Abstract
The selective formation of E- or Z-isomers is an important process in natural product metabolism. We show that the subunit composition of an enzyme can alter the geometrical composition of the enzymatic products. Hinokiresinol synthase, purified from Asparagus officinalis cell cultures, is responsible for the conversion of (7E,7′E)-4-coumaryl 4-coumarate to (Z)-hinokiresinol, the first step in norlignan formation. The protein is most likely a heterodimer composed of two distinct subunits, which share identity with members of the phloem protein 2 gene superfamily. Interestingly, each recombinant subunit of hinokiresinol synthase expressed in Escherichia coli solely converted (7E,7′E)-4-coumaryl 4-coumarate to the unnatural (E)-hinokiresinol, the E-isomer of (Z)-hinokiresinol. By contrast, a mixture of recombinant subunits catalyzed the formation of (Z)-hinokiresinol from the same substrate.
Keywords: asparagus, phloem protein 2, coumaryl coumarate, phenylpropanoid
Carbon-carbon double bonds, in both E- and Z-configurations, have been found in natural products including fatty acids, polyketides, terpenoids, and phenylpropanoids (1, 2). Different geometrical isomers differ in their physical, chemical, and biological properties. The control of geometrical isomerism has attracted much attention from chemists and biochemists, because this is a key step in natural product biosynthesis and is critically important in organic synthesis to produce only the desired isomer (3).
During natural product biosynthesis, usually either the E or Z double bond is formed selectively. For instance, in the biosynthesis of geraniol and nerol, geraniol (E-isomer) is formed directly from geranyl diphosphate, whereas nerol (Z-isomer) is formed from geraniol by isomerization catalyzed by an isomerase (2). cis-Prenyltransferases catalyze the synthesis of (Z)-prenyl chains, and trans-prenyltransferases catalyze the formation of (E)-prenyl chains, although both attach a common substrate, isopentenyl diphosphate, onto elongating prenyl chains (4). Overall, the selective formation of E or Z double bonds is controlled by distinct enzymes.
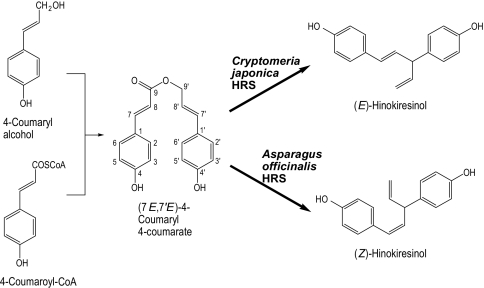
The norlignan is a class of phenylpropanoids, and a norlignan, hinokiresinol, has an E or Z double bond in its molecule (Fig. 1). Interestingly, (E)-hinokiresinol is distributed specifically in conifer heartwood, and the coloration of Chamaecyparis obtusa (Japanese cypress) heartwood is ascribed to (E)-hinokiresinol (5). (Z)-Hinokiresinol, however, is detected in herbaceous monocotyledons (6). Both hinokiresinols have antifungal activity and can be produced in response to stress, such as fungal infection and sapwood drying (7–9), suggesting that they are synthesized in vivo for plant protection.
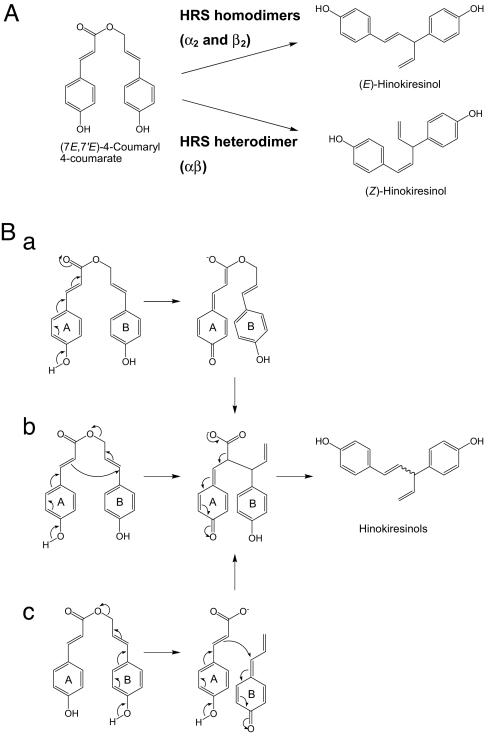
Fig. 1.
Biosynthetic pathway of hinokiresinols.
Previously, the mechanism of formation of E and Z double bonds in hinokiresinols was unknown. However, our findings have indicated that (Z)-hinokiresinol is derived from two nonidentical phenylpropane units (6). Subsequently, (Z)-hinokiresinol was found to be synthesized from a dimeric phenylpropanoid ester, (7E,7′E)-4-coumaryl 4-coumarate, by an enzyme preparation from elicitor-treated Asparagus officinalis cells (10). In addition, (E)-hinokiresinol was formed by an enzyme preparation isolated from Cryptomeria japonica (Japanese cedar) cells (11). Interestingly, the two enzymatic reactions proceed without any additional cofactors, and either (E)- or (Z)-hinokiresinol is formed selectively in each assay, directly from a common substrate, (7E,7′E)-4-coumaryl 4-coumarate. It is noteworthy that both reactions involve cleavage of the ester linkage, formation of a new carbon-carbon bond, and loss of a carbon atom, to produce a diphenylpentane (C17) carbon framework. Thus, the results above reveal that hinokiresinols are the first norlignans in the norlignan biosynthetic pathway (Fig. 1).
In this report, we show that hinokiresinol synthase (HRS), is involved in the formation of (Z)-hinokiresinol from (7E,7′E)-4-coumaryl 4-coumarate (Fig. 1). This protein is composed of two subunits encoded by distinct genes, which share identity with members of the phloem protein 2 (PP2) gene superfamily. Surprisingly, each recombinant subunit catalyzes the formation of (E)-hinokiresinol only. However, the mixture of both recombinant subunits shows (Z)-hinokiresinol synthase activity.
Results and Discussion
Purification and cDNA Cloning of HRS.
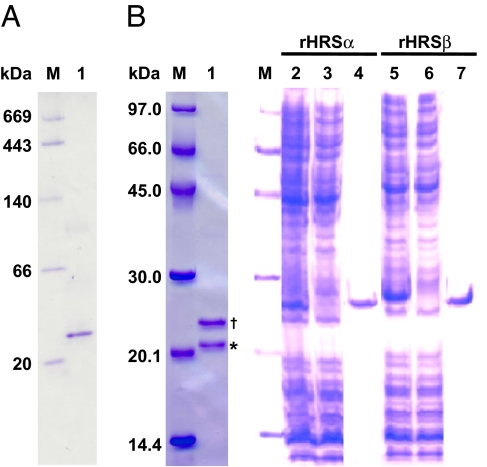
Initially, we carried out a 114-fold purification of HRS from cell-free extracts of elicitor-treated Asparagus cells, with a 1.7% yield by six-step column chromatography [supporting information (SI) Table 1]. Whereas a single band was seen by native PAGE (Fig. 2A, lane 1), SDS/PAGE analysis showed two polypeptide bands, corresponding to 21 and 23 kDa (Fig. 2B, lane 1). Although we attempted several times to obtain a large amount of purified enzyme, the amount we obtained was not sufficient to fully characterize this enzyme. Therefore, either band might have been a contaminant protein at this point, but we gel-purified both polypeptides by SDS/PAGE and submitted them to microsequencing.
Fig. 2.
Gel electrophoresis of natural HRS (purified from Asparagus), rHRSα, and rHRSβ. (A) Native PAGE. M, marker; 1, natural HRS. (B) SDS/PAGE. M, marker; 1, natural HRS; 2 and 5, lysate; 3 and 6, flow-through; 4 and 7, eluted from His-Bind resin. Note that the asterisk and dagger indicate HRSα and HRSβ, respectively.
The two polypeptides could not be sequenced without peptidase digestion, suggesting that the N termini of the natural proteins were blocked. Therefore, we digested them in the gel by using lysylendopeptidase after carboxymethylation, and the resulting peptide fragments were separated by reversed-phase HPLC. Seven peptide fragments from the 21-kDa polypeptide and two peptide fragments from the 23-kDa polypeptide were sequenced (SI Table 2). Several amino acids were substituted among peptide fragments from the 21-kDa polypeptide, suggesting the presence of isozymes, for example, 1a and 1b (SI Table 2). The 21- and 23-kDa polypeptides were later found to have similar amino acid sequences; thus, we have now designated the 21- and 23-kDa polypeptides HRSα and HRSβ, respectively.
Two degenerate oligonucleotide primers A and B, designed from these peptide fragments (3a and 6; SI Table 2), and an oligo(dT) primer were used for reverse transcriptase PCR amplification from cDNAs prepared from elicitor-treated Asparagus cells. A 675-bp fragment encoding the amino acid sequences identified from HRSα (1a, 2a, 3a, and 4; SI Table 2) and a 545-bp fragment encoding the amino acid sequences identified from HRSβ (5 and 6; SI Table 2) were cloned into the pCR2.1-TOPO vector. These amplified cDNA fragments were digested with SspI and EcoRI, producing 0.5-kb fragments that were used as probes for phage cDNA library screening. The identity between the probes was 61%, and no cross-hybridization of the probes was found during cDNA library screening. The inserts were PCR-amplified from single-plaque phages, cloned into pCR2.1-TOPO, and sequenced. The deduced amino acid sequences of the cloned genes were almost identical to the sequences obtained by microsequencing (Fig. 3A and SI Table 2).
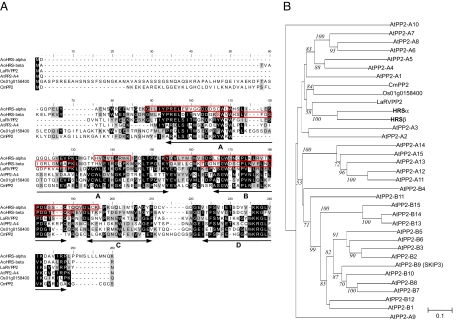
Fig. 3.
Amino acid sequence alignments and phylogenetic tree of HRS homologs. (A) Amino acid sequence alignments. White letters on black are identical amino acid residues; black letters on gray are similar. The peptide sequences obtained by microsequencing are shown in red boxes. The conserved motifs A through D of the PP2 domain are shown by arrows. GenBank accession numbers for these homologs are given in the parentheses: HRSα (AB358973), HRSβ (AB358974), LaRVPP2 (ABJ99589), AtPP2-A4 (NP_174654), Os01g0158400 (BAF03991), and CmPP2 (AAB24688). (B) Phylogenetic relationship of Arabidopsis PP2-like proteins (13) and HRS homologs. Only percentages of bootstrap values supported by >50% of the 1,000 replicates are indicated with italics.
The full-length cDNAs of HRSα and HRSβ are 998 bp and 906 bp long, with ORFs of 546 bp and 531 bp, encoding 182- and 177-aa residues, respectively. Their predicted molecular masses are 20.4 and 19.8 kDa, which are slightly lower than the apparent molecular masses estimated by SDS/PAGE (21 and 23 kDa; Fig. 2B). The ORFs of HRSα and HRSβ share 49% amino acid identity and 68% amino acid similarity.
HRSα and HRSβ Are Members of the PP2 Gene Superfamily.
A PSI-BLAST search identified several EST and protein sequences that were similar to HRSα and HRSβ (E value > 10−28). These sequences are annotated as PP2 or PP2-like genes. HRSα has 34%, 27%, 20%, and 19% amino acid identities to a functionally unknown Lycoris aurea lectin-like protein (LaRVPP2), Arabidopsis AtPP2-A4 (At1g33920), an Oryza sativa lectin-like protein (Os01g0158400), and Cucurbita maxima PP2 (CmPP2) (12), respectively. HRSβ shares 33%, 24%, 21%, and 18% amino acid identity with LaRVPP2, AtPP2-A4, Os01g0158400, and CmPP2, respectively (Fig. 3).
The Arabidopsis genome sequencing project has revealed that there are 30 PP2 and PP2-like proteins (Fig. 3B) with four PP2 motifs, A to D (Fig. 3A) (13). Moreover, the presence of many homologs containing these motifs within a vast variety of plant species, including angiosperms, gymnosperms, and mosses (13), indicates that the PP2 and PP2-like genes form a gene superfamily. Both HRSα and HRSβ contain these four motifs (Fig. 3A), indicating that HRSα and HRSβ are members of the PP2 gene superfamily. This finding is also supported by phylogenetic analysis, which shows that both HRSα and HRSβ belong to a clade of the gene superfamily (Fig. 3B).
Recombinant HRSα and HRSβ Catalyze the Formation of (E)-Hinokiresinol.
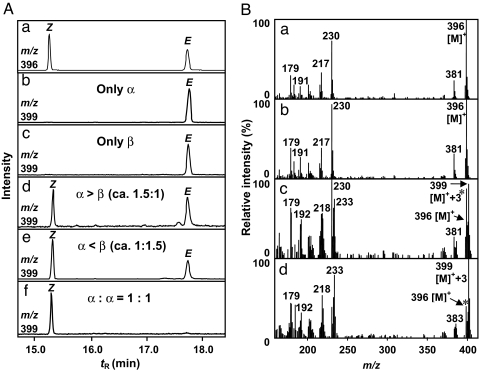
To confirm the catalytic function of HRSα and HRSβ, we expressed the ORFs individually in Escherichia coli and assayed hinokiresinol synthase activity. The recombinant proteins (referred to as rHRSα and rHRSβ) fused to poly(His) tags at the C termini were successfully purified on a His-Bind Resin column. The apparent molecular masses of rHRSα and rHRSβ subunits (both 25 kDa), as determined by SDS/PAGE, were slightly higher than the values calculated from their amino acid sequences (21.7 and 21.2 kDa) (Fig. 2B, lanes 4 and 7). The recombinant proteins were tested in vitro for activity toward a deuterium-labeled substrate, (7E,7′E)-4-[7′,9′,9′-2H3]coumaryl 4-coumarate. When either rHRSα or rHRSβ was incubated with the substrate, surprisingly, only (E)-[2H3]hinokiresinol was produced (Fig. 4Ab, Ac, and Bc). Thus, both HRSα and HRSβ are responsible for the formation of the norlignan carbon framework, involving cleavage of the ester linkage, new carbon-carbon bond formation, and loss of a carbon atom.
Fig. 4.
GC-MS analysis of hinokiresinol formed by recombinant HRS proteins as trimethylsilyl ethers. (A) Mass chromatograms. (a) Mixture of authentic (Z)- and (E)-hinokiresinols. (b) Incubation of trideuterated 4-coumaryl 4-coumarate with only rHRSα. (c) Incubation of trideuterated 4-coumaryl 4-coumarate with only rHRSβ. (d) Incubation of trideuterated 4-coumaryl 4-coumarate with rHRSα and rHRSβ (≈1.5:1). (e) Incubation of trideuterated 4-coumaryl 4-coumarate with rHRSα and rHRSβ (≈1:1.5). (f) Incubation of trideuterated 4-coumaryl 4-coumarate with rHRSα and rHRSβ (1:1). (B) Mass spectra. (a) Authentic unlabeled (E)-hinokiresinol. (b) Authentic unlabeled (Z)-hinokiresinol. (c) Enzymatically formed (E)-hinokiresinol from trideuterated 4-coumaryl 4-coumarate. (d) Enzymatically formed (Z)-hinokiresinol from trideuterated 4-coumaryl 4-coumarate. Note: m/z 396 is the molecular ion of unlabeled hinokiresinols and m/z 399 is the molecular ion of [2H3]hinokiresinols. The authentic unlabeled hinokiresinols (asterisks) were added in the experiments shown in Bc and Bd as carriers for quantification.
Both HRSα and HRSβ Are Required for (Z)-Hinokiresinol Synthase Activity.
It was very strange that (E)-hinokiresinol was formed by both rHRSα and rHRSβ, whereas natural HRS catalyzed the formation of only (Z)-hinokiresinol (10). We therefore hypothesized that (Z)-hinokiresinol formation may require both subunits in combination and assayed (Z)-hinokiresinol formation with a mixture of rHRSα and rHRSβ, because we found both HRSα and HRSβ in natural HRS (Fig. 2B, lane 1). After preincubation of the mixture of rHRSα and rHRSβ at 30°C for 2 min, (7E,7′E)-4-[7′,9′,9′-2H3]coumaryl 4-coumarate was transformed to (Z)-[2H3]hinokiresinol in addition to (E)-[2H3]hinokiresinol (Fig. 4 Ad and Ae). The product was identified by comparison of the mass spectrum and retention times with those of authentic samples.
As shown in Fig. 4 Ad and Ae, when the molar ratio of one of the recombinant proteins exceeded the other (that is, rHRSα > rHRSβ, and vice versa), (E)-[2H3]hinokiresinol formation was still observed, whereas when the ratio of rHRSα and rHRSβ was 1:1, strikingly, only (Z)-[2H3]hinokiresinol was formed (Fig. 4Af). These results demonstrate that both HRSα and HRSβ are required for (Z)-hinokiresinol synthase activity.
Natural HRS Is Most Likely a Heterodimer.
The results from the gel electrophoresis and enzymatic assays strongly support the possibility that natural HRS is composed of the distinct two subunits, HRSα and HRSβ. We further characterized the subunit composition of natural HRS, rHRSα, rHRSβ, and a 1:1 mixture of rHRSα and rHRSβ. Analysis by gel filtration chromatography showed that natural HRS, rHRSα, and rHRSβ were all 37–40 kDa (SI Fig. 6), indicating that these proteins exist as dimers. Thus, natural HRS is most likely a heterodimer (αβ), whereas rHRSα and rHRSβ are homodimers (α2 and β2). A 1:1 mixture of rHRSα and rHRSβ was also 37–40 kDa (SI Fig. 6) and the protein migration patterns on a native PAGE gel were similar among rHRSα, rHRSβ, and a 1:1 mixture of rHRSα and rHRSβ (SI Fig. 7). Given that both HRSα and HRSβ are required for (Z)-hinokiresinol synthase activity, and natural HRS consists of HRSα and HRSβ, it is strongly suggested that the subunit recombination between α2 and β2 to give αβ occurs in a 1:1 mixture of rHRSα and rHRSβ.
Biochemical Characterization of a Mixture of rHRSα and rHRSβ.
The kinetic properties of a 1:1 mixture of rHRSα and rHRSβ were determined by using (7E,7′E)-4-[7′,9′,9′-2H3]coumaryl 4-coumarate as substrate. The reaction period was 10 min, because the reaction was linear with time up to 20 min under the conditions used (SI Fig. 8A). The optimum pH, optimum temperature and Km values for a 1:1 mixture of rHRSα and rHRSβ were 6.0, 30°C and 0.44 μM, respectively (SI Fig. 8).
Evolution and Function of HRS.
Our data demonstrate that HRSα and HRSβ are members of the PP2 gene family. PP2 is one of the most abundant and enigmatic proteins in the phloem sap. PP2 and PP1 proteins constitute P protein, which plugs sieve plates to maintain turgor pressure within the sieve tube after injury to a sieve element, or which functions as a physical barrier against pathogens and pests (13). Most members of the PP2 gene superfamily have not been functionally characterized. Several PP2 proteins, including CmPP2, are known to have poly(GlcNAc)-binding activity (12). Recent in vitro studies have shown that PP2 interacts with a variety of RNAs and could be involved in the long-distance movement of viroids (14, 15). Moreover, more than half of the predicted Arabidopsis PP2-like proteins contain F boxes, which are typically involved in targeting proteins to the E3 ubiquitinylation degradation pathway, and some have TIR and AIG1 domains within the N-terminal region (13). Although diverse functions of the PP2 gene superfamily have been identified or proposed, an enzymatic activity, such as that of HRS, has not been reported. It may be speculated that a PP2-like protein, which is associated with plant protection, evolved to have the catalytic activity to produce antifungal hinokiresinols. Our finding that HRSα and HRSβ belong to the PP2 gene superfamily demonstrates a new biochemical function of this gene superfamily.
Molecular Mechanisms of Hinokiresinol Formation by HRS.
We revealed that HRS proteins catalyze the formation of hinokiresinols, although the formed geometrical isomers differ in terms of the subunit composition (Fig. 5A). (E)-Hinokiresinol was converted to (Z)-hinokiresinol by neither recombinant HRS proteins (M.Y., S.S., and T.U., unpublished observations) nor the crude enzymes from Asparagus and Cryptomeria cells (10, 11), indicating that hinokiresinols are directly formed from (7E,7′E)-4-coumaryl 4-coumarate. Thus, three hypothetical reaction mechanisms, namely, the ester enolate Claisen rearrangement (Fig. 5Ba), an intramolecular rearrangement (Fig. 5Bb), and a bimolecular coupling after an ester cleavage (Fig. 5Bc) (11, 16, 17) can be considered to participate in hinokiresinol formation from (7E,7′E)-4-coumaryl 4-coumarate. In mechanisms a and b, a hydroxyl group in the A aryl ring is requisite for the formation of the corresponding quinonemethide (Fig. 5 Ba and Bb). However, bimolecular coupling requires two hydroxyl groups in both A and B aryl groups (Fig. 5Bc), because both aryl groups need to become quinonemethides. To clarify the mechanism, further studies using several analogs lacking a free 4-hydroxyl group and inhibitors, as potential substrates, are needed.
Fig. 5.
Hinokiresinol formation by HRS proteins and the hypothetical reaction mechanisms. (A) HRS homodimers (α2 and β2) catalyze the formation of the E-isomer, and HRS heterodimer (αβ) catalyzes the formation of the Z-isomer. (B) The hypothetical reaction mechanism for hinokiresinol formation. (a) The ester enolate Claisen rearrangement. (b) Intramolecular rearrangement. (c) Bimolecular coupling after ester cleavage.
How does only HRS heterodimer catalyze the formation of (Z)-hinokiresinol? One possible answer might be that an active site lies at the interface between the two subunits, and only the active site cleft between α and β is suitable for Z-isomer production. X-ray crystallographic analysis combined with an enzymatic assay with potential substrates might answer this question.
In conclusion, we have identified the first genes governing norlignan biosynthesis, which involves formation of a new carbon-carbon bond. The subunit composition of HRS can control the stereoselective formation of (E)- and (Z)-hinokiresinol from (7E,7′E)-4-coumaryl 4-coumarate. Such a control mechanism is unique in natural product biosynthesis, because E or Z double-bond formation is usually controlled by distinct enzymes. By contrast, our data revealed that altering the subunit composition of an enzyme can alter the stereoselectivity in the double-bond formation that it catalyzes.
Materials and Methods
Plant Material.
The cell-suspension culture of A. officinalis cv. Accel was initiated and elicited with autoclaved Fusarium solani IFO4542 mycelia for 18 h as described (6, 10). The cells were collected by a strainer and washed with distilled water, and the excess water was absorbed with a paper towel. The cells were weighed and stored in liquid nitrogen until use.
Chemicals and Buffers.
(7E,7′E)-4-Coumaryl 4-coumarate and (7E,7′E)-4-[7′,9′,9′-2H2]coumaryl 4-coumarate were synthesized as described in ref. 10. (Z)-Hinokiresinol (6) and (E)-hinokiresinol (18) were isolated from A. officinalis and Chamaecyparis obtusa, respectively. Buffer A was composed of 20 mM Tris·HCl buffer (pH 7.8) containing 10% glycerol (vol/vol), 2 mM DTT, 1 mM sodium ethylenediaminetetraacetate (EDTA-Na2), 0.5 mM phenylmethanesulfonylfluoride (PMSF), and 2.5 mM 2-mercaptoethanol. Buffer B was identical except that the glycerol concentration was 5% (vol/vol). All buffers used for HPLC enzyme purification were purged with helium gas during chromatography. Oligonucleotide primers were synthesized by Espec Oligo Service. All other commercial reagents were obtained from Nacalaitesque or Wako Pure Chemicals, unless otherwise noted.
Enzyme Assay by HPLC.
The details of the enzyme assay by HPLC are described in SI Text.
Enzyme Purification.
All operations were carried out at 4°C. Frozen Asparagus cells (500–550 g) were pulverized by using a Waring blender, successively homogenized with a mortar and pestle for 20 min in the presence of polyvinylpolypyrrolidone (10% wt/wt of material), sea sand, and 1 liter of 0.1 M Tris·HCl (pH 7.8) buffer containing 0.5 mM PMSF, 5 mM sodium ascorbate, 10 mM DTT, and 1 mM EDTA-Na2. The slurry was filtered through four-layered gauze. The filtrate was centrifuged for 20 min at 8,500 × g and the protein in the supernatant was precipitated by adding ammonium sulfate (50% saturation). After centrifugation, additional ammonium sulfate was added to the supernatant so that the final concentration was 70% saturation. The suspension was left for 1 h and centrifuged at 8,500 × g for 30 min. The pellet was dissolved in buffer A containing 1 M ammonium sulfate. The undissolved residue was removed by centrifugation and the supernatant was applied to a Toyoperl Phenyl 650M column (Tosoh, 1.5 × 20 cm) that had been equilibrated with buffer A containing 1.8 M ammonium sulfate. The retained proteins were eluted with a linear gradient of ammonium sulfate (from 1.8 to 0 M). Fractions containing enzyme activity were combined and desalted by passing them through a Sephadex G-25 column (Amersham Pharmacia) that had been equilibrated with buffer B. The desalted sample was applied to a Cosmogel DEAE column (Nacalaitesque, 2 × 10 cm) equilibrated with buffer B, washed with a linear gradient of sodium acetate (from 0 to 0.1 M), and eluted with a linear gradient of NaCl (from 0 to 0.3 M). The fractions containing enzyme activity were combined and concentrated with a Centriprep 10 (Millipore) up to 5 ml, and the concentrated sample was passed through a HiLoad Superdex 200pg column (Amersham Pharmacia, 1.6 × 60 cm) equilibrated with 0.1 M Tris·HCl (pH 7.8) containing 2 mM DTT, 1 mM EDTA-Na2, 2.5 mM 2-mercaptoetanol, 0.5 mM PMSF, and 5% glycerol (vol/vol). The fractions indicating enzyme activity were collected and concentrated as above. The concentrated fraction was diluted five times with buffer B and applied to a Cosmogel DEAE (0.8 × 7.5 cm) column. The enzyme was eluted with a linear gradient of NaCl (from 0 to 0.3 M). The fractions showing enzyme activity were pooled and desalted by passing them through a Sephadex G-25 column as above. The desalted sample was applied to a MonoQ HR 5/5 column (Amersham Pharmacia) equilibrated with buffer B, and the enzyme was eluted in the same conditions used for the second Cosmogel DEAE column chromatography. The fraction containing enzyme activity was collected and applied again to a HiLoad Superdex 200pg column. The elution condition was the same as above. The fractions showing enzyme activity were combined, concentrated with a Centriprep 10 and an UltraFree 10000 (Millipore) up to 0.3 ml, and used for further characterization as the final purified preparation.
Gel Electrophoresis.
The details of gel electrophoresis are described in SI Text.
Microsequencing.
The details of microsequencing are described in SI Text.
Cloning of cDNAs Encoding HRSα and HRSβ.
Total RNA from Asparagus cells after 18 h elicitation (6) were isolated by using the method of Bugos et al. (19). mRNA was purified from total RNA by using a Poly(A) Quick mRNA Isolation Kit (Stratagene). A cDNA library was constructed from the mRNA by using the ZAP-cDNA Gigapack III Gold Cloning Kit (Stratagene) according to the manufacturer's instructions. Based on the partial amino acid sequences derived by microsequencing (SI Table 2), oligonucleotide primer A (5′-GTIGTIGGNGGNGAYGAYG-3′) and primer B (5′-GGIMGIYTIGGNATGGARGC-3′) were designed from the peptide fragments 3a and 6 as sense primers. cDNA was synthesized from the total RNA of the elicited Asparagus cells by using an oligo(dT) (5′-AGCTCGAGTTTTTTTTTTTTTTTTTT-3′) primer and SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer's recommendations. Degenerate PCR amplification was carried out by using the pairs of primer A and an oligo(dT) primer, or primer B and an oligo(dT) primer.
The amplified cDNA fragments were subcloned into the pCR2.1-TOPO (Invitrogen) vector, and the vector was transformed into E. coli TOP10 (Invitrogen) and sequenced. This enabled us to obtain two partial cDNA fragments (675 bp and 545 bp) encoding HRSα and HRSβ. The vectors harboring these inserts were digested with SspI and EcoRI, which released 0.5-kb fragments. The fragments were radiolabeled with [α-32P]dATP (3,000 μCi/mmol, Amersham) by using the DECAprime II kit (Ambion), and 1.2 × 105 plaque-forming units of phage for HRSα and 6.0 × 104 plaque-forming units of phage for HRSβ were screened by using the fragments as probes under high-stringency conditions (20). Phages from 12 positive plaques for HRSα and six positive plaques for HRSβ were recovered and submitted to second-round screening. The resulting single-plaque phages were recovered and the inserts were PCR-amplified by using T7 and T3 primers. The amplified fragments were cloned into pCR2.1-TOPO and fully sequenced to select a clone without the PCR-induced mutations. The cloned sequences were analyzed by means of a PSI-BLAST search on the National Center for Biotechnology Information (NCBI) web site (www.ncbi.nlm.nih.gov/) and ClustalW (21). A phylogenetic tree (Fig. 3B) was constructed by the neighbor-joining method (22).
Heterologous Expression of Recombinant HRSα and HRSβ in E. coli.
The ORFs of HRSα and HRSβ were PCR-amplified by using forward primers encoding an NdeI site plus gene-specific sequences beginning at the start codon (5′-TCATATGGATGGGCAACCAGAGCT-3′ and 5′-TCATATGGCTACTGTTGCCAAGGAG-3′), and reverse primers including the NotI site and gene-specific sequences beginning at the codon next to the stop codon (5′-TGCGGCCGCCTTTTGGTTCATCAGCAGAA-3′ and 5′-TGCGGCCGCACGGGGGAGTGGACGGAG-3′). PCR products were ligated into pCR2.1-TOPO vectors before introduction into E. coli TOP10 cells. The inserts were subcloned into pET23a (Novagen) to fuse a His-tag to the C terminus of HRS. Each construct was transformed into E. coli BL21(DE3)pLysS (Novagen). The bacteria were grown at 37°C in LB medium containing 100 mg/liter ampicillin. When A600 reached 0.6, 0.4 mM isopropyl thio-β-d-galactoside was added and incubated for an additional 4 h at 30°C to induce expression. After centrifugation at 2,000 × g for 10 min at 4°C, the pellet was resuspended in 4 ml of 20 mM Tris·HCl (pH 7.9) containing 0.5 mM NaCl and 5 mM imidazole. Cells were lysed on ice with a Branson Sonifier 250 (1 min × 3). Lysates were centrifuged at 10,000 × g for 10 min at 4°C and the supernatant containing soluble proteins was passed through a 0.45-μm Millex HV filter (Millipore) and applied to a column containing 1 ml of His-Bind Resin (Novagen). The column was washed successively with 6 ml of 20 mM Tris·HCl (pH 7.9) containing 0.5 mM NaCl and 60 mM imidazole (wash buffer), and 6 ml of wash buffer containing 100 mM imidazole. The bound protein was eluted with 6 ml of 20 mM Tris·HCl (pH 7.9) containing 0.5 mM NaCl and 1 M imidazole. The eluent was desalted with a Sephadex G-25 (Amersham Pharmacia) column and its purity was confirmed by SDS/PAGE. Each purified protein was stored in 50 mM Tris·HCl buffer (pH 7.5) containing 2 mM 2-mercaptoethanol and 30% glycerol at −20°C until use in the enzyme activity assay.
Recombinant Enzyme Assay.
Protein concentration was determined by using the Bradford method (16), with BSA as a standard. The typical reaction mixture (200 μl) for the (Z)-hinokiresinol synthase assay contained rHRSα (5 μg), rHRSβ (5 μg), 50 mM potassium phosphate buffer (pH 6.0) and 100 μM (7E,7′E)-4-[7′,9′,9′-2H3]coumaryl 4-coumarate. The reaction was initiated by the addition of substrate, and the reaction mixture was incubated at 30°C for 10 min. The reaction was terminated by extracting with 600 μl of ethyl acetate containing 1 μl of unlabeled (Z)-hinokiresinol (0.1 mg/liter). The ethyl acetate extracts were dried and dissolved in 4 μl of N,O-bis(trimethylsilyl)acetamide. After standing at 60°C for 40 min, an aliquot of the solution was subjected to GC-MS measurement and the products were identified and quantified, as described in ref. 6.
Biochemical Characterization of a Mixture of rHRSα and rHRSβ.
The details of the biochemical characterization of a mixture of rHRSα and rHRSβ are described in SI Text.
Estimation of the Molecular Masses of HRS Proteins.
The details of the estimation of the molecular masses of natural and recombinant HRS proteins are described in SI Text.
Supplementary Material
ACKNOWLEDGMENTS.
We thank M. Nakamura for help with maintenance of the Asparagus culture. This work was supported in part by Japan Society for the Promotion of Science Grants-in-Aid for Scientific Research 2660150, 16380116, and 18658069; by a grant from the New Energy and Industrial Technology Development Organization (Development of Fundamental Technologies for Controlling the Process of Material Production of Plants); by a grant from TOSTEM Foundation for Construction Materials Industry Promotion; and by MEXT Special Coordination Funds for Promoting Science and Technology: Integrated Research System for Sustainability Science (IR3S).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AB358973 (HRSα), AB358974 (HRSβ), ABJ99589 (LaRVPP2), NP_174654 (AtPP2-A4), BAF03991 (Os01g0158400), and AAB24688 (CmPP2)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0710357105/DC1.
References
- 1.Sankawa U. In: Comprehensive Natural Products Chemistry. Barton D, Nakanishi K, Meth-Cohn O, editors. vol. 1. Oxford: Elsevier; 1999. pp. 1–21. [Google Scholar]
- 2.Wise ML, Croteau R. In: Comprehensive Natural Products Chemistry. Barton D, Nakanishi K, Meth-Cohn O, editors. vol. 2. Oxford: Elsevier; 1999. pp. 97–153. [Google Scholar]
- 3.Clayden J, Greeves N, Warren S, Wothers P. Organic Chemistry. New York: Oxford Univ Press; 2001. [Google Scholar]
- 4.Kahrel Y, Takahashi S, Yamashita S, Koyama T. FEBS J. 2006;273:647–657. doi: 10.1111/j.1742-4658.2005.05097.x. [DOI] [PubMed] [Google Scholar]
- 5.Ohashi H, Hayashi H, Yamada M, Yasue M. Res Bull Fac Agr Gifu Univ. 1987;52:131–139. (Chem Abstr 1989, 110:92049p) [Google Scholar]
- 6.Suzuki S, Umezawa T, Shimada M. J Chem Soc, Perkin Trans. 2001;1:3252–3257. [Google Scholar]
- 7.Yamada T. Bull For For Prod Res Inst. 1998;69 [Google Scholar]
- 8.Imai T, Nomura M. J Wood Sci. 2005;51:537–541. [Google Scholar]
- 9.Yoshida K, Nishiguchi M, Hishiyama S, Kato A, Takahashi K. J Wood Sci. 2006;52:372–375. [Google Scholar]
- 10.Suzuki S, Nakatsubo T, Umezawa T, Shimada M. Chem Commun. 2002;1088 doi: 10.1039/b200217e. [DOI] [PubMed] [Google Scholar]
- 11.Suzuki S, Yamamura M, Shimada M, Umezawa T. Chem Commun. 2004;2838 doi: 10.1039/b409686j. [DOI] [PubMed] [Google Scholar]
- 12.Bostwick DE, Dannenhoffer JM, Skaggs MI, Lister RM, Larkins BA, Thompson GA. Plant Cell. 1992;4:1539–1548. doi: 10.1105/tpc.4.12.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diant S, Clark AM, Zhu Y, Vilaine F, Palauqui J, Kusiak C, Thompson GA. Plant Physiol. 2003;131:114–128. doi: 10.1104/pp.013086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomez G, Pallas V. Mol Plant-Microbe Interact. 2001;14:910–913. doi: 10.1094/MPMI.2001.14.7.910. [DOI] [PubMed] [Google Scholar]
- 15.Owens RA, Blackburn M, Ding B. Mol Plant—Microbe Interact. 2001;14:905–909. doi: 10.1094/MPMI.2001.14.7.905. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki S, Umezawa T. J Wood Sci. 2007;53:273–284. [Google Scholar]
- 17.Beracierta AP, Whiting DA. J Chem Soc Perkin Trans. 1978;1:1257–1263. [Google Scholar]
- 18.Takaku N, Choi DH, Mikame K, Okunishi T, Suzuki SJ, Ohashi H, Umezawa T, Shimada M. J Wood Sci. 2001;47:476–482. [Google Scholar]
- 19.Bugos RC, Chiang VL, Zhang XH, Campbell ER, Podila GK, Campbell WH. BioTechniques. 1995;19:734–737. [PubMed] [Google Scholar]
- 20.Hu WJ. Proc Natl Acad Sci USA. 1998;95:5407–5412. doi: 10.1073/pnas.95.9.5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Nucleic Acid Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saitu N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.