers and partially purified RML prions can seed amyloid formation of recMoPrP(89–230). (A and D) Typical traces depicting the kinetics of recMoPrP(89–230) amyloid formation in the presence of 0.4 M Gdn and PBS as monitored by ThT fluorescence over time. RecMoPrP(89–230) was incubated in solution with 2% preformed amyloid fibers (squares), RML prions precipitated by PTA from the brains of infected FVB mice (triangles), and PTA-precipitated brain homogenates from uninfected FVB mice (diamonds). As controls, no seed was added to the reactions (circles). (B and E) Seeding reactions with preformed fibrils (n = 4; ***, P < 0.001) or RML prions (n = 6; *, P < 0.05) significantly shortens the mean lag phase or period before ThT fluorescence rises above background. (C and F) ThT fluorescence signals at 2 h (C) and 6 h (F) of the reaction. Bars denote standard error (*, P < 0.05; ***, P < 0.001).
The presence of seeds in an amyloid-formation reaction was quantitatively detected either by observing a decrease in the mean lag phase of the reaction, compared with control samples, or an increase in the mean ThT signal during the period before negative control samples began to polymerize.
Prion-Infected Brain Homogenates Can Seed Amyloid Formation.
After the discovery that prions can be artificially created by polymerizing recPrP into amyloid (15), we asked whether prions from biological samples might be detected by their ability to seed amyloid formation in solutions of recPrP. In our initial studies, we found that brain homogenates prepared from rodents with experimental prion disease inhibited polymerization of recPrP into amyloid (data not shown). Additional experiments revealed that non-PrP impurities, such as serum albumin, tended to inhibit the reaction. Therefore, we partially purified prions from brain homogenates by using phosphotungstate (PTA) precipitation (16, 17), and then we used these partially purified prions to seed the amyloid polymerization reaction.
We purified RML prions from 500 μl of brain homogenate prepared from prion-infected FVB mice by using PTA in the presence of Sarkosyl. After washing the precipitate with the PTA/Sarkosyl solution, the pellet was resuspended in 150 μl of water, diluted to reduce the concentration of residual PTA and Sarkosyl, and added to an amyloid-formation reaction containing recMoPrP(89–230). The final concentration of the PTA-purified RML prions represented a 1:2,000 dilution of the resuspended pellet, corresponding to the equivalent of 0.017% brain homogenate, assuming no losses were sustained during the purification procedure. As a control, we performed the PTA precipitation protocol on brain homogenates of uninfected FVB mice.
We found that adding PTA-purified RML prions to the reaction accelerated amyloid formation, compared with controls (Fig. 1D). The mean lag phase for RML-seeded reactions was 6 h, a 50% reduction, compared with unseeded reactions (n = 6; P < 0.05) (Fig. 1E). Comparing ThT fluorescence for RML-seeded versus control reactions at 6 h, just before the control reactions began to form fibers, we found substantially greater signals from RML-seeded reactions (P < 0.05) (Fig. 1F).
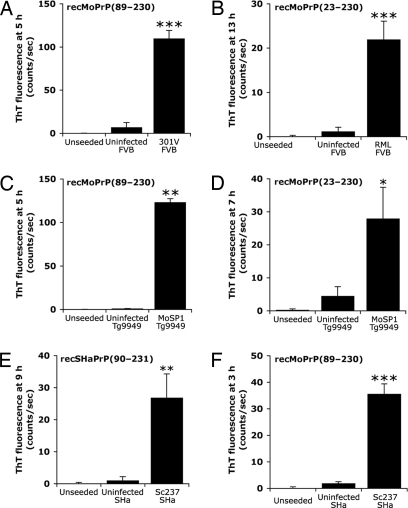
We next sought to determine whether the seeding effect observed with RML prions purified from infected FVB mice could be extended to other prion strains, including those isolated from Syrian hamsters, as well as to other recPrP substrates (Fig. 2). Using the PTA precipitation method, we partially purified 301V prions from FVB mice and MoSP1 prions from transgenic (Tg) mice overexpressing MoPrP(89–231), designated Tg9949 mice (18). The 301V strain was derived from bovine spongiform encephalopathy prions passaged through mice (19). MoSP1 results from serial passage of synthetic prions through Tg9949 mice (15, 20). Both 301V and MoSP1 efficiently seeded amyloid formation from recMoPrP(89–230) (Fig. 2 A and C). Using full-length recMoPrP(23–230) as the substrate for amyloid-formation reactions, we found that seeding was effective with both full-length RML prions from FVB mice (Fig. 2B) and truncated MoSP1 from Tg9949 mice (Fig. 2D). Seeds of purified Sc237 prions from infected Syrian hamsters (SHa) accelerated the formation of amyloid by using recombinant SHaPrP(90–231) (Fig. 2E). We tested the seeding reactions of nine other combinations of prion strains and recPrP substrates; all accelerated the formation of amyloid, compared with their respective controls (Table 1).
Fig. 2.
The amyloid seeding effect occurs with several different prion strains isolated from wild-type mice, Tg mice, and Syrian hamsters, and with several different recombinant PrP substrates. Each strain was purified by PTA precipitation of brain homogenates. The following prion strains successfully seeded amyloid formation from the following substrates: 301V for recMoPrP(89–230) (A); RML for full-length recMoPrP(23–230) (B); MoSP1 for both recMoPrP(89–230) (C) and full-length recMoPrP(23–230) (D); and Sc237 for both recSHaPrP(90–231) (E) and recMoPrP(89–230) (F). As controls, no seeds and uninfected, PTA-precipitated brain homogenates of the respective animals were added. For all experiments, seeding significantly reduced the lag phase compared with controls (n = 6; *, P < 0.01; **, P < 0.05; ***, P < 0.001). Bars denote standard error.
Table 1.
Prion strains used as seeds and recPrP substrates in amyloid-formation reactions
| Strain | Source* | recPrP Substrate | Result |
|---|---|---|---|
| RML | FVB | recMoPrP(89–230) | + |
| RML | FVB | recMoPrP(23–230) | + |
| RML | Tg9949 | recMoPrP(89–230) | + |
| RML | Tg9949 | recMoPrP(23–230) | + |
| RML | Tg4053 | recMoPrP(89–230) | + |
| RML | Tg4053 | recMoPrP(23–230) | + |
| MoSP1 | Tg9949 | recMoPrP(89–230) | + |
| MoSP1 | Tg9949 | recMoPrP(23–230) | + |
| 301V | FVB | recMoPrP(89–230) | + |
| 301V | FVB | recMoPrP(23–230) | + |
| Sc237 | SHa | recSHaPrP(90–231) | + |
| Sc237 | SHa | recMoPrP(89–230) | + |
| DY | SHa | recSHaPrP(90–231) | + |
| MoSP2 | Tg9949 | recMoPrP(89–230) | + |
| MoPrP,P101L | Tg(MoPrP,P101L)H | recMoPrP(89–230) | + |
| sCJD | Human | recHuPrP(90–231) | + |
*Source denotes the host animal from which prions were purified by PTA precipitation.
In many of the experiments in Fig. 2, we noted that reactions seeded with PTA-precipitated brain homogenates from uninoculated, control animals exhibited slightly higher ThT signals than their unseeded counterparts. Perplexed by this finding, we examined the effect that residual PTA and Sarkosyl may have on amyloid formation. We found that PTA is a strong inhibitor of amyloid formation, whereas Sarkosyl may inhibit or accelerate amyloid formation depending on its concentration (SI Fig. 7). Residual Sarkosyl from the precipitation procedure may be responsible for the slight increases in fluorescence observed in the negative control samples. Adding high concentrations of resuspended PTA pellets uniformly suppressed amyloid formation; this finding is likely because of the presence of PTA in the samples (SI Fig. 7A).
Promiscuous Seeding Occurs in the ASA.
We wanted to know whether the ASA would yield positive results only when the prions introduced arose from the same species as the recombinant protein used in the assay. Such species-specific seeding was reported for PrP amyloids of mouse and human sequences (13). We used the Sc237 prion strain isolated from Syrian hamsters by PTA precipitation to seed amyloid formation by using recMoPrP(89–230). To our surprise, we found that efficient seeding occurred (Fig. 2F).
Ultrasensitive Detection of Prions by Amyloid Seeding.
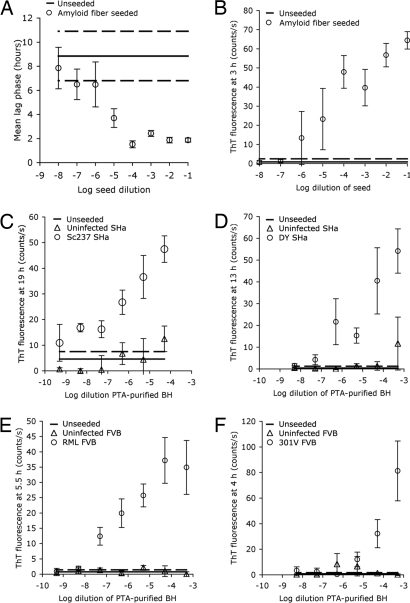
To determine the sensitivity of this assay, we used different concentrations of seeds in the amyloid-formation reactions (Fig. 3). By using preformed fibers of recMoPrP(89–230) as seeds, high concentrations of seed (10−1 to 10−4 dilutions) resulted in amyloid formation within 2.5 h. Diluting the seed to concentrations of <10−4 resulted in a gradual increase in lag phase until the lag phase became indistinguishable from unseeded reactions (Fig. 3A). However, we found that ThT fluorescence signals increased with higher seed concentrations in measurements made during early time periods (Fig. 3B). Background fluorescence from initial time point readings was subtracted to remove the seed's contribution.
Fig. 3.
Sensitivity of prion detection by amyloid seeding for preformed amyloid fibers or PTA-purified prions. (A) Preformed amyloid fibers were added to recMoPrP(89–230), and the mean lag phase for amyloid formation was measured. (B) Mean ThT fluorescence is shown as a function of the preformed seed concentration at 3 h. (C) Sc237 was added to recSHaPrP(90–231) in 10-fold serial dilutions, and the ThT fluorescence was measured at 19 h relative to controls. Total PrP in the Sc237 PTA pellet was calculated by ELISA; the 10−8 dilution contained ≈0.03 fg of PrP. (D–F) Titrations for DY prions by using recSHaPrP(90–231) as the substrate (D) and for RML prions (E) and 301V prions (F) by using recMoPrP(89–230) as substrates. For all panels, solid lines show unseeded control reactions. Bars and dashed line denote standard error.
Starting with a 1:20,000 dilution of Sc237 prions purified from infected Syrian hamster brains, we prepared up to six dilutions of seed, with each dilution 10-fold. These titrations of Sc237 prions were added to amyloid-formation reactions with recSHaPrP(90–231) as the substrate. As was the case for reactions seeded with preformed fibers, higher concentrations of Sc237 seed tended to increase the ThT fluorescence signal within the 19-h incubation period (Fig. 3C). The presence of Sc237 seeds accelerated amyloid formation; this acceleration was observed at seed concentrations as low as −8.3 log dilution. We quantified the total PrP in the resuspended Sc237 PTA pellet by capture ELISA relative to recombinant protein standards, and we found that the total PrP in the reactions with −8.3 log dilution of seed corresponds to ≈0.03 fg of PrP.
We also titrated hamster DY, mouse RML, and 301V prions (Fig. 3 D–F) and found that each strain had a different sensitivity limit for seeding reactions, with signals from DY detectable at −6.3 log dilution. RML was more readily detected than 301V, which only could be detected at levels above −4.3 log dilution.
ASA Detects Protease-Sensitive PrPSc (sPrPSc).
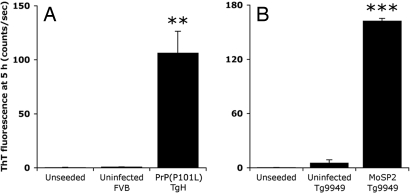
With many prion strains, PrPSc resists hydrolysis catalyzed by proteinase K (PK), whereas for other strains, PrPSc is sensitive to limited PK digestion. For example, prions arising spontaneously in Tg mice overexpressing the PrP(P101L) mutation, analogous to the mutation that causes one form of Gerstmann–Sträussler–Scheinker disease in humans, harbored infectivity when passaged to Tg(MoPrP,P101L)196 mice expressing lower levels of the same transgene (21). However, when brain homogenates from the Tg196 mice were digested with PK under standard conditions (20 μg/ml PK, 37°C, 1 h), no protease-resistant PrP was detected. By using the conformation-dependent immunoassay (CDI), sPrPSc was detected. Notably, these Tg196 mice displayed large numbers of PrP amyloid plaques in their brains (22). In humans with sporadic Creutzfeldt–Jakob disease (sCJD), as much as 90% of PrPSc was found to be protease-sensitive (17). Thus, we investigated whether the ASA could detect sPrPSc. Using PTA, we precipitated sPrPSc(P101L) from the brains of Tg(MoPrP,P101L)H mice expressing high levels of MoPrP(P101L) and added it to the amyloid-formation reactions. The sPrPSc(P101L) was able to seed amyloid formation even when the substrate consisted of the wild-type MoPrP sequence (Fig. 4A).
Fig. 4.
Two protease-sensitive prion strains, PrP(P101L) (A) and MoSP2 (B), are active in the amyloid seeding assay. PrP(P101L) and MoSP2 were purified by PTA precipitation of brain homogenates from Tg(MoPrP,P101L)H and Tg9949 mice, respectively. Both prion strains seeded amyloid formation of recMoPrP(89–230) within 5 h. Bars denote standard error (**, P < 0.01; ***, P < 0.001).
We also tested the ability of another protease-sensitive prion strain, MoSP2, to seed amyloid formation. MoSP2 prions were formed in the brains of Tg(MoPrP,89–231)9949 mice inoculated with unseeded recMoPrP(89–230) fibrils (20). It should be noted that MoSP2 differs from MoSP1, which arose from Tg9949 mice inoculated with seeded recMoPrP(89–230) fibers and exhibits protease resistance (15). When PTA-purified MoSP2 was added to amyloid-formation reactions, a substantial acceleration of the lag phase was observed (Fig. 4B and SI Fig. 8).
Detection of PrPSc in Human sCJD Samples.
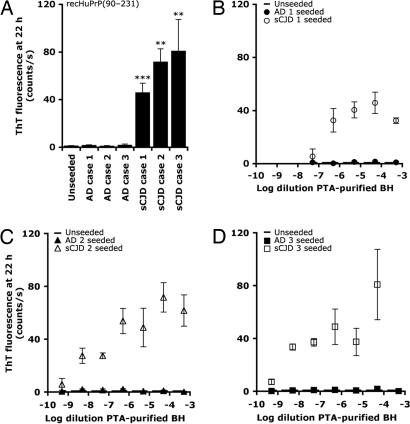
Next, we asked whether human prions in sCJD brain samples could be detected by the ASA (Fig. 5). Brain homogenates were prepared from postmortem tissue obtained from three individuals with sCJD. As a control, we prepared brain homogenates from individuals with Alzheimer's disease (AD). We applied the PTA-precipitation protocol used for mouse and hamster prions without modification. Using recHuPrP(90–231) as the substrate for the ASA, we found substantially higher ThT fluorescence at 22 h for sCJD samples, compared with AD samples or unseeded control (Fig. 5A). Reactions seeded with AD samples were indistinguishable from unseeded reactions.
Fig. 5.
The amyloid seeding assay can detect PrPSc precipitated from human sCJD brain homogenates. (A) ThT fluorescence at 22 h for three AD controls and three sCJD samples. For both AD and sCJD, resuspended PTA pellets were diluted 1:20,000. (B–D) Titrations of the resuspended PTA pellets to determine sensitivity (**, P <0.01; ***, P <0.001).
The resuspended PTA precipitates were titrated to determine the sensitivity of the ASA with human samples (Fig. 5 B–D). We observed a substantial seeding effect across four to six log dilutions of the samples, including those at −8.3 log dilution.
Discussion
We demonstrated that partially purified prions can act as seeds in amyloid-formation reactions and that this may be a useful method for detecting prions. The method robustly detected prions from every strain tested, including human sCJD, and was not restricted by the type of recPrP used. Statistically significant detection occurred at PrPSc levels as low as 0.03 fg for Sc237 prions; conservatively, the limit of detection for the ASA is at the femtogram level. The assay was even responsive to the presence of sPrPSc, which is otherwise difficult to detect. The absence of a seeding effect with AD samples (Fig. 5) indicates that the assay is specific for prions and not for amyloids in general that cause other neurodegenerative diseases. Therefore, this assay may find broad application in both the laboratory and clinic.
Among the many different methods for detecting prions, including the CDI (16), protein-misfolding cyclic amplification (PMCA) (23, 24), bioassays, ELISAs, and Western blots, this new assay offers some unique advantages (SI Table 2). Our ASA is able to detect protease-sensitive prions more readily than the CDI. Other techniques have been reported for the detection of sPrPSc after PTA precipitation but they are not currently in routine use and not as simple to apply as the ASA (25, 26). Bioassays, although extremely sensitive, require long incubation times (60–300 days) to yield results and, thus, are costly. The ASA returns results within 1 day.
Western blots and ELISAs both have limited sensitivities to PrPSc. Western blots rely on PK resistance, which precludes detection of sPrPSc, whereas ELISAs rely either on PK resistance or PrPSc-specific antibodies, which limit the sensitivity of the assays to the binding affinity of the antibody. Even when the most sensitive femtomolar-detection reagents are used, Western blots can detect, at best, 1,000-fold dilution of scrapie-infected brain homogenate. Because the ASA makes use of a seeding effect, the sensitivity is determined by the lowest concentration of seed that effectively accelerates amyloid formation. In this way, the amyloid-formation reaction is essentially an amplifier for the detected signal (ThT fluorescence). Use of the dye ThT also enhances the sensitivity of the assay because the background fluorescence at 442/482 nm (excitation/emission maxima) in the absence of amyloid is extremely low.
To amplify and detect prions, PMCA relies on sonication, which is inherently difficult to control. In our hands, PMCA results in a high percentage of false negatives and false positives, and it detects some strains more readily than others (data not shown). Others have recently shown that the PMCA reaction can spontaneously generate prions (27).
Given the inherent differences between brain samples obtained from humans and those obtained from laboratory animals, the detection of sCJD by the ASA attests to its robustness. With further optimization of the PTA precipitation step and ASA conditions, the limit of detection for human samples will likely be lower, raising the possibility that the assay could be applied to detect PrPSc in blood or other fluids where prions are scarce. The ASA may enable presymptomatic testing for sCJD diagnosis and offer a quantifiable biomarker to monitor during clinical trials.
We noted an absence of a species barrier in our amyloid seeding system (Fig. 2E). We previously showed that prion formation in transgenic mice that express both hamster and mouse PrP sequences is highly specific for the sequence that is homologous to the prion inoculum (7). At the time this work was undertaken, others reported the presence of species barriers in seeding amyloids by using recombinant amyloid fibers with sequences from humans and mice (13). More recently, promiscuous cross-seeding between mouse and hamster amyloids was reported (28). In a separate study (J. Stöhr, N. Weinmann, H. Wille, T. Kaimann, L. Nagel-Steger, E. Birkmann, G. Panza, S.B.P., M. Eigen, and D.R., unpublished data), it was shown that, under fibrillization conditions, the substrate PrP assumes a partially denatured structure and is in a monomer–dimer equilibrium with well defined contact sites between the molecules. The extent of denaturation and dimerization might determine in a sensitive manner whether substrate PrP addition to the PrPSc seed is specific for a particular structure or whether it only depends on general structural features of PrPSc. Although we do not understand how promiscuous cross-species seeding biophysically occurs in our system, it may prove to be a fortuitous discovery because it enables the detection of prions without necessitating the use of recombinant protein with the same sequence. We leveraged this advantage to detect sPrPSc(P101L) by using the wild-type recMoPrP(89–230) sequence (Fig. 4B).
We demonstrated that, under the conditions described, the assay is specific for sCJD relative to AD (Fig. 5). Whether this specificity arises from the PTA precipitation or the amyloid seeding step remains to be determined. We have not established whether adding Aβ amyloid fibers directly to our PrP amyloid-formation reactions results in seeding. Evidence indicates that all amyloids share some common structural motifs, including the fact that ThT is capable of binding >20 different types of amyloids (29). Notably, some antibodies have been reported to bind to β-sheet-rich motifs formed from several different proteins (30). Therefore, it is plausible that, under some conditions, cross-seeding could occur between Aβ and PrP.
The new ASA presented here provides a sensitive, rapid, and accurate means of detecting and characterizing prions. This assay should find broad applicability in the laboratory to supplement existing methods of identifying the presence of prions and differentiating prion strains, especially protease-sensitive strains. We demonstrated here that the ASA accurately detects prions from brain samples. With further refinement, the ASA also may find application for drug screening, as well as testing food and blood products for prions.
Materials and Methods
The production and purification of recombinant proteins are described in SI Materials and Methods.
Prion Strains.
The full-length RML prion strain propagated in Swiss mice was originally provided by W. Hadlow (Rocky Mountain Laboratory, Hamilton, MT) and was passaged in FVB mice obtained from Charles River Laboratories. MoSP1 prions were isolated from Tg(MoPrP,Δ23–88)9949/Prnp0/0 mice inoculated with seeded amyloid fibrils composed of recMoPrP(89–230) and then passaged through Tg9949 mice (20). MoSP2 was isolated from Tg9949 mice inoculated with unseeded amyloid fibrils composed of recMoPrP(89–230) (15). Tg(MoPrP,P101L)H mice are described elsewhere. The 301V prion strain was passaged through FVB mice (31). Sc237 prions were passaged intracerebrally through SHa as described (32). The DY prion strain was a gift from Richard Marsh (University of Wisconsin, Madison, WI) and was passaged through SHa (33).
Human sCJD and AD brain samples were obtained directly from donors. The disease status of each patient was evaluated by two neurologists at the University of California San Francisco Memory and Aging Center, which was confirmed postmortem by neuropathological analysis and, for sCJD cases, CDI testing of brain samples (17). All three AD cases had a clinical history of dementia. The three sCJD cases presented symptoms consistent with sCJD and expressed Met/Met at polymorphic PrP codon 129. Additional information about the human samples is provided in SI Table 3.
Preparation of Brain Homogenates.
To prepare 10% (wt/vol) brain homogenates, nine volumes of ice-cold PBS (supplemented with protease inhibitor mixture) (Roche) were added to brain tissue in a 50-ml tube. Brain tissue was homogenized on ice by using either needle extrusion through progressively smaller needles or bead beating (FastPrep FP120; Qbiogene). The sample was centrifuged at 500 × g for 5 min at room temperature (RT) to clarify samples. The supernatant was collected, the pellet was discarded, and aliquots were keep frozen at −80°C until use. Samples were thawed and diluted 1:1 with PBS/4% Sarkosyl to give 5% (wt/vol) brain homogenate.
PTA Precipitation.
PTA from a 10% (wt/vol) stock solution (pH 7.4) was added to brain homogenate to a final concentration of 0.5%. Typically, 500 μl of 10% brain homogenate was used. We incubated the sample with constant shaking (350 rpm) for 1 h at 37°C. After incubation, samples were centrifuged at 14,000 × g for 30 min at RT. We washed the pellet with 500 μl of PBS/2% Sarkosyl/protease inhibitor and then centrifuged the pellet again at 14,000 × g for 30 min at RT. We resuspended the pellet in 150 μl of water and then stored it at −80°C until use. For seeding experiments, 20 μl per well of a 1:200 dilution of the resuspended, PTA-precipitated pellet was added to reactions, which is equivalent to 0.017% brain homogenate.
Monitoring the Kinetics of in Vitro Amyloid Formation.
To monitor the formation of amyloid fibrils in our samples, we first dissolved lyophilized MoPrP(89–230) or MoPrP(23–230) in 6 M GdnHCl with a protein concentration of 5 mg/ml. The aliquot was kept frozen at −80°C until use. We thawed and diluted the stock protein solution to a final protein concentration of 50 μg/ml and incubated it in 1× PBS buffer (pH 7.0), 0.4 M GdnHCl, and 10 μM ThT in a reaction volume of 180 μl per well in 96-well plates (BD Falcon 353945; BD Biosciences). Because the protein is dissolved in 6 M Gdn, the final concentration of Gdn in the reaction is 0.46 M. One 3-mm glass bead (Fisher Scientific) was added to each well to increase agitation. For seeding experiments, resuspended PTA pellets were thawed for 10 min at RT and then vortexed. A volume of 2 μl of resuspended PTA pellet was diluted into 400 μl of water; 20 μl of the diluted sample was added to the well for a final volume of 200 μl. When indicated, 10-fold serial dilutions of this reaction mixture were prepared to evaluate the sensitivity of detection. Samples were normally run with six replicates because of variability in amyloid-formation kinetics. The 96-well plates were covered with sealing tape (235307; Fisher Scientific) and incubated at 37°C with continuous shaking on a plate reader (SpectraMax M2 or M5 fluorescence plate readers; Molecular Devices). The kinetics of fibril formation was monitored by top reading of fluorescence intensity every 5 min by using 444-nm excitation and 485-nm emission filters. We used two different instruments to collect the data presented here. All direct comparisons are between readings made on the same instrument. A step-by-step protocol and technical tips are included in SI Appendix 1.
Quantification of PrP in PTA Pellets.
For Sc237, a capture ELISA was performed to quantify PrP relative to a recombinant PrP ladder. The PTA pellet was denatured in 2 M Gdn for 2 h at room temperature. To D18-coated, capture ELISA plates (34), 10 μl of denatured PTA pellet and 90 μl of PBS/BSA were added to three wells, and the plate was incubated overnight at 4°C for capture. The plate was then washed three times with TBST, and 100 μl/well of the antibody-enzyme conjugate P-HRP was added. After a 1-h incubation at RT, the plate was washed seven times with TBST and developed with 100 μl of ABTS (KPLM). Absorbance was measured at 405 nm to determine soluble PrP in each well relative to the recombinant PrP ladder.
Supplementary Material
ACKNOWLEDGMENTS.
We thank the donors and their families for their contribution of the samples used in this study; Michael Geschwind, Bruce Miller, Stephen DeArmond, and Jiri Safar for the diagnosis of patients and characterization of sCJD and AD samples; the staff at the Hunters Point animal facility for providing brains from infected and noninfected animals; Ana Serban for providing recombinant proteins; and Holger Wille for helpful discussions of phosphotungstate precipitation. This work was supported by a Jane Coffin Childs Memorial Fund for Medical Resarch postdoctoral fellowship (to D.W.C.); a John Douglas French Alzheimer's Foundation postdoctoral fellowship (to Q.Z.); the G. Harold and Leila Y. Mathers Foundation; and National Institutes of Health Grants AG02132, AG10770, and AG021601 (to S.B.P.).
Footnotes
Conflict of interest statement: D.G., G.L., D.R., and S.B.P. have financial interest in InPro Biotechnology, Inc.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710152105/DC1.
References
- 1.Prusiner SB. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muchowski PJ. Neuron. 2002;35:9–12. doi: 10.1016/s0896-6273(02)00761-4. [DOI] [PubMed] [Google Scholar]
- 3.Ross CA, Poirier MA. Nat Med. 2004;10(Suppl):S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 4.Klatzo I, Gajdusek DC, Zigas V. Lab Invest. 1959;8:799–847. [PubMed] [Google Scholar]
- 5.Prusiner SB, McKinley MP, Bowman KA, Bolton DC, Bendheim PE, Groth DF, Glenner GG. Cell. 1983;35:349–358. doi: 10.1016/0092-8674(83)90168-x. [DOI] [PubMed] [Google Scholar]
- 6.DeArmond SJ, McKinley MP, Barry RA, Braunfeld MB, McColloch JR, Prusiner SB. Cell. 1985;41:221–235. doi: 10.1016/0092-8674(85)90076-5. [DOI] [PubMed] [Google Scholar]
- 7.Prusiner SB, Scott M, Foster D, Pan K-M, Groth D, Mirenda C, Torchia M, Yang S-L, Serban D, Carlson GA, et al. Cell. 1990;63:673–686. doi: 10.1016/0092-8674(90)90134-z. [DOI] [PubMed] [Google Scholar]
- 8.Wille H, Prusiner SB, Cohen FE. J Struct Biol. 2000;130:323–338. doi: 10.1006/jsbi.2000.4242. [DOI] [PubMed] [Google Scholar]
- 9.Baskakov IV, Legname G, Baldwin MA, Prusiner SB, Cohen FE. J Biol Chem. 2002;277:21140–21148. doi: 10.1074/jbc.M111402200. [DOI] [PubMed] [Google Scholar]
- 10.Bocharova OV, Breydo L, Parfenov AS, Salnikov VV, Baskakov IV. J Mol Biol. 2005;346:645–659. doi: 10.1016/j.jmb.2004.11.068. [DOI] [PubMed] [Google Scholar]
- 11.Leffers KW, Wille H, Stohr J, Junger E, Prusiner SB, Riesner D. Biol Chem. 2005;386:569–580. doi: 10.1515/BC.2005.067. [DOI] [PubMed] [Google Scholar]
- 12.Wetzel R. Acc Chem Res. 2006;39:671–679. doi: 10.1021/ar050069h. [DOI] [PubMed] [Google Scholar]
- 13.Baskakov IV. J Biol Chem. 2004;279:7671–7677. doi: 10.1074/jbc.M310594200. [DOI] [PubMed] [Google Scholar]
- 14.Rogers DR. Am J Clin Pathol. 1965;44:59–61. doi: 10.1093/ajcp/44.1.59. [DOI] [PubMed] [Google Scholar]
- 15.Legname G, Baskakov IV, Nguyen H-OB, Riesner D, Cohen FE, DeArmond SJ, Prusiner SB. Science. 2004;305:673–676. doi: 10.1126/science.1100195. [DOI] [PubMed] [Google Scholar]
- 16.Safar J, Wille H, Itri V, Groth D, Serban H, Torchia M, Cohen FE, Prusiner SB. Nat Med. 1998;4:1157–1165. doi: 10.1038/2654. [DOI] [PubMed] [Google Scholar]
- 17.Safar JG, Geschwind MD, Deering C, Didorenko S, Sattavat M, Sanchez H, Serban A, Vey M, Baron H, Giles K, et al. Proc Natl Acad Sci USA. 2005;102:3501–3506. doi: 10.1073/pnas.0409651102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Supattapone S, Muramoto T, Legname G, Mehlhorn I, Cohen FE, DeArmond SJ, Prusiner SB, Scott MR. J Virol. 2001;75:1408–1413. doi: 10.1128/JVI.75.3.1408-1413.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farquhar CF, Dornan J, Moore RC, Somerville RA, Tunstall AM, Hope J. J Gen Virol. 1996;77:1941–1946. doi: 10.1099/0022-1317-77-8-1941. [DOI] [PubMed] [Google Scholar]
- 20.Legname G, Nguyen H-OB, Baskakov IV, Cohen FE, DeArmond SJ, Prusiner SB. Proc Natl Acad Sci USA. 2005;102:2168–2173. doi: 10.1073/pnas.0409079102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsiao KK, Groth D, Scott M, Yang S-L, Serban H, Rapp D, Foster D, Torchia M, DeArmond SJ, Prusiner SB. Proc Natl Acad Sci USA. 1994;91:9126–9130. doi: 10.1073/pnas.91.19.9126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tremblay P, Ball HL, Kaneko K, Groth D, Hegde RS, Cohen FE, DeArmond SJ, Prusiner SB, Safar JG. J Virol. 2004;78:2088–2099. doi: 10.1128/JVI.78.4.2088-2099.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soto C, Anderes L, Suardi S, Cardone F, Castilla J, Frossard MJ, Peano S, Saa P, Limido L, Carbonatto M, et al. FEBS Lett. 2005;579:638–642. doi: 10.1016/j.febslet.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 24.Atarashi R, Moore RA, Sim VL, Hughson AG, Dorward DW, Onwubiko HA, Priola SA, Caughey B. Nat Methods. 2007;4:645–650. doi: 10.1038/nmeth1066. [DOI] [PubMed] [Google Scholar]
- 25.Birkmann E, Schafer O, Weinmann N, Dumpitak C, Beekes M, Jackman R, Thorne L, Riesner D. Biol Chem. 2006;387:95–102. doi: 10.1515/BC.2006.013. [DOI] [PubMed] [Google Scholar]
- 26.Birkmann E, Henke F, Weinmann N, Dumpitak C, Groschup M, Funke A, Willbold D, Riesner D. Vet Microbiol. 2007;123:294–304. doi: 10.1016/j.vetmic.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Deleault NR, Harris BT, Rees JR, Supattapone S. Proc Natl Acad Sci USA. 2007;104:9741–9746. doi: 10.1073/pnas.0702662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makarava N, Lee CI, Ostapchenko VG, Baskakov IV. J Biol Chem. 2007 doi: 10.1074/jbc.M704926200. in press. [DOI] [PubMed] [Google Scholar]
- 29.Chiti F, Webster P, Taddei N, Clark A, Stefani M, Ramponi G, Dobson CM. Proc Natl Acad Sci USA. 1999;96:3590–3594. doi: 10.1073/pnas.96.7.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 31.Farquhar CF, Dickinson AG. J Gen Virol. 1986;67:463–473. doi: 10.1099/0022-1317-67-3-463. [DOI] [PubMed] [Google Scholar]
- 32.Prusiner SB, Cochran SP, Alpers MP. J Infect Dis. 1985;152:971–978. doi: 10.1093/infdis/152.5.971. [DOI] [PubMed] [Google Scholar]
- 33.Bessen RA, Marsh RF. J Gen Virol. 1992;73:329–334. doi: 10.1099/0022-1317-73-2-329. [DOI] [PubMed] [Google Scholar]
- 34.Safar JG, Scott M, Monaghan J, Deering C, Didorenko S, Vergara J, Ball H, Legname G, Leclerc E, Solforosi L, et al. Nat Biotechnol. 2002;20:1147–1150. doi: 10.1038/nbt748. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.