Abstract
Although viruses of each of the 16 influenza A HA subtypes are potential human pathogens, only viruses of the H1, H2, and H3 subtype are known to have been successfully established in humans. H2 influenza viruses have been absent from human circulation since 1968, and as such they pose a substantial human pandemic risk. In this report, we isolate and characterize genetically similar avian/swine virus reassortant H2N3 influenza A viruses isolated from diseased swine from two farms in the United States. These viruses contained leucine at position 226 of the H2 protein, which has been associated with increased binding affinity to the mammalian α2,6Gal-linked sialic acid virus receptor. Correspondingly, the H2N3 viruses were able to cause disease in experimentally infected swine and mice without prior adaptation. In addition, the swine H2N3 virus was infectious and highly transmissible in swine and ferrets. Taken together, these findings suggest that the H2N3 virus has undergone some adaptation to the mammalian host and that their spread should be very closely monitored.
Keywords: avian, reassortant, interspecies transmission
The generation of a pandemic influenza virus first requires interspecies transmission, and the virus must then genetically adapt to the new host species (1, 2) via either point mutations (antigenic drift) or reassortment (antigenic shift); the latter is the exchange of gene segments between two different influenza viruses. Molecular epidemiology suggests that the 1918 Spanish flu pandemic was caused by a wholly avian H1N1 influenza virus that was introduced into humans (3). The 1957 (H2N2) and 1968 (H3N2) pandemic viruses were generated through genetic reassortment of human and avian strains acquiring the neuraminidase (NA) and/or HA and the polymerase basic 1 (PB1) gene from an avian virus and other genes from the previously circulating human virus (4). The reassortment may have occurred either in infected humans or in an intermediate host, e.g., swine or quail (5, 6), before human infection. Swine are referred to as a “mixing vessel” because of their susceptibility to both human and avian influenza viruses (7, 8). Therefore, reassortment of avian and mammalian influenza viruses in this intermediate host may produce new viruses that are transmissible to humans.
H2N2 influenza virus has not circulated in the human population for the past 40 years and is currently detected only in avian species (9–11). There are two distinct lineages of avian H2 influenza viruses. The Eurasian lineage is genetically more similar to human H2 viruses (12) than the American lineage. Nevertheless, some H2 viruses isolated from North American shorebirds carry HA of the Eurasian lineage, suggesting interregional transmission of the H2 gene (13). H2 subtypes are presently circulating in birds, especially migratory birds. Here we describe the isolation and characterization of H2N3 influenza A viruses from pigs with respiratory disease from two farms in the United States, a subtype not previously reported in swine. These H2N3 reassortant viruses contain genes derived from avian and swine influenza viruses. We also investigated the pathogenicity and transmissibility of the H2N3 isolates in different mammalian hosts. The H2N3 virus was able to replicate in pigs, mice, and ferrets and was transmitted among pigs and ferrets. Serologic evidence suggests that the virus continues to circulate in the affected swine production systems.
Results
Analysis of Clinical Samples.
In September 2006, the influenza virus A/Swine/Missouri/4296424/2006 (Sw/4296424) was isolated from several 5- to 6-week-old pigs with multifocal bronchopneumonia at a multisourced commercial swine nursery. Lung lesions included moderate, subacute to chronic, purulent bronchopneumonia and interstitial pneumonia with bronchiolitis and peribronchitis. Lung tissue was negative for porcine reproductive and respiratory syndrome virus (PRRSV), porcine circovirus type 2 (PCV2), and Mycoplasma hyopneumoniae but was positive for Streptococcus suis. Because of the characteristic influenza-like lesions and clinical signs of pneumonia, lung tissue homogenate was inoculated on Madin–Darby canine kidney (MDCK) cells. Cytopathic effects were detected on day 3 postinoculation (p.i.). The influenza virus nucleoprotein (NP) gene was detected in the infected cells by RT-PCR. The virus did not react with reference swine anti-sera (A/Sw/IA/1973 H1N1, A/Sw/TX/1998 H3N2, A/Sw/NC/2001 H1N1) in hemagglutination inhibition (HI) assays, and multiplex RT-PCR detected no H1N1 or H3N2 genes (14). The virus was submitted to the National Animal Disease Center (NADC) in February 2007 for subtyping and sequencing.
After the September isolate had been subtyped and sequenced (described below), a search of case records revealed that another “untypable” influenza isolate had been submitted in April 2006. A/Swine/Missouri/2124514/2006 (Sw/2124514) had been isolated from a 12-week-old pig with respiratory disease at another commercial grower–finisher swine farm. Lung lesions were histopathologically characteristic of swine influenza (severe, subacute inflammation of alveoli and bronchi with bronchiolar epithelial cell necrosis and metaplasia). The lung was negative for PRRSV, PCV2, and M. hyopneumonia but was positive for influenza A virus by RT-PCR (specific for the NP gene) and S. suis. The virus was submitted to the NADC in March 2007 for subtyping and sequencing.
Subtyping and Phylogenetic Analysis.
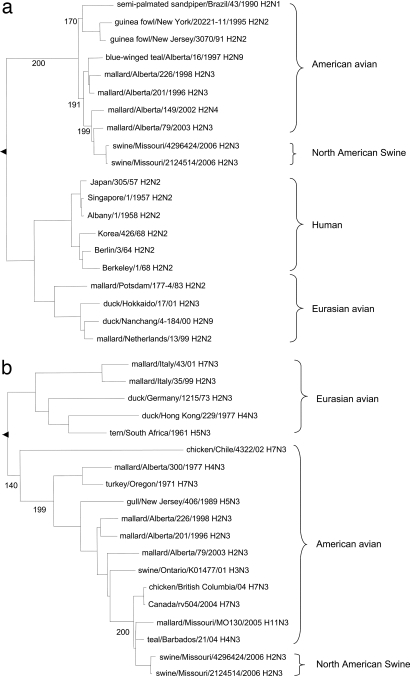
To identify and characterize both influenza viruses, nucleic acid sequencing and molecular and phylogenetic analysis were conducted. Both viruses were directly sequenced from low-passage isolates by using MDCK cells, and the sequences were confirmed after plaque purification and resequencing. They were identified as H2N3 viruses by nucleotide sequence and a BLAST search of the Influenza Sequence Database (www.flu.lanl.gov). The HA gene segment of Sw/4296424 most closely matched those of H2 viruses isolated from mallards in North America [up to 97.8% nucleotide sequence identity; supporting information (SI) Table 4]. Its NA segment was closely related to that of an H4N3 avian influenza virus (AIV) isolated from blue-winged teal (98.3% identity). With the exception of the polymerase acidic (PA) gene, its internal genes were derived from contemporary triple-reassortant swine influenza viruses currently found in the United States. These viruses carry internal genes from human (PB1), avian (PB2, PA), and swine [NP, matrix (M), nonstructural (NS)] influenza virus origin (SI Table 4). Its PA segment was 99.2% identical to that of the H6N5 AIV isolated from mallard ducks (SI Table 4). The Sw/2124514 and Sw/4296424 viruses showed 99.3–99.9% total nucleotide sequence identity (SI Table 5). Both isolates were repeatedly plaque cloned, retested, and confirmed by sequencing to belong to the H2N3 subtype. The H2N3 subtype was serologically confirmed by hemagglutination inhibition and neuraminidase inhibition assays. Phylogenetic analysis based on the HA and NA genes showed that these two viruses belong to the American avian lineage that is distinct from the Eurasian avian strains and the H2N2 viruses isolated from humans after the 1957 influenza pandemic (Fig. 1).
Fig. 1.
Phylogenetic trees of selected influenza virus H2 (a) and N3 (b) genes based on the nucleotide sequences of the ORFs. Horizontal distance is proportional to genetic distance. The trees are rooted to A/duck/Singapore/97 H5N3 (a) and A/tern/Astrakan/775/83 H3N3 (b). Numbers below nodes represent bootstrap values from 200 replicates.
Molecular Analysis of the HA and NA Surface Proteins.
Influenza A viruses contain two surface proteins: the HA is the receptor-binding and membrane-fusion glycoprotein, and the NA is a receptor-destroying enzyme. The viral HA is a critical factor of host species specificity of influenza viruses (15). To characterize residues within HA that may be associated with adaptation of an avian virus to the mammalian host, we compared the amino acid sequences of swine HAs with those of the putative reference avian viruses. Molecular comparison of the HA molecules of the two swine H2N3 isolates revealed that they differ from the putative reference H2N3 virus isolated from mallards by six common amino acid substitutions (D36N, Q226L, T274I, V316I, L419I, and L506V) (SI Table 6). The substitution Q226L was found in both swine H2N3 isolates, whereas position 228 contained G, identical to the avian consensus sequence (Table 1) (16). In contrast, human HA molecules of H2 subtype contain 226L and 228S, whereas early human H2 isolates contain 226L and 228G (Table 1), similar to the swine isolates. Positions 36N, 274I, 316I, and 419I are unique to the two swine H2N3 isolates (SI Table 6), whereas the respective positions in human and avian isolates depicted in Fig. 1a are 36D, 274T, 316V, and 419L. For the influenza isolates depicted in Fig. 1a, position 506V is conserved among human, two swine H2N3 isolates, and the avian isolates, except for A/mallard/Alberta/2004 (H2N3) as shown in SI Table 6. Two common amino acid changes in the NA amino acid sequence of both swine isolates were found when compared with the reference H4N3 virus isolated from blue-winged teal: H47Y and H253Y (SI Table 7). The position 47Y in both swine H2N3 isolates is the same as the respective amino acid in Eurasian avian isolates depicted in Fig. 1b; conversely, the position in North American avian isolates is 47H. The position 253Y is unique to the swine H2N3 isolates, and the position 253H is conserved in Eurasian and North American avian isolates depicted in Fig. 1b. Interestingly, Sw/4296424 (H2N3), isolated 5 months later than Sw/2124514 (H2N3), had two additional substitutions (P162S and L321V) in the HA molecule, and had three additional substitutions (V30I, I49T, and A135T) in the NA molecule when compared with the HA and NA of Sw/2124514 (SI Tables 6 and 7). The position 30I (Sw/4296424) in the NA molecule is similar to Eurasian isolates, whereas the position 30V (Sw/2124514) is conserved in Northern American avian isolates.
Table 1.
Comparison of amino acids in HA receptor-binding site of human, avian, and swine H2 influenza virus isolates
| Virus strains | HA receptor-binding residues |
|||||
|---|---|---|---|---|---|---|
| 138/148 | 190/200 | 194/204 | 225/235 | 226/236 | 228/238 | |
| Avian consensus | A | E | L | G | Q | G |
| Mallard/2003/H2N3 | A | E | L | G | Q | G |
| Sw/4296424 | A | E | L | G | L | G |
| Sw/2124514 | A | E | L | G | L | G |
| Human consensus | A | E | L | G | L | G/S |
| Davis/1/57 | A | E | L | G | L | G |
| Albany/7/57 | A | E | L | G | L | G |
| RI/5+/57 | A | E | L | G | L | S |
| Albany/6/58 | A | E | L | G | L | S |
| Ohio/2/59 | A | E | L | G | L | S |
| Berlin/3/64 | A | E | L | G | L | S |
Under HA receptor-binding residues, the H3 numbering system is used for numbers before the slash, and the H2 numbering system is used for numbers after the slash. The Davis/1/57 and Albany/7/57 viruses were isolated earlier in the human H2 pandemic when compared to the other H2 human viruses listed here.
Pathogenicity and Transmissibility of H2N3 Swine Influenza Viruses in Pigs.
To investigate the extent of swine adaptation of the H2N3 virus, we investigated its pathogenicity in this host by inoculating 20 4-week-old pigs with 2 × 106 50% tissue culture infective dose (TCID50) of the Sw/4296424 virus. Only one H2N3 virus was chosen, because of the high identity between the two isolates. Twelve control pigs were mock-inoculated with noninfectious cell culture supernatant. We assessed transmissibility by cohousing 10 age-matched contact pigs with the inoculated pigs, starting on day 3 p.i. All pigs used for the study were seronegative at day 0 for antibodies against swine influenza H1N1, H1N2, H2N3, and H3N2 viruses by HI assay. Five inoculated pigs and three control pigs were euthanized for necropsy on days 3, 5, and 7 p.i. The 10 contact pigs and 5 virus-inoculated pigs were serologically tested by HI assay with H2N3 on day 24 after contact or day 27 p.i., respectively. No acute respiratory signs were observed. Necropsy revealed severe macroscopic lung lesions (plum-colored, consolidated areas) in inoculated pigs but revealed none in control pigs (Table 2). The histopathologic score (0–3) expressing the extent of damage to lung architecture was >2 in inoculated pigs (Table 2). Lungs from inoculated pigs euthanized on day 3, 5, or 7 p.i. exhibited mild to moderate interstitial pneumonia and acute to subacute necrotizing bronchiolitis with slight lymphocytic cuffing of bronchioles and vessels (Fig. 2). Virus was titrated in bronchoalveolar lavage fluid (BALF) and isolated from nasal swab samples. Virus titers in the lung ranged from 104.3 to 106.5 TCID50/ml on days 3 and 5 p.i. (SI Table 8) and were negative on day 7 p.i. In the H2N3 inoculated group, virus was isolated from nasal swab samples in 25% (5 of 20) of pigs on day 3, 67% (10 of 15) on day 5, and 20% (2 of 10) on day 7 p.i.; in the contact group, 10% (1 of 10) of samples were positive on days 5 and 7 after contact. In contrast, 100% (10 of 10) of the contact pigs were seropositive after 24 days of contact with inoculated pigs (SI Table 9). Some control pigs had an occasional small focus of mild interstitial pneumonia (Table 2), but they were negative for swine influenza virus infection. All pigs were negative for PRRSV and M. hyopneumoniae by PCR. Our results indicate that the H2N3 virus is pathogenic in pigs and is transmissible among pigs.
Table 2.
Macroscopic and microscopic pneumonia in pigs inoculated with H2N3 virus Sw/4296424 or mock-inoculated
| Group | Lung lesion score, % | Histopathologic score (0–3) |
|---|---|---|
| H2N3 day 3 | 27.57 ± 7.36 | 2.23 ± 0.23 |
| Control day 3 | 0.00 ± 0.00 | 0.33 ± 0.09 |
| H2N3 day 5 | 21.86 ± 2.90 | 2.37 ± 0.11 |
| Control day 5 | 0.00 ± 0.00 | 0.56 ± 0.06 |
| H2N3 day 7 | 21.57 ± 5.02 | 2.07 ± 0.25 |
| Control day 7 | 0.00 ± 0.00 | 0.22 ± 0.11 |
Values are the mean ± SEM.
Fig. 2.
Microscopic lung sections from control and infected pigs. (a) Bronchiole in the lung of a control pig inoculated with noninfectious cell culture supernatant. Note the regular outline of the pseudostratified columnar epithelium. (b) Necrotizing bronchiolitis in the lung of a pig 3 days after inoculation with H2N3 swine influenza virus. The epithelial lining of the airway is focally disrupted by sloughing of necrotic infected cells and early reactive proliferation of the remaining epithelium. The lumen contains sloughed epithelial cells and mixed leukocytes. A small number of lymphocytes are seen infiltrating subepithelial and peribronchiolar connective tissue.
Pathogenicity of H2N3 Swine Influenza Viruses in Mice.
To test the ability of the H2N3 Sw/4296424 virus to replicate in mice, we inoculated 6- to 7-week-old BALB/c mice intranasally with 102-106 TCID50. Mice inoculated with 104 TCID50 or more showed signs of disease (e.g., labored breathing, rough fur, weight loss, and lethargy) (SI Table 10). Seventy-five percent of mice that received 106 TCID50 died, but there were no deaths at lower doses. Viral RNA was detected by real-time RT-PCR (17) in the lungs of mice after inoculation with 106 or 105 TCID50 (SI Table 10). Histopathologically, the H2N3 virus induced multiple or coalescing foci of interstitial pneumonia and proliferative alveolitis characterized by prominent pneumocyte hypertrophy and infiltration of alveolar walls with a mixed population of macrophages, lymphocytes, and neutrophils (SI Fig. 3). Some alveolar lumens contained fibrin clots and light mixed leukocytic exudates. Taken together, these findings indicate that H2N3 is pathogenic in mice without previous adaptation.
Transmissibility of H2N3 Swine Influenza Virus in Ferrets.
To cause a pandemic, an emergent influenza A virus must infect humans and be efficiently transmitted among humans. To investigate the potential of the reassortant H2N3 virus to transmit in mammalian systems, we used the ferret contact model (18). Three 18-week-old ferrets, housed in separate cages, were inoculated with 102.5 TCID50 of the H2N3 virus Sw/2124514. After 24 h, one contact animal was placed in each cage. Nasal washes were taken on days 1, 4, and 7 p.i., and virus was titrated in embryonated eggs. Virus was detected in all inoculated and contact ferrets, but none showed obvious clinical signs (Table 3). These results indicate that the H2N3 influenza virus infected ferrets and was transmitted via contact efficiently.
Table 3.
Virus titers in nasal washes from H2N3 (Sw/2124514)-inoculated and contact ferrets
| Animal | Virus titer, EID50/ml |
HI titer |
|||
|---|---|---|---|---|---|
| Day 1 | Day 4 | Day 7 | Day 21 | Day 28 | |
| Inoculated no. 333 | 104.8 | 105.5 | <101.0 | 1:160 | 1:160 |
| Contact no. 362 | <101.0 | 106.3 | 106.5 | 1:320 | 1:320 |
| Inoculated no. 366 | 106.0 | 105.5 | <101.0 | 1:320 | 1:160 |
| Contact no. 334 | <10 | 105.8 | 107.0 | 1:320 | 1:320 |
| Inoculated no. 368 | 105.8 | 104.5 | <101.0 | 1:160 | 1:320 |
| Contact no. 364 | <101.0 | 106.8 | 106.3 | 1:320 | 1:320 |
Serological Investigation of H2N3 Swine Influenza Viruses in Outbreak Farms.
To further investigate the spread of the H2N3 viruses, we conducted a limited serological survey of animals associated with the two affected production systems. In the first study, in spring 2007, serum samples were taken from sows from four farms that provided piglets to the nursery farms during the September 2006 outbreak. Ninety percent (54 of 60) were seropositive for the presence of antibodies to Sw/4296424 (SI Table 11). A number of the tested animals were present at the time of the index case, and it is unclear whether they were infected at that time or whether they were infected subsequently. The data do, however, show that the virus was present at both sow and nursery farms and that the virus efficiently transmitted between animals. All sows in this operation had antibody titers >1:40 to H1N1 and H3N2 swine influenza viruses, because they had been previously vaccinated with a bivalent H1N1 and H3N2 killed-influenza vaccine.
Serum samples were also collected in spring 2007 from 30 sows and 90 weaned pigs associated with the April 2006 outbreak, and they were tested for the presence of antibodies to Sw/2124514 by using the HI assay. Of the 30 sows and 90 weaned pigs sampled, 1 of 30 and 26 of 90 were seropositive (SI Table 11), respectively.
Discussion
In this report we characterized reassortant H2N3 viruses isolated from pigs in the United States. Molecular and phylogenetic analysis revealed that the HA, NA, and PA gene segments are similar to those of AIVs of the American lineage, whereas other gene segments are similar to those of contemporary swine influenza viruses that are triple-reassortant viruses containing human, avian, and swine influenza virus genes. In addition to their potential impact on animal health, these H2N3 viruses have intrinsic properties that make them of considerable concern to public health. These properties include the following: (i) they belong to the H2 subtype as did the 1957 human pandemic strain that disappeared in 1968 (hence, individuals born subsequent to 1968 have little preexisiting immunity to this subtype); (ii) they are circulating in swine, a host shown experimentally to select for mammalian virus traits (7); (iii) they have receptor binding site changes associated with increased affinity for α2,6Gal-linked sialic acid viral receptors; and (iv) they are able to replicate and transmit in swine and ferrets via contact. The latter two points suggest that the swine H2N3 viruses have undergone adaptation to the mammalian host and as such have the ability for sustained transmission. Reinforcing this possibility is the finding in one of the production systems that young pigs born at least 6 months after the index case were seropositive for the virus. Although it is not clear whether the seropositivity in the young animals was due to infection or maternal antibodies, these data suggest that the virus continues to circulate within the affected production systems. The fact that the H2N3 viruses are known to have infected two independent swine production systems and that serologic studies suggest they continue circulating is in contrast to other reports of a wholly AIV infection in North American swine, in which infections have appeared to be self-limiting (19, 20).
Although the genetic factors that are associated with successful zoonotic transmission of influenza viruses remain largely unknown, receptor-binding properties are likely involved. In avian H2 and H3 influenza viruses, HA receptor-binding-site residues corresponding to codon positions 138, 190, 194, 225, 226, and 228 (using the H3 numbering system) are highly conserved (16). In human H2 and H3 viruses, leucine and serine substitutions at residues 226 and 228, respectively, have been shown to accompany their adaptation from avian to human hosts (21). For example, changing the human virus H3 residue L226 to Q226 dramatically changes the receptor-binding specificity of the virus from mammalian to avian virus-like (22). The substitution Q226L was found in both new swine H2N3 isolates, whereas position 228 retains G, which is typical of AIVs (Table 1). This same 226L/228G combination was observed in the first viruses of the 1957 H2N2 human pandemic. The later pandemic strains contained 228S after full adaptation to humans (23). In addition to the genetic signatures, biologic evidence also suggests that the H2N3 viruses have undergone adaptation to mammalian hosts as evidenced by replication in mice, swine, and ferrets, with efficient transmission via contact in the latter two. Strong support for the importance of receptor-binding changes on transmissibility of influenza viruses has come from studies of the 1918 H1N1 pandemic strain. Investigators using genetically reconstructed virus were able to show that substitution of only two amino acids in the receptor-binding site of this virus was enough to abolish transmission among ferrets (24). Although the evidence that receptor-binding changes are required for the successful adaptation of avian viruses in humans is strong, it should also be noted that this trait on its own is not sufficient. Classical swine H1N1 influenza viruses, for example, have a preference for α2,6Gal-linked sialic acid viral receptors (7). Despite this preference and a number of self-limiting human infections, these viruses have not successfully established in the human population, suggesting that other host range barriers exist, reducing the transmission of swine influenza viruses between humans. An examination of the other viral proteins shows that a number of substitutions are present when H2N3 isolates were compared with influenza viruses isolated from swine or mallard ducks, although their significance is unknown (SI Tables 12–17).
Although the original source of the H2N3 virus is unclear, both farms use surface water collected in ponds for cleaning barns and watering animals. Considering the swine were housed in barns that prevented intrusion of birds, especially waterfowl, it seems probable the avian virus was introduced into the animals via water, a mode of transmission that has been described before (19, 20, 25). As more reports emerge linking the use of untreated pond water to the transmission of AIVs to swine, the risks associated with this practice must be fully evaluated. The appearance of the two highly identical H2N3 viruses in both production systems is unexplained because there is no known relationship between the farms in terms of sharing of equipment, of common feed or water source, or of the movement of animals, workers, or veterinarians.
Pigs are purported to be a mixing vessel for avian and human influenza viruses because their tracheal epithelial cells carry receptors for both human and avian influenza viruses (7). Supporting this theory is the documentation of genetic reassortment between avian- and human-like influenza viruses in Italian pigs (8). In this light, pigs have often been implicated in the emergence of human pandemic strains. More recent evidence has, however, shown that similar receptor expression is also available in the human and quail host (6, 26), and the direct evidence that human pandemic viruses are generated in swine is ambiguous. Nevertheless, our results provide further evidence for the potential of swine to promote reassortment between different influenza viruses, and the genetic and biologic properties of the H2N3 viruses described suggest that it would be prudent to establish vigilant surveillance in pigs and in workers who have occupational exposure.
Materials and Methods
Analysis of Clinical Samples.
In April 2006, an outbreak of respiratory disease occurred in pigs at a commercial grower–finisher swine farm. At necropsy, the attending veterinarian observed gross lesions of pneumonia and submitted formalin-fixed and unfixed sections of lung tissue to the Minnesota Veterinary Diagnostic Laboratory (MVDL). At the MVDL, the formalin-fixed tissue was routinely processed for histopathology. Bronchial swab samples from the unfixed lung tissue were suspended in 2 ml of PBS and tested for M. hyopneumoniae by PCR (27). Unfixed sections (≈5 g) of lung were cultured aerobically for bacteria by inoculation on MacConkey, colistin–nalidixic acid, brilliant green, and blood agar plates with and without nicotinamide adenosine dinucleotide (NAD) factor (Staphylococcus epidermidis nurse colonies). In parallel, unfixed sections of lung (≈10 g) were homogenized in Eagle's minimal essential medium (MEM) containing 4% BSA, 15 μg/100 ml trypsin, and an antibiotic mixture of neomycin, gentamicin, penicillin, streptomycin, and amphotericine B and were cultivated on MDCK cells. RNA and DNA were also isolated from the homogenate for diagnostic tests for influenza virus nucleoprotein (RT-PCR), PRRSV ORF 6 (RT-PCR), and PCV2 ORF 2 (PCR).
For virus isolation, 10% lung homogenates were centrifuged for 10 min at 640 × g. The supernatant was passed through 0.45-μm filters to remove any bacterial contamination and was inoculated on monolayers of MDCK cells in 24-well plates. The MDCK cells were maintained in Eagle's MEM containing 1 μg/ml TCPK-trypsin and 0.3% bovine albumin. The plates were incubated at 37°C in a CO2 incubator and were observed daily. After cytopathic effects were observed, infected cells were lysed by freezing and thawing, and virus was serotyped by HI assays with turkey erythrocytes.
In September 2006, another outbreak of respiratory disease occurred in 5- to 6-week-old pigs at a different multisourced commercial swine nursery. Again, gross lesions were consistent with pneumonia, and lung tissues were submitted to the MVDL for testing as described above.
HI Assays.
HI assays were performed for serologic subtyping of H2N3 viruses to determine seroconversion and to test convalescent serum samples collected from the various swine herds associated with the outbreak. Sera were heat-inactivated at 56°C, treated with a 20% suspension of kaolin (Sigma–Aldrich) to eliminate nonspecific inhibitors, and adsorbed with 0.5% turkey red blood cells. The sera were tested for antibodies against H2N3 swine influenza viruses and reference strains of swine influenza (A/Swine/IA/1973 H1N1, A/Swine/TX/98 H3N2, and A/Sw/NC/2001 variant H1N1) virus by HI assay (28). The ferret sera were tested to determine seroconversion for H2N3 virus.
DNA Sequencing, Phylogenetic Analysis, and Subtype Determination.
Viral RNA was prepared from 200 μl of virus suspension with the RNeasy Mini Kit (Qiagen) as directed by the manufacturer. Two-step RT-PCR was conducted by using universal primers as reported in refs. 29 and 30. Each gene segment was amplified under standard conditions. PCR products were purified by using a QIAamp Gel extraction kit (Qiagen) and sequenced by using an ABI 3730 DNA Analyzer (Applied Biosystems). Multiple sequence alignments were made by using CLUSTAL W (31), and phylogenetic trees were generated by using the neighbor-joining algorithm in the PHYLIP version 3.57C software package (32). A Megablast search of the Influenza Sequence Database was performed. The viral subtype determined by sequencing was compared with those from GenBank. The isolate was plaque-purified, retested, and again subtyped by RT-PCR and sequencing (plaques were uniform in appearance, and two plaques from each isolate were chosen for amplification and sequencing).
Experiments in Pigs.
Pigs were obtained from a healthy herd that was free of swine influenza virus and PRRSV. All animal experiments were in compliance with the Institutional Animal Care and Use Committee of the NADC. The inoculation protocol has been described in ref. 33. Briefly, 20 4-week-old cross-bred pigs were inoculated intratracheally with 2 × 106 TCID50 per pig of Sw/4296424 virus prepared in MDCK cells. Four-week-old contact pigs were commingled with inoculated pigs on day 3 p.i. to study transmission efficiency. Twelve control pigs were inoculated with noninfectious cell culture supernatant. Five of 20 inoculated pigs and 3 of 12 control pigs were euthanized on days 3, 5, and 7 p.i., respectively. The remaining five pigs from the inoculated group and three control pigs were euthanized on day 27 p.i. and were analyzed for seroconversion. Nasal swabs were taken on days 0, 3, 5, and 7 p.i., placed in 2 ml of MEM, and stored at −80°C. Blood was collected from all inoculated, contact, and control pigs on days 0, 3, 5, 7, and 14 p.i. Blood was also collected from contact pigs on day 24 after contact and from the remaining five inoculated and control pigs on day 27 p.i. and was analyzed for seroconversion. Each lung was lavaged with 50 ml of MEM to obtain BALF.
Viral load in BALF was determined in a 96-well plate as described in ref. 33. Briefly, 10-fold serial dilutions of each sample were made in serum-free MEM supplemented with TPCK-trypsin and antibiotics. Each dilution (100 μl) was plated on PBS-washed confluent MDCK cells in 96-well plates. Plates were evaluated for cytopathic effects after 24 to 48 h. At 48 h, plates were fixed with 4% phosphate-buffered formaldehyde and immunocytochemically stained with a monoclonal antibody to influenza A nucleoprotein (34). The TCID50/ml was calculated for each sample by the method of Reed and Muench (35).
Virus was isolated from nasal swab samples stored at −80°C by thawing and vortexing each sample for 15 sec, centrifuging it for 10 min at 640 × g, and passing the supernatant through 0.45-μm filters to reduce bacterial contamination. An aliquot of 100 μl was plated on confluent, PBS-washed MDCK cells in 48-well plates. After incubation for 1 h at 37°C, 500-μl serum-free MEM supplemented with 1 μg/ml TPCK trypsin and antibiotics was added. All wells were evaluated for cytopathic effects after 48–72 h. Subsequently, plates were fixed with 4% phosphate-buffered formaldehyde and stained as described above.
BALF was tested for the presence of PRRSV and M. hyopneumoniae by diagnostic PCR assays. For PRRSV, the total RNA was isolated from each sample by using the RNeasy Mini Kit (Qiagen). One microgram of the extracted RNA and a primer pair specific for ORF 7 of PRRSV were used in real-time PCR as described in ref. 36. DNA was extracted from BALF for detecting M. hyopneumoniae as described in ref. 36.
Examination of Lungs of Experimental Pigs.
At necropsy, lungs were removed in toto. A single experienced veterinarian recorded the percentage of gross lesions of lobes showing the purple–red consolidation typical of swine influenza virus infection. A mean value was determined for the seven pulmonary lobes of each animal (33). Tissue samples from the trachea, the right cardiac pulmonary lobe, and other affected lobes were fixed in 10% buffered formalin, routinely processed, and stained with hematoxylin and eosin for histopathologic examination. Lung sections were given a score of 0 to 3 to reflect the severity of bronchial epithelial injury (33) according to the following criteria: 0.0: no significant lesions; 1.0: a few airways showing epithelial damage and light peribronchiolar lymphocytic cuffing, often accompanied by mild focal interstitial pneumonia; 1.5: more than a few airways affected (up to 25%), often with mild focal interstitial pneumonia; 2.0: 50% of airways affected, often with interstitial pneumonia; 2.5: ≈75% of airways affected, usually with significant interstitial pneumonia; 3.0: >75% of airways affected, usually with interstitial pneumonia. A single experienced pathologist scored all slides and was blind to the treatment groups.
Experiments in Mice.
Six- to 7-week old BALB/c mice, bred in the mouse facility of the NADC in Ames, IA, were used for infectivity experiments. All experiments were in compliance with the Institutional Animal Care and Use Committee of the NADC. Animals were weighed and anesthetized with isoflurane USP (Phoenix Pharmaceutical) before intranasal inoculation with 102-106 TCID50 of H2N3 virus (Sw/4296424) in a volume of 50 μl. Weight was recorded once daily, and general health status was observed twice daily. After onset of disease, general health status was observed three times per day. Surviving mice were euthanized on day 14 p.i., and the lungs were collected. The right lung was stored in an Eppendorf tube at −80°C for virus detection, and the left lung was fixed in 10% formalin for histopathologic analysis. Virus detection was done in a 10% tissue homogenate in PBS (homogenized twice for 1 min in a Mini BeadBeater-8; Biospec Products). The homogenate was centrifuged at 640 × g for 5, min and the supernatant was transferred to 1.5-ml reaction tubes for RNA isolation. Real-time RT-PCR was used to detect viral RNA as described in ref. 17.
Experiments in Ferrets.
Influenza-negative ferrets were obtained through the ferret breeding program and were housed at St. Jude Children's Research Hospital in compliance with the St. Jude Children's Research Hospital Animal Care and Use Committee. Infection and transmissibility of H2N3 was tested in six 18-week-old ferrets. Three ferrets were inoculated intranasally with 102.5 TCID50 of H2N3 (Sw/2124514). Twenty-four hours p.i., one naïve contact animal was introduced into the cage of each inoculated animal. Nasal washes were collected on days 1, 4, and 7 p.i., and virus was titrated in embryonated eggs. Egg 50% infective dose (EID50) values were calculated by the Reed–Muench method (35). Seroconversion was determined after 21 days by determining the serum neutralizing antibody titers of the inoculated and contact ferrets, as described in ref. 37.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Sharon Naron and Fred Tatum for critical review of the manuscript; Deb Clouser, Michelle Harland, Kevin Hassall, Trudy Tatum, Hannah Polashek, Deb Adolphson, Brian Pottebaum, and Jason Huegel for animal studies and technical assistance; the DNA Sequence Unit and Histopathology Core Unit at the National Animal Disease Center for their assistance; and veterinarians Jim Lowe (Carthage Veterinary Services, Ltd, Carthage, IL) and Erin Johnson (Boehringer Ingelheim Vetmedica, Inc., Albers, IL) for their help and assistance. This project has been funded in part with federal funds from the National Institute Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract HHSN266200700005C, and by Center for Disease Control and Prevention Grant U01 CI000357-01.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. EU258935–EU258942 (A/Sw/2124514) and EU258943–EU258950 (A/Sw/4296424)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0710286104/DC1.
References
- 1.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lipatov AS, Govorkova EA, Webby RJ, Ozaki H, Peiris M, Guan Y, Poon L, Webster RG. J Virol. 2004;78:8951–8959. doi: 10.1128/JVI.78.17.8951-8959.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taubenberger JK. Proc Am Philos Soc. 2006;150:86–112. [PMC free article] [PubMed] [Google Scholar]
- 4.Kawaoka Y, Krauss S, Webster RG. J Virol. 1989;63:4603–4608. doi: 10.1128/jvi.63.11.4603-4608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scholtissek C. Virus Genes. 1995;11:209–215. doi: 10.1007/BF01728660. [DOI] [PubMed] [Google Scholar]
- 6.Wan H, Perez DR. Virology. 2006;346:278–286. doi: 10.1016/j.virol.2005.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ito T, Couceiro JN, Kelm S, Baum LG, Krauss S, Castrucci MR, Donatelli I, Kida H, Paulson JC, Webster RG, et al. J Virol. 1998;72:7367–7373. doi: 10.1128/jvi.72.9.7367-7373.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castrucci MR, Donatelli I, Sidoli L, Barigazzi G, Kawaoka Y, Webster RG. Virology. 1993;193:503–506. doi: 10.1006/viro.1993.1155. [DOI] [PubMed] [Google Scholar]
- 9.Liu JH, Okazaki K, Bai GR, Shi WM, Mweene A, Kida H. Virus Genes. 2004;29:81–86. doi: 10.1023/B:VIRU.0000032791.26573.f1. [DOI] [PubMed] [Google Scholar]
- 10.Munster VJ, Baas C, Lexmond P, Waldenstrom J, Wallensten A, Fransson T, Rimmelzwaan GF, Beyer WE, Schutten M, Olsen B, et al. PLoS Pathog. 2007;3:e61. doi: 10.1371/journal.ppat.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krauss S, Walker D, Pryor SP, Niles L, Chenghong L, Hinshaw VS, Webster RG. Vector Borne Zoonotic Dis. 2004;4:177–189. doi: 10.1089/vbz.2004.4.177. [DOI] [PubMed] [Google Scholar]
- 12.Schafer JR, Kawaoka Y, Bean WJ, Suss J, Senne D, Webster RG. Virology. 1993;194:781–788. doi: 10.1006/viro.1993.1319. [DOI] [PubMed] [Google Scholar]
- 13.Makarova NV, Kaverin NV, Krauss S, Senne D, Webster RG. J Gen Virol. 1999;80(Pt 12):3167–3171. doi: 10.1099/0022-1317-80-12-3167. [DOI] [PubMed] [Google Scholar]
- 14.Choi YK, Goyal SM, Joo HS. J Vet Diagn Invest. 2002;14:62–65. doi: 10.1177/104063870201400113. [DOI] [PubMed] [Google Scholar]
- 15.Neumann G, Kawaoka Y. Emerg Infect Dis. 2006;12:881–886. doi: 10.3201/eid1206.051336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matrosovich MN, Gambaryan AS, Teneberg S, Piskarev VE, Yamnikova SS, Lvov DK, Robertson JS, Karlsson KA. Virology. 1997;233:224–234. doi: 10.1006/viro.1997.8580. [DOI] [PubMed] [Google Scholar]
- 17.Richt JA, Lager KM, Clouser DF, Spackman E, Suarez DL, Yoon KJ. J Vet Diagn Invest. 2004;16:367–373. doi: 10.1177/104063870401600501. [DOI] [PubMed] [Google Scholar]
- 18.Herlocher ML, Elias S, Truscon R, Harrison S, Mindell D, Simon C, Monto AS. J Infect Dis. 2001;184:542–546. doi: 10.1086/322801. [DOI] [PubMed] [Google Scholar]
- 19.Karasin AI, Brown IH, Carman S, Olsen CW. J Virol. 2000;74:9322–9327. doi: 10.1128/jvi.74.19.9322-9327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karasin AI, West K, Carman S, Olsen CW. J Clin Microbiol. 2004;42:4349–4354. doi: 10.1128/JCM.42.9.4349-4354.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Connor RJ, Kawaoka Y, Webster RG, Paulson JC. Virology. 1994;205:17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- 22.Rogers GN, Paulson JC, Daniels RS, Skehel JJ, Wilson IA, Wiley DC. Nature. 1983;304:76–78. doi: 10.1038/304076a0. [DOI] [PubMed] [Google Scholar]
- 23.Matrosovich M, Tuzikov A, Bovin N, Gambaryan A, Klimov A, Castrucci MR, Donatelli I, Kawaoka Y. J Virol. 2000;74:8502–8512. doi: 10.1128/jvi.74.18.8502-8512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tumpey TM, Maines TR, Van Hoeven N, Glaser L, Solorzano A, Pappas C, Cox NJ, Swayne DE, Palese P, Katz JM, et al. Science. 2007;315:655–659. doi: 10.1126/science.1136212. [DOI] [PubMed] [Google Scholar]
- 25.Brown JD, Swayne DE, Cooper RJ, Burns RE, Stallknecht DE. Avian Dis. 2007;51:285–289. doi: 10.1637/7636-042806R.1. [DOI] [PubMed] [Google Scholar]
- 26.Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. Nature. 2006;440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 27.Calsamiglia M, Pijoan C, Trigo A. J Vet Diagn Invest. 1999;11:246–251. doi: 10.1177/104063879901100307. [DOI] [PubMed] [Google Scholar]
- 28.Palmer DF, Coleman MT, Dowdle WR, Schild GC. Advanced Laboratory Techniques for Influenza Diagnosis. Washington, DC: United States Department of Health, Education, and Welfare; 1975. pp. 51–52. [Google Scholar]
- 29.Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. Arch Virol. 2001;146:2275–2289. doi: 10.1007/s007050170002. [DOI] [PubMed] [Google Scholar]
- 30.Ma W, Gramer M, Rossow K, Yoon KJ. J Virol. 2006;80:5092–5096. doi: 10.1128/JVI.80.10.5092-5096.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson JD, Higgins DG, Gibson TJ. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Felsenstein J. PHYLIP. Seattle: Department of Genetics, University of Washington; 1993. Version 3.5c. [Google Scholar]
- 33.Richt JA, Lager KM, Janke BH, Woods RD, Webster RG, Webby RJ. J Clin Microbiol. 2003;41:3198–3205. doi: 10.1128/JCM.41.7.3198-3205.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kitikoon P, Nilubol D, Erickson BJ, Janke BH, Hoover TC, Sornsen SA, Thacker EL. Vet Immunol Immunopathol. 2006;112:117–128. doi: 10.1016/j.vetimm.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 35.Reed L, Muench H. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 36.Lekcharoensuk P, Lager KM, Vemulapalli R, Woodruff M, Vincent AL, Richt JA. Emerg Infect Dis. 2006;12:787–794. doi: 10.3201/eid1205.051060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Govorkova EA, Webby RJ, Humberd J, Seiler JP, Webster RG. J Infect Dis. 2006;194:159–167. doi: 10.1086/505225. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.