Abstract
In human lung adenocarcinomas harboring EGFR mutations, a second-site point mutation that substitutes methionine for threonine at position 790 (T790M) is associated with approximately half of cases of acquired resistance to the EGFR kinase inhibitors, gefitinib and erlotinib. To identify other potential mechanisms that contribute to disease progression, we used array-based comparative genomic hybridization (aCGH) to compare genomic profiles of EGFR mutant tumors from untreated patients with those from patients with acquired resistance. Among three loci demonstrating recurrent copy number alterations (CNAs) specific to the acquired resistance set, one contained the MET proto-oncogene. Collectively, analysis of tumor samples from multiple independent patient cohorts revealed that MET was amplified in tumors from 9 of 43 (21%) patients with acquired resistance but in only two tumors from 62 untreated patients (3%) (P = 0.007, Fisher's Exact test). Among 10 resistant tumors from the nine patients with MET amplification, 4 also harbored the EGFRT790M mutation. We also found that an existing EGFR mutant lung adenocarcinoma cell line, NCI-H820, harbors MET amplification in addition to a drug-sensitive EGFR mutation and the T790M change. Growth inhibition studies demonstrate that these cells are resistant to both erlotinib and an irreversible EGFR inhibitor (CL-387,785) but sensitive to a multikinase inhibitor (XL880) with potent activity against MET. Taken together, these data suggest that MET amplification occurs independently of EGFRT790M mutations and that MET may be a clinically relevant therapeutic target for some patients with acquired resistance to gefitinib or erlotinib.
Keywords: lung adenocarcinoma, XL880
Somatic mutations in exons encoding the tyrosine kinase domain of the epidermal growth factor receptor (EGFR) are found in a proportion of lung adenocarcinomas (1). Nearly 90% of these mutations occur as either multinucleotide in-frame deletions in exon 19 that eliminate four amino acids (LREA), or as a single missense mutation that substitutes arginine for leucine at position 858 (L858R). Both genetic lesions are associated with increased sensitivity of lung adenocarcinomas to the selective EGFR kinase inhibitors, gefitinib (Iressa) and erlotinib (Tarceva) (2–4). Multiple prospective trials have demonstrated an ≈75% response rate for patients whose tumors harbor these mutations (5).
Unfortunately, lung cancers with drug-sensitive EGFR mutations that initially respond to gefitinib or erlotinib eventually develop acquired resistance (6, 7). In approximately half of cases, tumor cells obtained after disease progression contain a second-site mutation in the EGFR kinase domain (8–12). The most common (>90%) lesion involves a C → T change at nucleotide 2369 in exon 20, which substitutes methionine for threonine at position 790 (T790M). Other mechanisms that contribute to resistance to EGFR inhibitors, either in the absence or presence of the T790M mutation, remain to be established.
To determine whether lung cancers that acquire resistance to either gefitinib or erlotinib display additional and/or specific genetic alterations that might play a role in disease progression, we performed high-resolution genomic analysis (aCGH) of tissue samples from 12 patients whose tumors initially responded but subsequently progressed while on these drugs. We compared these results with those obtained from genomic analysis of lung adenocarcinomas with EGFR mutations resected from 38 patients who were never treated with kinase inhibitors. Among three genomic loci with recurrent differences in CNAs between the two groups, we focused on one that encompasses the gene encoding the MET tyrosine kinase. Using several molecular and cellular techniques, we verified the aCGH findings and then extended our studies to additional EGFR mutant tumors. We also examined the activity of MET protein in available EGFR mutant lung adenocarcinoma cell lines and studied drug responses in one cell line (NCI-H820) found to contain an EGFR drug-sensitive mutation (an exon 19 deletion), an EGFR drug-resistance mutation (T790M), and MET amplification.
Results
Characterization of the Cancer Genome in Lung Adenocarcinomas from Patients with Acquired Resistance to EGFR Kinase Inhibitors.
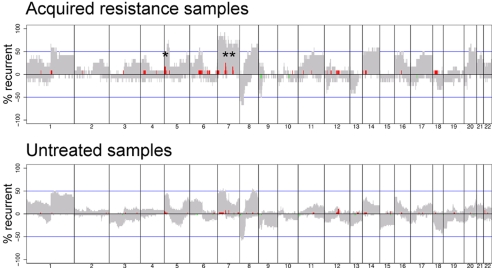
We obtained 12 tumor DNA samples from 12 patients with lung adenocarcinomas containing EGFR mutations and documented disease progression after prolonged treatment on gefitinib or erlotinib. We then subjected the DNAs to aCGH, using a 60-mer oligonucleotide array platform (Agilent). We analyzed fluorescence ratios of scanned images of the arrays to identify statistically significant changes in copy number using a version of the circular binary segmentation algorithm (13). The overall pattern of large-scale genomic events was consistent with previous high-resolution genomic profiles of human lung cancer (14, 15) (Fig. 1Upper).
Fig. 1.
Recurrence of chromosomal alterations found in EGFR mutant lung adenocarcinomas from patients with acquired resistance to EGFR tyrosine kinase inhibitors (n = 12) or from untreated patients (n = 38). Shown is the percentage of samples with CNAs after data segmentation (y axis) plotted for each probe evenly aligned along the x axis in chromosome order. The gray areas denote counts of chromosomal gain and loss defined by log2 ratios ±0.2. Amplifications or deletions having >2-fold change in copy number, defined by log2 ratios ±1.0, are shown by bright red and bright green lines, respectively. Asterisks denote amplifications that occurred in more than one sample in the acquired resistance cohort.
Specific Recurrent CNAs Identified in Tumor Samples from Patients with Acquired Resistance vs. Those from Untreated Resected EGFR Mutant Tumors.
We next compared results from tumors with acquired resistance to those obtained from a separate aCGH analysis of 38 mutant EGFR lung adenocarcinomas resected from patients who had never received treatment with kinase inhibitors. DNA from the untreated tumors was analyzed by using 44K Agilent chips (16). The recurrent genomic gains and losses in these samples appeared grossly similar to the acquired resistance set (Fig. 1 Lower).
After mode-centering, comparison of the two sets (at the location of each of the 44K probes; see Materials and Methods) revealed three major loci of recurrent CNAs unique to samples from patients with acquired resistance (Fig. 1 Upper and Table 1). One locus, at 7p11-12, includes EGFR and was amplified compared with the untreated set in 3 of the 12 tumor samples (nos. 5, 6, and 10a). These results are consistent with the notion that EGFR mutation and amplification occur frequently in tumors from patients with acquired resistance (11). A second locus, at an interval encompassing 7q31.2, was found in two samples (nos. 6 and 10a) [supporting information (SI) Fig. 4]. The gene encoding MET lies in this interval and encodes a receptor tyrosine kinase implicated in the development, maintenance, and progression of cancers in both animals and humans (reviewed in ref. 17). The third CNA occurred on 5p15.2–15.3 and was found in two samples (nos. 5 and 10a); candidate genes in this region remain to be identified (Table 1). We did not observe any genomic deletions that were significantly overrepresented in either treated or untreated groups.
Table 1.
Genomic loci with significant copy number changes in 12 EGFR mutant tumor samples from patients with acquired resistance compared with 38 EGFR mutant tumor samples from untreated patients
| Cytoband | MCR | CNA | Above threshold, no. | RefSeq genes, no. | Target gene |
|---|---|---|---|---|---|
| 5p15.2–15.3 | 8.2–14.6 | >8 | 2 | 7 | Unknown |
| 7p11–12 | 53.8–55.5 | >8 | 3 | 4 | EGFR |
| 7q31.2 | 114.8–116.4 | >12 | 2 | 6 | MET |
Data were obtained from aCGH chips as described in the Materials and Methods. Loci were listed if they displayed a log2 ratio >1, corresponding to a copy number >4 (P = 0.05, Fisher's exact test). MCR, minimal common region (Mb); CNA, maximum copy number alteration; RefSeq, reference sequence according to the National Center for Biotechnology Information.
Genomic Gains on Chromosome 7 in Tumor Cells from Patients with Acquired Resistance.
We next examined the individual aCGH profiles of the region of interest on chromosome 7 at higher resolution in all of the samples. Eleven of the samples showed broad gains of chromosome 7, including the region of EGFR (data not shown). Samples 5, 6, and 10a displayed further focal amplification at the locus encompassing EGFR, and samples 6 and 10a had additional focal amplifications at the locus, including MET (SI Fig. 4). None of these samples had focal amplification of the gene encoding hepatocyte growth factor (HGF), the ligand for MET, located at 7q21.1. An additional tumor sample from patient number 10 (10b, a metastatic lymph node) also displayed focal amplifications at both EGFR and MET.
To determine the proportion of drug-resistant tumor cells with amplified MET, we assessed MET gene copy number per cell by dual-color fluorescent in situ hybridization (FISH) in the one tumor sample (no. 6) for which we had sufficient viable tumor cells for analysis. Cells were labeled with probes that hybridized to the centromere of chromosome 7 (CEP7; green) or to MET (red). We found that the tumor sample comprised a mixed population of cells. All were polysomic for chromosome 7. However, although some cells displayed equal numbers of copies of CEP7 and MET (averaging 4–6 copies), others harbored greater numbers of copies of MET than CEP7 (SI Fig. 5). Taken together with the aCGH results on the same tumor sample, these data suggest that tumor cells in patient no. 6 have ≈4–6 copies of chromosome 7 with additional focal amplifications of the region containing MET in approximately half of the cells.
Confirmation of MET Status by Quantitative PCR and Analysis of Additional Patient Tumor Samples.
To confirm further the results from the aCGH studies, we next performed quantitative “real-time” PCR (qPCR) to determine the status of MET in DNA samples from four independent cohorts of tumor samples. As a control gene, we selected one [(MTHFR (5,10-methylenetetrahydrofolate reductase; located at 1p36.3)] from a genomic region that showed no CNA in any sample by aCGH and is not subject to germ-line copy number polymorphism.
In the first cohort—the tumor samples already analyzed by aCGH—quantitative PCR results confirmed the results (Table 2, nos. 1–12). We then tested four additional tumors from our own patients with acquired resistance (Table 2, nos. 13–16) and three additional drug-resistant tumors from Taiwan (Table 2, nos. 17–19). In total, in these tumor samples from 19 patients, four displayed MET amplification (e.g., a fold change relative to MTHFR > 1.5). Here, we chose a ratio of MET:MTHFR > 1.5 to define MET amplification based on corresponding data from FISH and aCGH analysis of tumor cells from patient no. 6 (SI Fig. 5) and of cell lines (see below and Table 2).
Table 2.
EGFR mutation and MET status of lung adenocarcinoma cell lines and tumor samples from patients with acquired resistance to EGFR inhibitors
| Patient | 1° EGFR mutation | T790M | MET F.C. | Drug | Duration, months |
|---|---|---|---|---|---|
| Ref. | NA | NA | 1.0 | NA | NA |
| H820 | Del E746-E749 | Y | 2.7 | NA | NA |
| PC-9 | Del E746-A750 | N | 0.8 | NA | NA |
| 1 | Del L747-E749; A750P | Y | not amp. by aCGH | erl. | 26 |
| 2 | Del L747-P753 | N | 1.8 | erl. | 22 |
| 3 | Del E746-A750 | N | 0.6* | gef. | 11 |
| 4 | Del E746-A750 | N | not amp. by aCGH | gef. | 9 |
| 5 | L858R | N | 0.8* | gef. | 17 |
| 6 | Del E746-A750 | N | 1.8* | erl. | 9 |
| 7 | Del E746-T751insA | N | 0.6* | erl. | 10 |
| 8 | Del L747-S752insQ | N | not amp. by aCGH | erl. | 32 |
| 9 | L858R | N | not amp. by aCGH | gef., erl. | >25 |
| 10a | Del L747-T751; K754E | Y | 1.8* | gef. | 11 |
| 10b | Del L747-T751; K754E | N | 3.5* | gef. | 11 |
| 11 | L858R | Y | not amp. by aCGH | gef. | 47 |
| 12 | L858R | N | not amp. by aCGH | gef. | 15 |
| 13 | L858R | Y | not amp. by aCGH | erl. | 17 |
| 14 | Del E746-A750 | Y | 0.5 | gef., erl. | 31 |
| 15 | Del E746-A750 | N | 0.8 | gef., erl. | 32 |
| 16 | Del E746-A750 | N | 0.5 | erl. | 24 |
| Ref. | NA | NA | 1.0 | NA | NA |
| H820 | Del E746-E749 | Y | 2.2 | NA | NA |
| 17 | Del E746-A750 | Y | 1.1 | erl. | 6 |
| 18 | Del E746-A750 | N | 1.1 | gef. | 5 |
| 19 | Del E746-A750 | N | 2.8 | gef., erl. | 27 |
EGFR mutation status was determined as described in the Materials and Methods; the absence or presence of the drug-resistance EGFRT790M mutation is indicated by a Y (yes) or N (no). For patient no. 10, two individual samples (10a and 10b) were examined. For MET fold change, values are given relative to MTHFR as assessed by qPCR, described in the Materials and Methods, with DNA from a reference sample (Ref.) and H820 cells included in each set. Samples with MET amplification are in boldface. None of the seven samples for which only aCGH was performed showed MET amplification [″not amplified (amp.) by aCGH″]. Samples for which both qPCR and aCGH were performed are marked with an asterisk. Tumor samples 6, 10a, and 10b all showed focal MET amplification by aCGH (see SI Fig. 4) and by qPCR. Tumor no. 6 also displayed MET amplification by FISH (see text and SI Fig. 5). F.C., fold change; NA, not applicable; Del, deletion; ins; insertion; erl., erlotinib; gef., gefitinib. Time in months patient was on kinase inhibitor treatment when re-biopsy was performed.
To extend these findings to another independent cohort, we performed qPCR analysis on DNA from EGFR mutant tumors obtained from 24 Taiwanese patients with acquired resistance to gefitinib (SI Table 3, nos. 20–43). Matched pretreatment tumor DNA samples were available for comparison. We detected MET amplification in tumors from five patients. In four of these samples, MET amplification was found in only posttreatment samples, suggesting that selection for cells with MET amplification occurred while these patients were on gefitinib. Note that here, we used a more stringent criterion for MET amplification (MET:MTHFR ratio > 5) because the samples were tested with an independent protocol (see Materials and Methods and SI Table 3 legend) for which concurrent aCGH or FISH data were unavailable.
In two patients (SI Table 3, nos. 30 and 32), MET amplification was detected in tumor samples obtained before treatment. In one of these patients (no. 32), the untreated sample was a surgically resected lung tumor. Disease recurred more than three years later, and the posttreatment specimen was derived from omentum 15 months after an initial response to gefitinib. The other patient (no. 30) was diagnosed with a CT-guided lung biopsy and had a confirmed partial radiographic response on gefitinib. The “acquired resistance” specimen was obtained when pleural fluid developed 8 months after starting gefitinib. These results could be due to genetic heterogeneity within individual tumors and/or tumor heterogeneity within individual patients. The observed amplification of MET in some tumors after treatment with TKIs could be attributed to selection of subpopulations of cells with MET amplification.
When data (using qPCR, aCGH, and/or FISH) from the multiple independent cohorts were combined, we found MET amplification in 9 of 43 patients with acquired resistance, compared with 2 of 62 untreated patients (P = 0.007, Fisher's Exact test). The common EGFRT790M resistance mutation was found in 20 of 43 (46.5%) patients with acquired resistance. Interestingly, 4 of the 10 tumor samples (from nine patients) with MET amplification harbored the EGFRT790M mutation as well. Thus, tumors with acquired resistance to gefitinib or erlotinib may exhibit amplification of MET in the absence or presence of a second-site drug-resistant mutation in EGFR. There was no correlation between increased copies of MET and type of primary drug-sensitive EGFR mutation (exon 19 deletion vs. exon 21 point mutation) or duration of drug treatment (data not shown).
An Established Lung Adenocarcinoma Cell Line Contains an Exon 19 Deletion Associated with Drug-Sensitivity, an Exon 20 Point Mutation Associated with Drug-Resistance, and Increased Copies of MET.
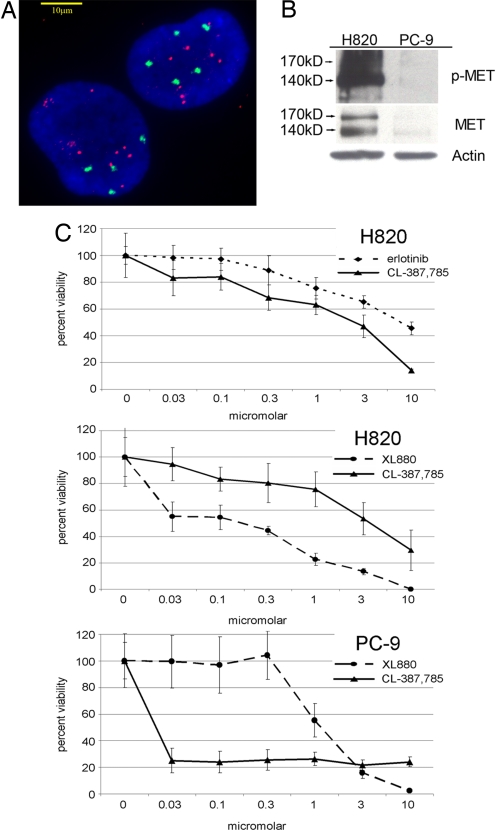
We performed qPCR analysis of MET in lung adenocarcinoma cell lines with EGFR mutations. Surprisingly, we found that one cell line—H820—contained not only drug-sensitive (del E746-E749) and drug-resistant (T790M) EGFR kinase domain mutations (data not shown) but also MET amplification (Table 2). Consistent with these results, we found by using FISH that the majority of H820 cells harbored 4–6 copies of chromosome 7 (CEP7) and 7–9 copies of MET (Fig. 2A). As judged by aCGH, large regions of chromosome 7 were amplified to varying degrees but no specific focal amplifications were observed at loci containing either EGFR or MET (data not shown).
Fig. 2.
MET status and sensitivity of H820 cells to the MET inhibitor, XL880. (A) H820 cells display, as assessed by dual-color FISH analysis (see Materials and Methods), ≈4–6 copies of chromosome 7 (CEP7), with additional focal amplifications at loci containing MET. The cells shown are representative of the whole population. Scale bar: 10-μm. CEP7 probe, green; MET probe, red; nuclei, blue (DAPI). (B) Lysates from H820 and PC-9 cells were immunoblotted by using anti-phospho-(p)-MET (Y1234/5), anti-total MET, and anti-actin antibodies. H820 cells harbor an exon 19 deletion (E746-E749), a T790M mutation, and amplified no MET, whereas PC-9 cells harbor an exon 19 deletion (E746-A750), no T790M mutation, and no MET amplification. (C) Growth inhibition curves of H820 and PC-9 cells treated with MET and EGFR inhibitors at various concentrations. Error bars indicate one standard deviation from six replicates.
To determine whether the high number of copies of MET caused increased MET enzymatic activity, we used a surrogate kinase assay in which immunoblots of H820 cell lysates were probed with polyclonal antibodies that recognize MET phosphorylated at tyrosine residues (Y1234/5). These phosphorylation sites are located in the activation loop and are indicative of kinase activation (Fig. 2B). Total MET protein was measured by immunoblotting with an anti-MET monoclonal antibody. Both antibodies recognize the 170-kDa precursor form of the enzyme and the mature 140-kDa beta subunit. The smaller protein is the product of proteolytic cleavage of the larger precursor and also contains the tyrosine kinase catalytic domain (18). We compared results with H820 cells to those with PC-9 cells, a lung cancer line that harbors an exon 19 deletion (E746-A750) of EGFR but no T790M mutation and no MET amplification (Table 2). Extracts from PC-9 cells contained relatively small amounts of total and phosphorylated MET protein. By contrast, H820 cell extracts displayed much greater amounts of MET protein, much of which was phosphorylated at Y1234/5 (Fig. 2B).
Sensitivity of H820 Cells to a MET Inhibitor but Not to EGFR Inhibitors.
To explore further the functional consequences of MET amplification in EGFR mutant tumor cells, we used fluorescence-based growth inhibition assays to measure the sensitivity of H820 cells to erlotinib, CL-387,785 [an irreversible EGFR inhibitor known to overcome T790M-mediated resistance (10, 19, 20)], and XL880. The third compound abolishes MET and VEGFR2 tyrosine kinase activity at subnanomolar levels and also inhibits the PDGFRβ, KIT, FLT3, TIE-2, and RON kinases in vitro with low nM potency (21, 22).
H820 cells were relatively insensitive to erlotinib, because the concentration of drug required to inhibit the growth of 50% of the cells (IC50) was ≈10 micromolar (Fig. 2C Top). The IC50 of CL-387,785 was also in the micromolar range (Fig. 2C Top and Middle). By contrast, H820 cells displayed sensitivity to XL880 in the nanomolar range (Fig. 2C Middle). At these concentrations of XL880, the kinase activity of MET was inhibited, as measured by examination of H820 cell lysates from XL880-treated cells using the surrogate kinase assays described above (Fig. 3Left). By comparison, PC-9 cells were insensitive to XL880 but were much more sensitive to CL-387,785 (Fig. 2C Bottom). These results suggest that MET inhibitors like XL880 may be able to overcome MET-mediated resistance to EGFR kinase inhibitors, even in cells that harbor the T790M mutation.
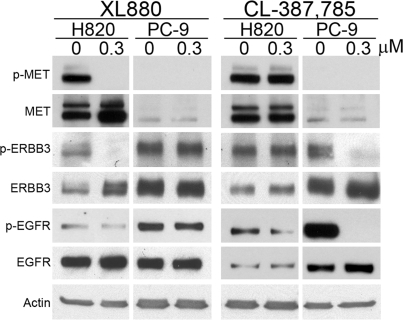
Fig. 3.
ERBB3 signaling in H820 cells depends on MET status. Lysates from H820 and PC-9 cells, either untreated (DMSO-only control; 0 μM) or treated with 0.3 μM XL880 or 0.3 μM CL-387,785, were immunoblotted by using anti-phospho-(p)-MET (Y1234/5), anti-total MET, anti-phospho-(p)-ERBB3 (Y1289), anti-total ERBB3, anti-phospho-(p)-EGFR (Y1092), anti-total EGFR, and anti-actin antibodies.
Dependence of ERBB3 Signaling on MET in H820 Cells.
During the course of the work described here, others reported that MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling (23). ERBB3 is a heterodimeric partner for EGFR that mediates phosphoinositide 3-kinase activity in gefitinib-sensitive lung cancer cell lines (24). We found that XL880 treatment strongly inhibited the phosphorylation of ERBB3 in H820 cells but not in PC-9 cells. Conversely, treatment with CL-387,785 strongly inhibited phosphorylation of ERBB3 in PC-9 cells but not in H820 cells (Fig. 3).
To determine whether the inhibition of ERBB3 phosphorylation in H820 cells treated with XL880 was due specifically to inhibition of the MET kinase, we transfected MET-specific siRNAs into H820 and PC-9 cells to knockdown expression of the protein. After transfection with MET siRNAs, both phospho-MET and total-MET were virtually undetectable by immunoblotting of H820 cell extracts (SI Fig. 6). In concordance with the results obtained with XL880, phospho-ERBB3 levels were also reduced in H820 cells after treatment with the MET siRNAs. By contrast, we observed no effect on phospho-ERBB3 status in similarly treated PC-9 cells and no effect on MET or ERBB3 status in cells treated with siRNAs against a control gene (GAPD) (SI Fig. 6). Thus, ERBB3 signaling appears to depend on MET protein in H820 cells, even though these cells harbor EGFR mutations.
We also measured the effect of siRNA knockdown of MET on the growth of H820 cells. In multiple experiments, the number of viable cells remaining 72 h after treatment with MET siRNAs was reduced, whereas treatment with GAPD siRNA had no effect. However, the effect of MET siRNA was modest (between 80 and 90% of controls in both total cell count and growth inhibition assays; data not shown), less than that seen with the kinase inhibitor, XL880. Thus, in H820 cells, XL880 may inhibit other kinases in addition to MET that affect cell viability. Alternatively, the effect of kinase inhibition by XL880 is different from the effect of siRNA-mediated knockdown of MET, because the latter probably diminishes MET activity more slowly.
Tumor Samples with MET Amplification Lack MET Mutations.
Gain-of-function mutations of MET have been discovered in both sporadic and inherited forms of human renal papillary carcinomas (25–27). The majority of mutations are located in exons that encode the kinase domain of the receptor (17). In the samples with MET amplification and adequate DNA for analysis (samples 2, 6, 10a, 10b, and H820 cells), we sequenced coding regions for the MET kinase domain (exons 15–21) and did not find any somatic mutations (data not shown). In addition, we did not identify any somatic mutations outside the MET kinase domain (exons 3–14) in H820 cells (data not shown).
Discussion
Mutations that substitute methionine for threonine at position 790 in the EGFR kinase domain have been found in ≈50% of lung adenocarcinomas from patients with acquired resistance to the EGFR inhibitors, gefitinib and erlotinib (refs. 8–12 and this article). This knowledge has led to the identification of alternative EGFR inhibitors that can overcome T790M-mediated resistance in vitro and potentially in patients (19, 20). However, other mechanisms could additionally contribute to disease progression in these patients.
In this study, we used high-resolution genome-wide profiling of EGFR mutant tumor samples before and after treatment to implicate the MET proto-oncogene as an additional therapeutic target in patients with acquired resistance to gefitinib or erlotinib. MET encodes a heterodimeric transmembrane receptor tyrosine kinase composed of an extracellular alpha-chain disulfide-bonded to a membrane spanning beta-chain (18, 28). Binding of the receptor to its ligand, hepatocyte growth factor/scatter factor, induces receptor dimerization, triggering conformational changes that activate MET tyrosine kinase activity. MET activation can have profound effects on cell growth, survival, motility, invasion, and angiogenesis (17). Dysregulation of MET signaling has been shown to contribute to tumorigenesis in a number of malignancies. For example, activating mutations have been associated with both sporadic and inherited forms of human papillary renal carcinomas (25–27). In addition, gastric carcinomas have high-level MET amplification (29), and some other cancers display aberrant transcriptional up-regulation of MET (30).
In our studies of various EGFR mutant lung adenocarcinoma samples, we found MET to be amplified in 9 of 43 (21%) patients with acquired resistance vs. 2 of 62 (3%) patients unexposed to EGFR kinase inhibitors. In a separate genomic analysis of 371 primary lung adenocarcinoma samples and 242 matched normal controls, MET amplification was not identified as a significant recurrent focal event (31). Thus, although MET amplification can be found in lung cancers (32, 33), it does appear to be a rare event in lung adenocarcinomas never treated with EGFR kinase inhibitors.
The presence of MET amplification in combination with gain-of-function drug-sensitive EGFR mutations could together lead to cellular changes that confer enhanced fitness to cells bearing both alterations. The EGFR T790M resistance mutation could further potentiate the growth properties of such tumor cells, because the oncogenic activity of EGFR kinase mutant alleles is enhanced by the T790M change (34). Consistent with this notion, 40% of the samples with MET amplification in this study harbored the T790M mutation. Furthermore, we found that an existing lung adenocarcinoma cell line—H820 cells—harbored an EGFR mutation associated with drug-sensitivity (E746-E749), an EGFR mutation associated with drug-resistance (T790M), and MET amplification. Notably, these cells were isolated from a patient who did not undergo any prior treatment with gefitinib or erlotinib. Thus, these cells may not represent “acquired resistance” per se; however, their existence does demonstrate that all these genetic lesions can occur within the same cells.
MET amplification could lead to EGFR inhibitor resistance by activating ERBB3 signaling (23). Our data using XL880, a small molecule that inhibits MET kinase activity, and siRNAs that knockdown MET expression, suggest that in H820 cells, ERBB3 signaling depends highly on MET and not EGFR activity. This interaction between EGFR, MET, and ERBB3 in H820 cells appears to be different from that observed in an EGFR mutant lung adenocarcinoma cell line (HCC827 GR) selected for gefitinib-resistance in vitro (23). In those resistant cells, treatment with a MET inhibitor alone did not affect ERBB3 phosphorylation. Such discrepancies may be explained by the highly different ways in which the cell lines were derived.
Recently, small-molecule MET inhibitors have shown promise as anti-cancer therapy in phase I trials (22). Our in vitro data demonstrate that the MET inhibitor, XL880, is more effective at inhibiting the viability of lung adenocarcinoma cells with EGFRT790M and MET amplification than either reversible (erlotinib) or irreversible (CL-387,785) EGFR inhibitors. Collectively, these findings suggest that compounds like XL880 could play a significant role in the treatment of patients whose EGFR mutant lung adenocarcinomas have developed acquired resistance to existing EGFR inhibitors as a result of increased copy numbers of MET.
Materials and Methods
Tissue Procurement.
Tumor specimens were obtained through protocols approved by the Institutional Review Boards of Memorial Sloan–Kettering Cancer Center, Aichi Cancer Center Central Hospital, New York University Medical Center, Chang-Gung Memorial Hospital, and National Taiwan University Hospital. All patients gave informed consent.
Mutational Analyses.
Genomic DNA was extracted from tumor specimens, and primers for EGFR (exons 18–24) analyses were as published in refs. 4 and 35. PCR-RFLP assays for exon 19 deletions and L858R and T790M missense mutations were performed as published in refs. 9 and 36. All mutations were confirmed at least twice from independent PCR isolates, and sequence tracings were reviewed in the forward and reverse directions by visual inspection. Primers for MET sequencing are listed in SI Table 4.
aCGH Profiling.
Genomic DNA was extracted from tumor samples, using standard techniques. Normal genomic DNA (Promega) was used as a reference for all samples. DNA was digested and labeled by random priming using Bioprime reagents (Invitrogen) and Cy3- or Cy5-dUTP. Labeled DNA was hybridized to Agilent 244K CGH arrays for acquired resistance samples, and 44K CGH arrays for untreated samples. For additional details, see SI Methods.
Quantitative Real-Time PCR.
See SI Methods for details.
Cell Lines and Viability Assays.
NCI-H820 cells were developed by A. Gazdar (University of Texas Southwestern Medical Center). PC-9 cells were a gift from M. Ono (Kyushu University, Fukuoka, Japan). Cells were grown in RPMI supplemented with FBS. Growth inhibition assays were performed with the CellTiter-Blue cell viability kit (Promega), per the manufacturer's instructions. All assays with H820 cells were performed at least three independent times; for PC-9 cells, all assays were performed at least two independent times (see SI Methods). The complete aCGH dataset is available at http://cbio.mskcc.org/Public/Bean_etal_PNAS_2007.
Nomenclature.
Two numbering systems are used for EGFR. The first denotes the initiating methionine in the signal sequence as amino acid −24. The second, used here, denotes the methionine as amino acid +1. There are also two mRNA transcripts for MET that differ by an in-frame deletion of 54 nt (28, 37); because the isoform lacking these 54 nt appears to be the most abundant isoform (38), we consider full-length MET protein to consist of 1,390 (not 1,408) aa.
Immunoblotting.
See methods and supporting text in ref. 4 for details on cell lysis and immunoblotting reagents. At least three independent experiments were performed for all analyses. See SI Methods for a list of the antibodies used.
FISH.
FISH was performed according to established protocols (see SI Methods).
siRNA.
siGenome ON-TARGETplus SMARTpool MET (L-003156) and ON-TARGETplus siCONTROL GAPD pool (D-001830) (Dharmacon) were used according to the manufacturer's instructions. All siRNA transfections were performed three independent times (see SI Methods).
Supplementary Material
ACKNOWLEDGMENTS.
We thank K. Politi, C. Sawyers, and H.E.V. for helpful discussions; T. Mitsudomi (Aichi Cancer Center Hospital, Nagoya, Japan) for providing DNA samples; Z. Zeng and M. Weiser (both of Memorial Sloan–Kettering Cancer Center) for providing the MET BAC probe for FISH; Exelixis for providing XL880; and all of the patients and/or their family members who consented for tissue acquisition. This work was supported by University of Texas Specialized Programs of Research Excellence in Lung Cancer National Cancer Institute Grants P50CA75907 (to A.G.), U01-CA84999 (to W.G.), and R21-CA115051 (to V.M.); Taiwan National Science Council Grants NSC95-2314-B-002-113-MY3 (to J.-Y.S.) and NSC95-2314-B-002-227-MY3 (to C.H.Y.); Chang-Gung Medical Research Fund Grant CMRPG 350031 (to W.-C.C.); National Health Research Institutes Grant 96-A1-MG-PP-04-014 (to S.F.H.); the Doris Duke Charitable Foundation (W.P.), the Thomas G. Labrecque Foundation (W.G. and W.P.), Joan's Legacy: The Joan Scarangello Foundation to Conquer Lung Cancer (W.P.); National Insitutes of Health Grants K08-CA097980 and R01-CA121210 (to W.P.); the Jodi Spiegel Fisher Cancer Foundation (W.P.); the Carmel Hill Fund (W.P.); and funds from the Miner Family.
Note.
During the course of the work described here, others published similar findings after identifying MET as a candidate resistance gene by using an in vitro resistance modeling approach (23). In that study, MET amplification was detected in 4 of 18 (22%) lung cancers that had become resistant to gefitinib or erlotinib. Two of the samples examined here—nos. 10a and 10b—were also analyzed in that study (i.e., patient 12); results were mostly concordant.
Footnotes
Conflict of interest statement: V.M. and W.P. are part of a pending patent application on EGFR T790M mutations.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710370104/DC1.
References
- 1.Pao W, Miller VA. J Clin Oncol. 2005;23:2556–2568. doi: 10.1200/JCO.2005.07.799. [DOI] [PubMed] [Google Scholar]
- 2.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 3.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 4.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, et al. Proc Natl Acad Sci USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riely GJ, Politi KA, Miller VA, Pao W. Clin Cancer Res. 2006;12:7232–7241. doi: 10.1158/1078-0432.CCR-06-0658. [DOI] [PubMed] [Google Scholar]
- 6.Riely GJ, Pao W, Pham D, Li AR, Rizvi N, Venkatraman ES, Zakowski MF, Kris MG, Ladanyi M, Miller VA. Clin Cancer Res. 2006;12:839–844. doi: 10.1158/1078-0432.CCR-05-1846. [DOI] [PubMed] [Google Scholar]
- 7.Jackman DM, Yeap BY, Sequist LV, Lindeman N, Holmes AJ, Joshi VA, Bell DW, Huberman MS, Halmos B, Rabin MS, et al. Clin Cancer Res. 2006;12:3908–3914. doi: 10.1158/1078-0432.CCR-06-0462. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, Halmos B. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 9.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwak EL, Sordella R, Bell DW, Godin-Heymann N, Okimoto RA, Brannigan BW, Harris PL, Driscoll DR, Fidias P, Lynch TJ, et al. Proc Natl Acad Sci USA. 2005;102:7665–7670. doi: 10.1073/pnas.0502860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balak MN, Gong Y, Riely GJ, Somwar R, Li AR, Zakowski MF, Chiang A, Yang G, Ouerfelli O, Kris MG, et al. Clin Cancer Res. 2006;12:6494–6501. doi: 10.1158/1078-0432.CCR-06-1570. [DOI] [PubMed] [Google Scholar]
- 12.Kosaka T, Yatabe Y, Endoh H, Yoshida K, Hida T, Tsuboi M, Tada H, Kuwano H, Mitsudomi T. Clin Cancer Res. 2006;12:5764–5769. doi: 10.1158/1078-0432.CCR-06-0714. [DOI] [PubMed] [Google Scholar]
- 13.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Biostatistics (Oxford) 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 14.Tonon G, Wong KK, Maulik G, Brennan C, Feng B, Zhang Y, Khatry DB, Protopopov A, You MJ, Aguirre AJ, et al. Proc Natl Acad Sci USA. 2005;102:9625–9630. doi: 10.1073/pnas.0504126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao X, Weir B. A., LaFramboise T, Lin M, Beroukhim R, Garraway L, Beheshti J, Lee JC, Naoki K, Richards WG, et al. Cancer Res. 2005;65:5561–5570. doi: 10.1158/0008-5472.CAN-04-4603. [DOI] [PubMed] [Google Scholar]
- 16.Broderick SR, Chitale D, Motoi N, Gong Y, Pao W, Venkatraman E, Rusch V, Brennan C, Gerald W, Ladanyi M. J Clin Oncol. 2007;25:7686. [Google Scholar]
- 17.Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Nat Rev Mol Cell Biol. 2003;4:915–925. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- 18.Giordano S, Ponzetto C, Di Renzo MF, Cooper CS, Comoglio PM. Nature. 1989;339:155–156. doi: 10.1038/339155a0. [DOI] [PubMed] [Google Scholar]
- 19.Carter TA, Wodicka LM, Shah NP, Velasco AM, Fabian MA, Treiber DK, Milanov ZV, Atteridge CE, Biggs WH, Edeen PT, et al. Proc Natl Acad Sci USA. 2005;102:11011–11016. doi: 10.1073/pnas.0504952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi S, Ji H, Yuza Y, Meyerson M, Wong KK, Tenen DG, Halmos B. Cancer Res. 2005;65:7096–7101. doi: 10.1158/0008-5472.CAN-05-1346. [DOI] [PubMed] [Google Scholar]
- 21.Eder JP, Heath E, Appleman L, Shapiro G, Wang D, Malburg L, Zhu AX, Leader T, Wolanski A, LoRusso P. J Clin Oncol. 2007;25:3526. [Google Scholar]
- 22.LoRusso P, Appleman L, Heath E, Malburg L, Zhu AX, Carey M, Shapiro GI, Eder JP. Eur J Cancer Suppl. 2006;4:141. [Google Scholar]
- 23.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, et al. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 24.Engelman JA, Janne PA, Mermel C, Pearlberg J, Mokuhara T, Fleet C, Cichowski K, Johnson BE, Cantley LC. Proc Natl Acad Sci USA. 2005;102:3788–3793. doi: 10.1073/pnas.0409773102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeffers M, Schmidt L, Nakaigawa N, Webb CP, Weirich G, Kishida T, Zbar B, Vande Woude GF. Proc Natl Acad Sci USA. 1997;94:11445–11450. doi: 10.1073/pnas.94.21.11445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt L, Duh FM, Chen F, Kishida T, Glenn G, Choyke P, Scherer SW, Zhuang Z, Lubensky I, Dean M, et al. Nat Genet. 1997;16:68–73. doi: 10.1038/ng0597-68. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt L, Junker K, Nakaigawa N, Kinjerski T, Weirich G, Miller M, Lubensky I, Neumann HP, Brauch H, Decker J, et al. Oncogene. 1999;18:2343–2350. doi: 10.1038/sj.onc.1202547. [DOI] [PubMed] [Google Scholar]
- 28.Park M, Dean M, Kaul K, Braun MJ, Gonda MA, Vande Woude G. Proc Natl Acad Sci USA. 1987;84:6379–6383. doi: 10.1073/pnas.84.18.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smolen GA, Sordella R, Muir B, Mohapatra G, Barmettler A, Archibald H, Kim WJ, Okimoto RA, Bell DW, Sgroi DC, et al. Proc Natl Acad Sci USA. 2006;103:2316–2321. doi: 10.1073/pnas.0508776103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsuda M, Davis IJ, Argani P, Shukla N, McGill GG, Nagai M, Saito T, Lae M, Fisher DE, Ladanyi M. Cancer Res. 2007;67:919–929. doi: 10.1158/0008-5472.CAN-06-2855. [DOI] [PubMed] [Google Scholar]
- 31.Weir BA, Woo MS, Getz G, Perner S, Ding L., Beroukhim R, Lin WM, Province MA, Kraja A, Johnson L, et al. Nature. 2007 10.1038. [Google Scholar]
- 32.Lutterbach B, Zeng Q, Davis LJ, Hatch H, Hang G, Kohl NE, Gibbs JB, Pan BS. Cancer Res. 2007;67:2081–2088. doi: 10.1158/0008-5472.CAN-06-3495. [DOI] [PubMed] [Google Scholar]
- 33.Choong NW, Ma PC, Salgia R. Exp Opin Ther Targets. 2005;9:533–559. doi: 10.1517/14728222.9.3.533. [DOI] [PubMed] [Google Scholar]
- 34.Godin-Heymann N, Bryant I, Rivera MN, Ulkus L, Bell DW, Riese DJ, II, Settleman J, Haber DA. Cancer Res. 2007;67:7319–7326. doi: 10.1158/0008-5472.CAN-06-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shih JY, Gow CH, Yu CJ, Yang CH, Chang YL, Tsai MF, Hsu YC, Chen KY, Su WP, Yang PC. Int J Cancer. 2006;118:963–969. doi: 10.1002/ijc.21458. [DOI] [PubMed] [Google Scholar]
- 36.Pan Q, Pao W, Ladanyi M. J Mol Diagn. 2005;7:396–403. doi: 10.1016/S1525-1578(10)60569-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodrigues GA, Naujokas MA, Park M. Mol Cell Biol. 1991;11:2962–2970. doi: 10.1128/mcb.11.6.2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.