Abstract
Liver receptor homolog 1 (LRH-1), an orphan nuclear receptor, is highly expressed in liver and intestine, where it is implicated in the regulation of cholesterol, bile acid, and steroid hormone homeostasis. Among the proposed LRH-1 target genes in liver are those encoding cholesterol 7α-hydroxylase (CYP7A1) and sterol 12α-hydroxylase (CYP8B1), which catalyze key steps in bile acid synthesis. In vitro studies suggest that LRH-1 may be involved both in stimulating basal CYP7A1 and CYP8B1 transcription and in repressing their expression as part of the nuclear bile acid receptor [farnesoid X receptor (FXR)]-small heterodimer partner signaling cascade, which culminates in small heterodimer partner binding to LRH-1 to repress gene transcription. However, in vivo analysis of LRH-1 actions has been hampered by the embryonic lethality of Lrh-1 knockout mice. To overcome this obstacle, mice were generated in which Lrh-1 was selectively disrupted in either hepatocytes or intestinal epithelium. LRH-1 deficiency in either tissue changed mRNA levels of genes involved in cholesterol and bile acid homeostasis. Surprisingly, LRH-1 deficiency in hepatocytes had no significant effect on basal Cyp7a1 expression or its repression by FXR. Whereas Cyp8b1 repression by FXR was also intact in mice deficient for LRH-1 in hepatocytes, basal CYP8B1 mRNA levels were significantly decreased, and there were corresponding changes in the composition of the bile acid pool. Taken together, these data reveal a broad role for LRH-1 in regulating bile acid homeostasis but demonstrate that LRH-1 is either not involved in the feedback regulation of bile acid synthesis or is compensated for by other factors.
LIVER RECEPTOR HOMOLOG-1 (LRH-1; also called NR5A2, α-fetoprotein transcription factor, FTZ-F1-related factor, and CYP7A1 promoter binding factor) is an orphan member of the nuclear receptor superfamily that is highly expressed in liver, exocrine pancreas, intestine, and ovary (1,2,3; reviewed in4). LRH-1 belongs to a nuclear receptor subfamily that includes steroidogenic factor 1 and fushi tarazu factor 1. Members of this subfamily regulate target gene transcription by binding as monomers to DNA response elements with consensus sequence 5′-PyCAAGGPyCPu-3′ (4). LRH-1 has high constitutive transcriptional activity that can be regulated by phosphorylation (5). Although various phospholipids have been shown to bind in the ligand-binding pocket of human LRH-1 (6,7,8), the physiological relevance of these interactions remains uncertain.
In the intestine, LRH-1 binding sites have been identified in the regulatory regions of a number of genes involved in cholesterol and bile acid homeostasis, including those encoding the ATP-binding cassette (ABC) transporters ABCG5 and ABCG8, the organic solute transporters (OST) α and β, and the ileal apical sodium-dependent bile acid transporter (ASBT; also known as solute carrier family 10, member A2) (9,10,11). LRH-1 is also implicated in the regulation of other intestinal processes including epithelial cell renewal and the synthesis of glucocorticoids (12,13). Heterozygous Lrh-1+/−-mice have decreased epithelial cell proliferation and are susceptible to chemical-induced intestinal inflammation (13,14).
LRH-1 binding sites have also been identified in the regulatory regions of many genes involved in cholesterol and bile acid homeostasis in liver (4). Among these are the genes encoding cholesterol 7α-hydroxylase (CYP7A1), which catalyzes the first and rate-limiting step in bile acid synthesis, and cholesterol 12α-hydroxylase (CYP8B1), which catalyzes a step in the conversion of chenodeoxycholic acid to cholic acid (15). In vitro studies have suggested that LRH-1 has dual effects on CYP7A1 and CYP8B1 expression (16,17). First, it enhances basal transcription of these genes. Second, it is involved in the feedback repression of CYP7A1 and CYP8B1 through a signaling cascade involving the farnesoid X receptor (FXR), a nuclear bile acid receptor, and small heterodimer partner (SHP) (16,17). Activation of FXR by bile acids induces expression of SHP, an atypical orphan nuclear receptor lacking a DNA-binding domain that functions as a strong transcriptional repressor through interactions with other nuclear receptors and transcription factors (18; reviewed in Ref. 19). Because SHP binds efficiently to LRH-1 and inhibits its transcriptional activity in vitro, it is proposed that this interaction causes CYP7A1 and CYP8B1 repression (16,17). Whereas the roles of FXR and SHP in this signaling cascade have been demonstrated in vivo using gene knockout mice (20,21,22), Lrh-1−/− mice die during early embryogenesis, precluding their use in the analysis of LRH-1 function in adult tissues (23). Interestingly, heterozygous Lrh-1+/− mice have elevated CYP7A1 and CYP8B1 mRNA levels (23,24), suggesting that the dominant effect of LRH-1 on Cyp7a1 and Cyp8b1 transcription is the recruitment of SHP, not the induction of their basal activity. LRH-1 overexpression studies in mice have yielded contrasting outcomes: whereas CYP7A1 mRNA levels were increased in LRH-1 transgenic mice (23), overexpression of LRH-1 using an adenoviral delivery system caused a strong decrease in CYP7A1 and CYP8B1 mRNA levels (24). Given these mixed results, the precise role of LRH-1 in the regulation of bile acid synthesis remains unclear.
In this report, we describe the generation and characterization of mice deficient for LRH-1 in either hepatocytes or the intestinal epithelium. These studies demonstrate roles for LRH-1 in regulating bile acid homeostasis in both tissues. However, they also unexpectedly show that LRH-1 is not essential for FXR-mediated feedback repression of bile acid synthesis.
RESULTS
Generation of Tissue-Specific LRH-1-Deficient Mice
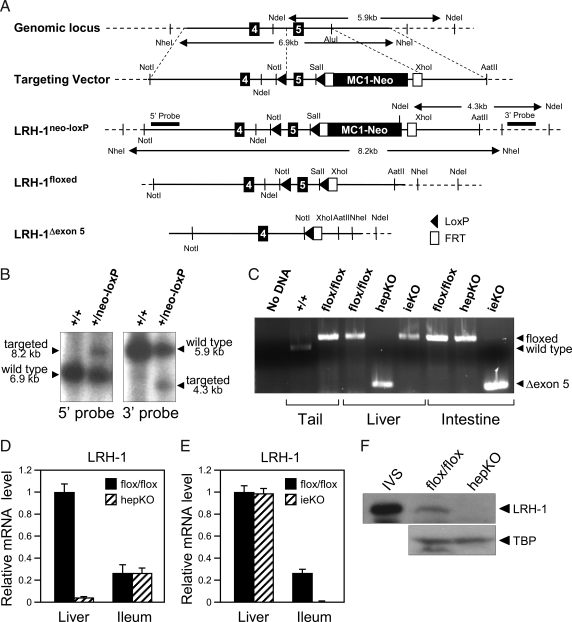
To circumvent the early embryonic lethality caused by complete LRH-1 deficiency, mice were generated in which exon 5 of Lrh-1, which encodes the second zinc finger of the DNA-binding domain, was flanked by loxP sites (Fig. 1, A–C). The resulting Lrh-1flox/flox mice were crossed against either albumin-cre or villin-cre mice to generate knockout mice lacking an intact Lrh-1 gene in either hepatocytes (hepKO) or intestinal epithelium (ieKO), respectively (Fig. 1B). As predicted, real-time quantitative PCR (RT-qPCR) assays performed with primers directed against the floxed region revealed a marked decrease in LRH-1 mRNA in liver and intestinal epithelium of hepKO and ieKO mice, respectively (Fig. 1, D and E). The residual LRH-1 mRNA containing exon 5 in liver of hepKO mice (Fig. 1D) is likely due to Lrh-1 expression in nonhepatocytes. Whereas an LRH-1 transcript was detected by RT-PCR in liver of hepKO mice and intestine of ieKO mice using primers flanking exon 5, cloning and sequencing of this transcript from both tissues revealed a stop codon created by deletion of the floxed region that precludes production of functional protein (supplemental Fig. 1, published as supplemental data on The Endocrine Society’s Journals Online web site at http://mend.endojournals.org). Consistent with the mRNA data, there was a marked decrease in LRH-1 protein in liver extracts prepared from hepKO mice (Fig. 1F). For unknown reasons, we were unable to detect LRH-1 protein in intestinal extracts prepared from either control or ieKO mice (data not shown). Based on these data and the gene expression data shown below, we conclude that LRH-1 is efficiently eliminated in hepatocytes and intestinal epithelium of hepKO and ieKO mice, respectively.
Figure 1.
Generation of Mice Deficient for LRH-1 in Liver or Intestine
A, Schematic representation of the targeting strategy to disrupt Lrh-1. The schematic shows exons 4 and 5 of Lrh-1, the targeting vector, and the targeted Lrh-1 allele both before and after successive removal of the MC1-neocassette with FLPe recombinase and exon 5 with cre recombinase. LoxP sites (solid arrowheads), FLPe recombinase target sites (Frt; open boxes), selected restriction enzyme sites and the 5′- and 3′-probes used for Southern blots are indicated. B, Southern blot analysis of DNA derived from wild-type (+/+) or targeted (+/neo-loxP) ES cells digested with NdeI or NheI and probed with 5′- and 3′-probes. Positions of wild-type and targeted alleles and their sizes are shown. C, PCR analysis of Lrh-1 alleles using DNA prepared from tail, liver, and intestine of Lrh-1+/+, Lrh-1flox/flox, hepKO, or ieKO mice as indicated. A control PCR reaction performed with no DNA is shown on the left. The positions of the PCR products generated by the wild-type Lrh-1 allele and the Lrh-1flox/flox (floxed) and deleted exon 5 Lrh-1 (Δexon 5) alleles are indicated by arrows on the left. D and E, RT-qPCR analysis done using a primer set that recognizes Lrh-1 exon 5 and cDNA from liver and ileum of either Lrh-1flox/flox and hepKO mice (D) or Lrh-1flox/flox and ieKO mice (E). F, Western blot analysis using in vitro synthesized LRH-1 (IVS) or nuclear extracts prepared from livers of Lrh-1flox/flox and hepKO mice and antibodies against LRH-1 or TATA-binding protein (TBP). The LRH-1 band migrates at approximately 62 kDa.
There were no overt abnormalities in the hepKO and ieKO mice nor were there changes in tissue morphology seen in hematoxylin and eosin-stained sections of liver or small intestine (data not shown). Analysis of plasma parameters did not reveal significant changes in triglyceride, free fatty acid, glucose, bile acid or alanine aminotransferase concentrations in hepKO or ieKO mice (Table 1). Plasma cholesterol and aspartate aminotransferase levels were modestly but significantly decreased in hepKO and ieKO mice, respectively (Table 1). Hepatic cholesterol and triglyceride concentrations were not significantly decreased in either the hepKO and ieKO mice (Table 1).
Table 1.
Plasma and Liver Parameters in LRH-1-Deficient Mice
| Parameter | flox/flox | hepKO | P Value | flox/flox | ieKO | P Value |
|---|---|---|---|---|---|---|
| Cholesterol (mg/dl) | 156 ± 4.0 | 125 ± 2.4 | 0.033 | 109 ± 3.5 | 115 ± 6.3 | 0.70 |
| Triglycerides (mg/dl) | 140 ± 6.8 | 122 ± 4.4 | 0.42 | 90.8 ± 2.4 | 97.8 ± 5.2 | 0.57 |
| Free fatty acids (mmol/liter) | 0.71 ± 0.01 | 0.74 ± 0.02 | 0.64 | 0.58 ± 0.01 | 0.57 ± 0.02 | 0.81 |
| Bile acids (μmol/liter) | 5.5 ± 0.5 | 5.3 ± 0.1 | 0.88 | 4.2 ± 0.2 | 4.5 ± 0.2 | 0.61 |
| Glucose (mmol/liter) | 17.1 ± 0.5 | 16.1 ± 0.4 | 0.57 | 17.6 ± 0.3 | 19.7 ± 0.8 | 0.27 |
| Aspartate aminotransferase (U/liter) | 43 ± 0.8 | 37.8 ± 0.5 | 0.055 | 44 ± 0.2 | 41.3 ± 0.4 | 0.033 |
| Alanine aminotransferase (U/liter) | 40.3 ± 1.5 | 38.4 ± 0.9 | 0.69 | 34.6 ± 0.4 | 33.3 ± 0.7 | 0.46 |
| Hepatic cholesterol (mg/g) | 2.8 ± 0.1 | 2.7 ± 0.1 | 0.34 | 2.1 ± 0.15 | 2.5 ± 0.15 | 0.080 |
| Hepatic triglyceride (mg/g) | 10.3 ± 1.7 | 7.0 ± 0.6 | 0.092 | 9.8 ± 1.5 | 9.0 ± 1.5 | 0.73 |
Plasma parameters were measured in mice fasted for 4 h (n = 4–6 mice per group). Data are shown ± sem
Effects of LRH-1 Deficiency in Hepatocytes
The consequences of eliminating LRH-1 in hepatocytes on the expression of a panel of genes involved in bile acid and cholesterol homeostasis were examined by RT-qPCR using mRNA prepared from livers of hepKO and control Lrh-1flox/flox mice. Although CYP7A1 mRNA levels trended upward in hepKO mice, this difference was not statistically significant (Table 2). In contrast, CYP8B1 mRNA levels were decreased approximately 10-fold in the hepKO mice, demonstrating differential effects of LRH-1 on Cyp7a1 and Cyp8b1 expression. Significant decreases were also seen in the mRNAs encoding SHP, FXR, the bile acid biosynthetic enzyme CYP27A1, the scavenger receptor B1 (SCARB1), and the transporters ABCG5, ABCG8, bile salt export pump (BSEP), multidrug resistance transporter 2 (also known as ABCB4), multidrug resistance protein (MRP2; also known as ABCC2), MRP3 (also known as ABCC3), and the sodium-dependent bile acid cotransporter (NTCP; also known as solute carrier family 10, member A1) (Table 2). Thus, LRH-1 deficiency affects expression of a number of genes involved in cholesterol and bile acid homeostasis in liver. There were no significant changes in the levels of mRNAs encoding apolipoprotein A1, CYP7B1, 3-hydroxy-3-methylglutaryl-coenzyme A synthase 1, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, and hepatocyte nuclear factor 4α (HNF4α).
Table 2.
Liver Gene Expression in Mice Deficient for LRH-1 in Hepatocytes
| Gene | hepKO /flox/flox | P Value | Change |
|---|---|---|---|
| ABCG5 | 0.41 ± 0.05 | 0.001 | ↓ |
| ABCG8 | 0.38 ± 0.03 | 0.001 | ↓ |
| APOA1 | 0.88 ± 0.07 | 0.21 | |
| BSEP | 0.71 ± 0.08 | 0.037 | ↓ |
| CYP27A1 | 0.82 ± 0.03 | 0.030 | ↓ |
| CYP7A1 | 1.6 ± 0.2 | 0.14 | |
| CYP7B1 | 1.6 ± 0.4 | 0.15 | |
| CYP8B1 | 0.090 ± 0.012 | 0.002 | ↓ |
| FXR | 0.53 ± 0.10 | 0.031 | ↓ |
| HMGCR | 0.80 ± 0.16 | 0.38 | |
| HMGCS1 | 0.71 ± 0.09 | 0.25 | |
| HNF4α | 0.92 ± 0.07 | 0.61 | |
| MDR2 | 0.46 ± 0.03 | <0.001 | ↓ |
| MRP2 | 0.60 ± 0.05 | <0.001 | ↓ |
| MRP3 | 0.15 ± 0.03 | <0.001 | ↓ |
| NTCP | 0.42 ± 0.03 | <0.001 | ↓ |
| SHP | 0.033 ± 0.006 | <0.001 | ↓ |
| SCARB1 | 0.50 ± 0.02 | <0.001 | ↓ |
Data are shown ± sem (n = 6–7 mice per group). Significant changes are indicated by arrows. HMGCR, HMG-CoA reductase; HMGCS1, HMG-CoA synthase 1.
We also examined the effect of LRH-1 deficiency in hepatocytes on gene expression in ileum. There was a significant increase in FXR mRNA and a decrease in fibroblast growth factor 15 (FGF15) mRNA, which encodes a hormone secreted from the small intestine that is required for FXR-mediated feedback repression of Cyp7a1 (25,26) (Table 3).
Table 3.
Intestinal Gene Expression In Mice Deficient for LRH-1 in Hepatocytes
| Gene | hepKO /flox/flox | P Value | Change |
|---|---|---|---|
| ABCG5 | 1.0 ± 0.03 | 0.63 | |
| ABCG8 | 1.5 ± 0.2 | 0.080 | |
| ASBT | 2.3 ± 0.7 | 0.17 | |
| FGF15 | 0.49 ± 0.13 | 0.047 | ↓ |
| FXR | 2.0 ± 0.2 | 0.002 | ↑ |
| HNF4α | 1.2 ± 0.1 | 0.18 | |
| IBABP | 1.0 ± 0.1 | 0.84 | |
| OSTα | 1.6 ± 0.2 | 0.058 | |
| OSTβ | 0.74 ± 0.02 | 0.15 | |
| SHP | 0.65 ± 0.26 | 0.52 | |
| SCARB1 | 1.8 ± 0.3 | 0.077 |
Data are shown ± sem (n = 6–7 mice per group). Significant changes are indicated by arrows.
Effects of LRH-1 Deficiency in Intestinal Epithelium
We next examined the effect of eliminating LRH-1 in the intestinal epithelium on the expression of genes involved in bile acid and cholesterol homeostasis. mRNA was prepared from the ileal epithelium of ieKO and control Lrh-1flox/flox mice. SHP mRNA was reduced 10-fold in ileum of ieKO mice (Table 4). The absence of LRH-1 also caused significant reductions in ileal bile acid-binding protein (IBABP) and FGF15 mRNA levels (Table 4). These genes have not been previously described as regulated by LRH-1. There were also trends toward decreased expression of Asbt, Ostα, and Ostβ, which have been shown to be regulated directly by LRH-1 in vitro (10,11).
Table 4.
Intestinal Gene Expression in Mice Deficient for LRH-1 in Intestinal Epithelium
| Gene | ieKO /flox/flox | P Value | Change |
|---|---|---|---|
| ABCG5 | 1.2 ± 0.1 | 0.39 | |
| ABCG8 | 1.2 ± 0.5 | 0.49 | |
| ASBT | 0.55 ± 0.14 | 0.073 | |
| FGF15 | 0.49 ± 0.08 | 0.041 | ↓ |
| FXR | 1.2 ± 0.1 | 0.30 | |
| HNF4α | 1.2 ± 0.1 | 0.16 | |
| IBABP | 0.19 ± 0.05 | 0.010 | ↓ |
| OSTα | 0.56 ± 0.09 | 0.098 | |
| OSTβ | 0.60 ± 0.09 | 0.058 | |
| SHP | 0.10 ± 0.04 | 0.002 | ↓ |
| SCARB1 | 0.78 ± 0.18 | 0.37 |
Data are shown ± sem (n = 7–8 mice per group). Significant changes are indicated by arrows.
No significant changes were seen in hepatic gene expression in ieKO mice although there was a strong trend toward decreased FXR mRNA (Table 5). Although there was a 2-fold decrease in FGF15 mRNA in the intestine of ieKO mice, there was no corresponding increase in Cyp7a1 expression (Table 5).
Table 5.
Liver Gene Expression in Mice Deficient for LRH-1 in Intestinal Epithelium
| Gene | ieKO/flox/flox | P Value |
|---|---|---|
| ABCG5 | 1.1 ± 0.2 | 0.75 |
| ABCG8 | 1.0 ± 0.2 | 0.99 |
| APOA1 | 1.0 ± 0.2 | 0.87 |
| BSEP | 1.3 ± 0.1 | 0.062 |
| CYP27A1 | 0.90 ± 0.17 | 0.60 |
| CYP7A1 | 0.75 ± 0.21 | 0.56 |
| CYP7B1 | 1.2 ± 0.4 | 0.69 |
| CYP8B1 | 1.0 ± 0.1 | 0.88 |
| FXR | 0.79 ± 0.11 | 0.053 |
| HMGCR | 1.4 ± 0.5 | 0.48 |
| HMGCS1 | 0.85 ± 0.14 | 0.61 |
| HNF4α | 1.2 ± 0.1 | 0.21 |
| MDR2 | 1.1 ± 0.2 | 0.41 |
| MRP2 | 1.1 ± 0.2 | 0.75 |
| MRP3 | 0.84 ± 0.11 | 0.37 |
| NTCP | 1.1 ± 0.1 | 0.64 |
| SHP | 0.81 ± 0.12 | 0.20 |
| SCARB1 | 1.2 ± 0.2 | 0.38 |
Data are shown ± sem (n = 7–8 mice per group). HMGCR, HMG-CoA reductase; HMGCS1, HMG-CoA synthase 1.
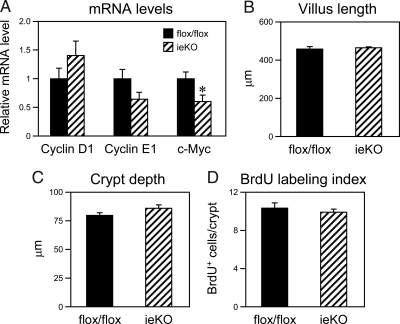
LRH-1 is abundant in the crypts of both the small and large intestine, and Lrh-1+/− mice have reduced intestinal proliferation, decreased crypt depth, and decreased mRNA levels of cyclin D1, cyclin E1, and c-myc (12). Consistent with this previous study, c-myc mRNA levels were decreased in duodenum of ieKO mice, and cyclin E1 mRNA levels trended lower (Fig. 2A). However, there were no significant changes in villus length, crypt depth, bromodeoxyuridine (BrdU) labeling index, or mRNA levels of cyclin D1 in the duodenum of ieKO mice (Fig. 2, A–D). These data raise the possibility that LRH-1 deficiency in both the epithelial and stromal compartments is required to blunt proliferation in the intestine. Alternatively, there may be a compensatory proliferative response in the intestinal epithelium of ieKO mice.
Figure 2.
LRH-1 Deficiency in Intestinal Epithelium Does Not Affect Proliferation in Duodenum
A, mRNA levels of the indicated genes were analyzed by RT-qPCR using intestinal epithelium derived from Lrh-1flox/flox and ieKO mice (n = 7–8 mice per group). *, P < 0.05. B–D, Villus length (B), crypt depth (C), and BrdU labeling index (D) were measured in duodenum of Lrh-1flox/flox and ieKO mice.
Effects of LRH-1 Deficiency on the Bile Acid Pool
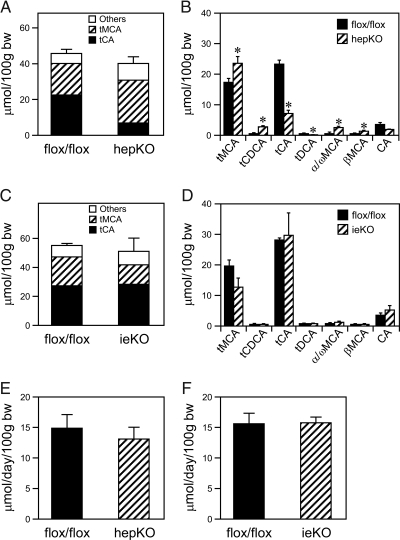
The effect of LRH-1 deficiency on bile acid pool size and composition was analyzed. In hepKO mice, there was no change in the bile acid pool size but a marked change in its composition (Fig. 3, A and B). Compared with control mice, bile from hepKO mice contained significantly higher concentrations of tauromuricholic acids, taurochenodeoxycholic acid, α/ω-muricholic acid, and β-muricholic acid and lower concentrations of taurocholic acid and taurodeoxycholic acid (Fig. 3B). This altered profile is consistent with the decreased CYP8B1 mRNA levels in hepKO mice (Table 2). ieKO mice had no significant changes in either bile acid pool size or composition (Fig. 3, C and D). No change was seen in fecal bile acid excretion in either the hepKO or ieKO mice (Fig. 3, E and F).
Figure 3.
Effects of LRH-1 Deficiency in Liver or Intestine on Bile Acid Pool Size, Composition, and Excretion
A–D, Bile acid pool size and composition were measured in either hepKO (A and B) or ieKO mice (C and D) and corresponding Lrh-1flox/flox mice. tMCA, Tauromuricholic acid; tCDCA, taurochenodeoxycholic acid; tCA, taurocholic acid; tDCA, taurodeoxycholic acid; α/ωMCA, combined α/ω-muricholic acid; βMCA, β-muricholic acid; CA, cholic acid. All bile acid species shown in supplemental Table 1 were measured but only those at concentrations more than 1 μmol/100 g body weight (bw) are shown. *, P < 0.05. E and F, Fecal bile acid excretion was measured in either hepKO (E) or ieKO mice (F) and corresponding Lrh-1flox/flox mice. Data are the mean ± sem (n = 4–6 mice per group).
Effects of Activating FXR on Gene Expression in hepKO and ieKO Mice
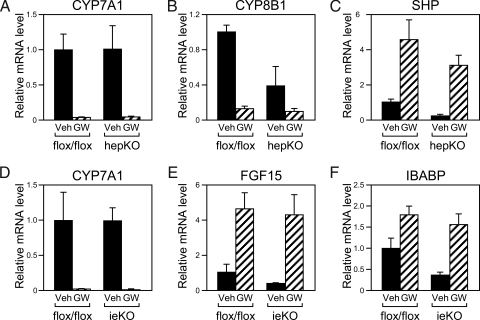
Hepatic LRH-1 is proposed to play a central role in FXR-mediated repression of Cyp7a1 and Cyp8b1 (16,17). To address this hypothesis directly, hepKO and control Lrh-1flox/flox mice were administered the potent, selective FXR agonist GW4064 for 14 h, at which point the mice were killed and CYP7A1 and CYP8B1 mRNA levels were measured by RT-qPCR. Surprisingly, Cyp7a1 was repressed as efficiently in hepKO mice as in control mice (Fig. 4A). Cyp8b1 was also repressed by GW4064 in hepKO mice, although the fold repression was reduced relative to that seen in control mice due to the decrease in basal mRNA levels in the hepKO mice (Fig. 4B). GW4064 also efficiently repressed Cyp7a1 and Cyp8b1 expression in livers of ieKO mice (Fig. 4D and data not shown).
Figure 4.
Effect of FXR Activation on Gene Expression in Tissue-Specific LRH-1 Deficient Mice
hepKO, ieKO, and control Lrh-1flox/flox mice were treated with vehicle (Veh) or GW4064 (GW) for 14 h and RNA prepared from liver and ileum. RT-qPCR was used to measure mRNA levels of CYP7A1 (A and D), CYP8B1 (B), and SHP (C) in liver and FGF15 (E) and IBABP (F) in ileum. Data are the mean ± sem (n = 3–4 mice per group).
The efficient repression of Cyp7a1 by GW4064 in hepKO mice was particularly surprising in light of their low hepatic SHP mRNA levels (Table 2). However, GW4064 administration caused marked increases in hepatic SHP mRNA in hepKO mice (Fig. 4C). Fgf15 and Ibabp were also efficiently induced in ileum of ieKO mice despite their reduced basal expression (Fig. 4, E and F). Thus, although LRH-1 contributes to the basal expression of Shp, Ibabp, and Fgf15, it is not required for their efficient induction by FXR.
DISCUSSION
LRH-1 is implicated in the regulation of many genes involved in cholesterol and bile acid homeostasis (4). On the CYP7A1 gene, LRH-1 is proposed to serve both as a positive-acting basal transcription factor and as a docking site for the transcriptional repressor SHP, which is induced by bile acids through an FXR-dependent mechanism (16,17). The role of SHP in repressing Cyp7a1 has been validated in vivo using Shp knockout mice (21,22). Although LRH-1 was identified in an unbiased screen as a major transcription factor bound to the CYP7A1 promoter (2) and several studies have shown that LRH-1 can regulate CYP7A1 either in vitro or in vivo (16,17,23,24,27), a comprehensive analysis of LRH-1 function in vivo has been hampered by the lack of Lrh-1 knockout mice. In this report, we describe the generation and characterization of mice deficient for LRH-1 in hepatocytes and intestinal epithelium.
A surprising outcome of our studies was that LRH-1 deficiency in hepatocytes had no significant effect on either basal Cyp7a1 expression or its repression by FXR. This was unexpected given that LRH-1 binds to the murine Cyp7a1 promoter as measured by chromatin immunoprecipitation assays (28), and LRH-1 haploinsufficiency or overexpression affects hepatic CYP7A1 mRNA levels in mice (23,24). There are at least two possible explanations for these data. First, LRH-1 may not regulate Cyp7a1 or play a relatively minor role. Alternatively, there may be a redundant factor or compensatory response in the hepKO mice that maintains Cyp7a1 regulation in the absence of LRH-1. One possible candidate for mediating a compensatory response is HNF4α, which binds to the same composite response element in the CYP7A1 promoter as LRH-1 and, like LRH-1, is repressed by SHP in vitro (29,30). In vitro studies have shown that bile acids can down-regulate CYP7A1 transcription by reducing HNF4α activity (31). Moreover, mice deficient for HNF4α in liver have decreased CYP7A1 mRNA and protein levels during the dark cycle (32). Although no significant changes in HNF4α mRNA levels were seen in hepKO mice, additional studies will be needed to determine the role of HNF4α and its relationship with LRH-1 in regulating both basal expression and FXR-mediated repression of CYP7A1. Additional studies will also be required to determine whether our findings in mice are relevant in other species, including humans. There are well-established species differences in the regulation of CYP7A1, especially with regard to its modulation by cholesterol (33,34,35,36,37,38,39,40). Thus, LRH-1 may be important for the regulation of CYP7A1 in other species.
In contrast to Cyp7a1, basal Cyp8b1 expression was significantly reduced in mice deficient for LRH-1 in liver. As predicted, the decrease in Cyp8b1 expression was accompanied by decreased concentrations of taurocholic acid and increased levels of tauromuricholic acids in the bile acid pool. As in the case of Cyp7a1, FXR-mediated repression of Cyp8b1 still occurred in the hepKO mice, although the magnitude of the decrease was reduced. These data demonstrate an important role for LRH-1 in determining the composition of the bile acid pool through effects on Cyp8b1 expression. They also reveal marked differences in the regulation of Cyp7a1 and Cyp8b1. The differential regulation of Cyp7a1 and Cyp8b1 was also recently highlighted by Kim et al. (26), who showed that Cyp7a1 is repressed more efficiently than Cyp8b1 by FGF15.
In the hepKO mice, there were significant decreases in hepatic SHP, CYP27A1, SCARB1, ABCG5, ABCG8, BSEP, multidrug resistance transporter 2, MRP2, MRP3, and sodium-dependent bile acid cotransporter mRNA levels. Several different mechanisms may contribute to their decreased expression. First, LRH-1 may regulate these genes directly. In the case of Shp, Mrp3, Abcg5, Abcg8, Scarb1, and Bsep, sites that bind LRH-1 have been identified in the gene-regulatory regions (9,16,17,28,41,42). The decreased expression of a subset of these genes may be due also to changes in the activity of FXR, which directly regulates the Shp, Bsep, and Mrp2 genes (16,17,43,44). FXR mRNA levels were decreased approximately 2-fold in livers of hepKO mice consistent with the previous finding that LRH-1 binds to the Fxr promoter (28). Furthermore, it was previously shown that cholic acid is an important endogenous agonist for FXR in mice (45). In hepKO mice, the reduction in CYP8B1 mRNA and resulting decrease in cholic acid may contribute to the reduced hepatic expression of genes that are regulated by FXR. Thus, decreases in both FXR protein levels and its endogenous ligands could contribute to the altered expression of FXR target genes in hepKO mice.
LRH-1 also appears to have both direct and indirect effects on gene expression in ileum. In ieKO mice, SHP, IBABP, and FGF15 mRNA levels were significantly decreased in ileum, and OSTα, OSTβ, and ASBT mRNA levels trended lower. Whereas Shp, Ostα, Ostβ, and Asbt have been shown previously to be regulated by LRH-1 in vitro (10,11,16,17), Ibabp and Fgf15 have not. It remains to be determined whether intestinal LRH-1 regulates Ibabp and Fgf15 directly or indirectly. Notably, FGF15 mRNA was also decreased in ileum of mice deficient for LRH-1 in liver. Because FGF15 is regulated strongly by FXR (25,46), this decrease may be due to the aforementioned reduction in FXR agonist activity in the hepKO mice. However, because Shp and Ibabp are also FXR target genes (16,17,47), it is surprising that their mRNA levels were not also significantly decreased in the hepKO mice.
While this paper was in preparation, Mataki et al. (48) reported the characterization of mice in which exons 4 and 5 of Lrh-1 were deleted in hepatocytes using a tamoxifen-inducible cre. Although many of their findings with respect to the effect of LRH-1 deficiency on hepatic gene expression and bile acid composition are similar to ours, they did not investigate the effect of LRH-1 deficiency on feedback regulation of bile acid synthesis nor did they generate mice lacking LRH-1 in the intestinal epithelium. Notably, they reported a significant decrease in the bile acid pool size and a significant increase in fecal bile acid excretion in their LRH-1-deficient mice that were not seen in the current study. Moreover, they did not see the modest decrease in plasma cholesterol that we observed in the hepKO mice. The basis for these differences is not clear but could relate to either the different strategies used to disrupt Lrh-1 or the different background strains. Regarding the decreased plasma cholesterol levels in our hepKO mice, this is a surprising finding in light of the lack of change in CYP7A1 mRNA levels and decreased hepatic expression of ABCG5 and ABGG8, which promote cholesterol secretion into the bile. At present we do not have a molecular explanation for the effect of LRH-1 deficiency on cholesterol homeostasis.
In summary, we show that LRH-1 affects the expression of a number of genes in liver and intestine that are involved in cholesterol and bile acid homeostasis. Among these are several that were not previously linked to LRH-1, including Cyp27a1 in liver and Ibabp and Fgf15 in ileum. These findings reveal a broader role for LRH-1 in regulating bile acid homeostasis than was previously appreciated. We also demonstrate that LRH-1 plays a major role in determining the composition of the bile acid pool through effects on Cyp8b1 expression. Surprisingly, however, LRH-1 is not essential for FXR-mediated feedback regulation of Cyp7a1. Additional studies will be required to determine whether LRH-1 has no role in the feedback regulation of bile acid synthesis or whether there are redundant factors or a compensatory response that maintains Cyp7a1 regulation in the absence of LRH-1.
MATERIALS AND METHODS
Animal Procedures
All animal experiments were approved by the Institutional Animal Care and Research Advisory Committee of the University of Texas Southwestern Medical Center. Animals were housed in a pathogen-free and a temperature-controlled environment with 12-h light, 12-h dark cycles and fed standard irradiated rodent chow (TD7913; Harlan Teklad, Madison, WI) and water ad libitum. All animal experiments were done with 8- to 12-wk-old male mice killed between 1000 and 1200 h. For measurement of bile acid pool size and composition, mice were fasted 4 h before euthanasia. For the intestinal proliferation study, BrdU (100 mg/kg in PBS) was injected ip 2 h before euthanasia. For experiments with GW4064, mice were administrated GW4064 (100 mg/kg, in 1% methylcellulose, 1% Tween 80) or vehicle by oral gavage and killed 14 h later.
Generation of LRH-1 Deficient Mice
High-fidelity PCR amplification of 129SvEv genomic DNA was used to generate an approximately 3.7-kb long arm including exon 4 and parts of introns 3 and 4, an approximately 0.8-kb targeting arm including exon 5 and parts of introns 4 and 5, and an approximately 1.6-kb short arm including intron 5. These fragments were assembled in the pMC1neo PolyA vector (Stratagene, La Jolla, CA) containing a neomycin resistance cassette and loxP and FRT sites. AatII-linearized DNA was electroporated into 129SvEv-derived embryonic stem (ES) cells. ES cells were screened for targeted recombination by Southern blot analysis using 5′- and 3′-probes and DNA digested with NheI or NdeI, respectively. Probes were generated by PCR amplification using the following primers: 5′-probe forward primer, CCATGGTGGATTTGGTTCTC; reverse primer, TGTAGCATAAGTTGGTCCGG; 3′-probe forward primer, GGCTGTTCTTGCTACTTGAG; reverse primer, CAGGTGCACTGTATGTAGCT. Two independently derived ES cell clones were injected into C57BL/6J blastocysts to produce chimeric mice that transmitted the modified Lrh-1 locus. The neomycin resistance cassette was removed by crossing Lrh-1flox/+ mice with R26::FLPe transgenic mice (49). Hepatocyte and intestinal epithelium-specific LRH-1 deficiency was achieved by breeding Lrh-1flox/flox mice with albumin-cre (50) or villin-cre (51) transgenic mice. hepKO and ieKO mice and their corresponding control Lrh-1flox/flox mice were maintained on mixed C57BL6/129 backgrounds. To confirm homologous recombination and tissue-specific deletion of exon 5 of Lrh-1, genomic DNA was extracted from tail, liver, and intestine, and PCR analysis was performed using the following primers: forward primer 1, CGATGTCCCTACTGTCGA; reverse primer 1, CGCAGCATTCTTCGGCAG; forward primer 2, CATAAGGGCTCAGTGGCAC; reverse primer 2, CTTCACTGGCTGCCAAGCTG.
Morphometry and Proliferation Measurements
The small intestine was cut into three segments of equal length representing the duodenum, jejunum, and ileum. Each section was cut transversely, fixed with 10% formalin, paraffin embedded, sectioned, and stained with hematoxylin and eosin. For villus and crypt measurements, duodenum sections were photographed using a Nikon DXM1200F camera and Nikon Eclipse 80i microscope at ×X and ×20 magnification, and 10 villi and crypts per mouse (n =11–12 mice per group) were measured using MetaVue software (Meta Imaging Series 6.1; Lewisville, TX). To determine proliferation indices, BrdU incorporation was detected by immunohistochemistry. Sections were deparaffinized and rehydrated. DNA was denatured by 2 n HCl for 1 h at 37 C and neutralized by immersing sections two times in 0.1 m borate buffer (pH 8.5) for 5 min each. Sections were permeabilized by 0.3% Triton for 5 min, incubated in 1.5% horse serum for 30 min, and incubated with antimouse BrdU antibody (Roche Molecular Biochemicals, Indianapolis, IN) at a 1:25 dilution overnight at 4 C. Biotinylated horse antimouse IgG was added at a dilution 1:200 for 30 min at room temperature followed by fluorescein isothiocyanate-conjugated streptavidin at a 1:50 dilution for 30 min at room temperature. Ten crypts per mouse were analyzed for BrdU incorporation (n =4 mice per group).
Liquid Chromatography/Mass Spectrometry Analysis of Bile Acids
Gall bladder, liver, and intestines were removed and placed into 50 ml of EtOH. The tissues in solution were spiked with 50 μg of chenodeoxycholic acid-D4 (C/D/N Isotopes; Pointe-Claire, Canada) and extracted. Samples were minced and boiled for 1 h to reduce the EtOH volume to approximately 30 ml. Samples were filtered through no. 2 Whatman paper, and the volume was brought to 50 ml using a volumetric flask. One milliliter of the extract was centrifuge filtered using a polyvinylidine difluoride membrane (Millipore Corp., Bedford, MA) before analysis. Bile acids were quantified by liquid chromatography/mass spectrometry (Agilent Technologies, Palo Alto, CA) with electrospray ionization in negative ion mode by modification of a published procedure (52). Briefly, samples were loaded onto a precolumn (Zorbax C8, 4.6 × 12.5 mm, 5 μm; Agilent) at 1 ml/min for 1 min with water containing 5 mm NH4Ac and then backflushed onto the analytical column at 0.4 ml/min (Eclipse XDB-C18, 4.6 × 50 mm, 5 μm; Agilent). The mobile phase consisted of methanol/0.024% formic acid (A) and water/10 mm NH4Ac/0.024% formic acid (B). The following gradient was run for a total run time of 30 min: 0–15 min, 70–80% (A); 15–17 min, 80% (A); 17–20 min, 80–95% (A); 20–26 min, 95% (A). MS parameters were set as follows: gas temperature, 350 C; nebulizer pressure, 30 pounds per square inch gauge; drying gas (nitrogen), 12 liters/min; capillary voltage, 4000 V; fragmentor voltage, 200 V. Under these conditions it is not possible to resolve the closely related taurine-cojugated trihydroxy bile acids, α-, β-, and ω-muricholic acid (MCA). Likewise, unconjugated α- and ω-MCA cannot be resolved. For this reason, the pair of unconjugated MCAs is reported as α/ω-MCA and the three taurine-conjugated bile acids are reported as tauromuricholic acid. Selective ion monitoring was used to detect the conjugated and unconjugated bile acids (supplemental Table 1, published as supplemental data on The Endocrine Society’s Journals Online web site at http://mend.endojournals.org). Quantification was performed based on peak areas using external calibration curves of standards prepared in methanol. CDCA-D4 was used to calculate the recovery of bile acids after extraction relative to a blank control.
Fecal Bile Acid Excretion
Fecal excrement was collected from individually housed mice over a continuous 72-h period. Bile acids were extracted and quantified as previously described (53).
RT-qPCR Analysis
Total RNA was extracted from liver and scraped intestine using RNA STAT-60 (Tel-Test, Inc., Friendswood, TX). After DNase I (Roche Molecular Biochemicals) treatment, RNA was reverse transcribed into cDNA with random hexamers using the SuperScript II First-Strand Synthesis System (Invitrogen, Carlsbad, CA). Primers for each gene were designed using Primer Express Software (Applied Biosystems, Foster City, CA) and were validated as previously described (54). Primer sequences are shown in supplemental Table 2. RT-qPCR reactions contained 25 ng of cDNA, 150 nm of each primer, and 5 μl of SYBR GreenER PCR Master Mix (Invitrogen) and were carried out in triplicate using an Applied Biosystems Prism 7900HT instrument. Relative mRNA levels were calculated using either the comparative CT or standard curve methods normalized to cyclophilin or 18S RNA, respectively.
Immunoblot Analysis
Nuclear extracts were prepared from liver using hypotonic buffer [10 mm HEPES (pH 7.5), 10 mm KCl, 1.5 mm MgCl2, 1.5 mm dithiothreitol, Complete Protease Inhibitor Cocktail (Roche Diagnostic)] and nuclear lysis buffer (50 mm Tris-HCl, pH 7.5; 150 mm NaCl; 0.1% Triton X-100; Complete Protease Inhibitor Cocktail). Nuclear extract (50 μg) was resolved on a 10% sodium dodecyl sulfate-polyacrylamide gel and transferred to a polyvinylidine difluoride membrane (Amersham Pharmacia Biotech, Piscataway, NJ). The membrane was probed with guinea pig polyclonal anti-LRH-1 antibody against mouse LRH-1 residues 318–560. Recombinant LRH-1 protein was generated as previously described (55). The LRH-1 antibody was used at a dilution of 1:500 at 4 C overnight followed by a secondary horseradish peroxidase-conjugated antibody at a dilution of 1:10,000. The membrane was reprobed with antibody against TATA-binding protein (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) as a loading control. Proteins were detected by Super Signal West Femto chemiluminescense substrate (Pierce Chemical Co., Rockford, IL).
Plasma and Liver Parameters
Vena cava blood was collected and transferred into Li-heparin tubes (Sarstedt, Newton, NC). Samples were centrifuged at 4000 rpm at 4 C for 10 min, and total plasma cholesterol, triglycerides, glucose, aspartate aminotransferase, and alanine aminotransferase were measured using a Vitros 250 automated analyzer (Johnson & Johnson, New Brunswick, NJ). Plasma free fatty acids were measured with colorimetric assay kits (Roche). Total plasma bile acids were measured using enzymatic assay kits (Diagnostic Chemicals Ltd., Charlottetown, Prince Edward Island, Canada). Hepatic cholesterol and triglyceride concentrations were measured as previously described (56) except that Triton X-100 was used in place of Triton X-114 and the kits used to measure cholesterol and triglyceride were from Roche and Trinity Biotech (Jamestown, NY), respectively.
Statistical Analyses
Minitab Release 14 software (Minitab, Inc., State College, PA) was used. Values are expressed as mean ± sem. Significant differences of two groups were evaluated using a two-tailed, unpaired Student’s t-test. Multiple groups were analyzed by one-way ANOVA followed by Fisher’s least significant difference test.
Supplementary Material
Acknowledgments
We thank Dr. James Richardson and the Molecular Pathology Core Laboratory for their assistance with histology; Dr. Raymond MacDonald for the LRH-1 antisera; Drs. Patrick Maloney and Tim Willson for GW4064; and Dr. Takeshi Inagaki, Angie Bookout, Laura Molyneux, and other members of the Kliewer/Mangelsdorf laboratory for assistance with mouse experiments.
Footnotes
This work was supported by National Institutes of Health Grant DK067158 (to S.A.K.) and grants from the Welch Foundation (to D.J.M. and S.A.K.) and the Howard Hughes Medical Institute (to D.R.S., C.L.C., Y.Z., D.J.M.). D.J.M. is an Investigator of the Howard Hughes Medical Institute.
Disclosure Summary: Y.K.L., D.R.S., C.L.C., M.C., L.P., Y.Z., and R.E.H. have nothing to declare. B.G. is employed by, and has equity in, GlaxoSmithKline. D.J.M. owns equity in Exelixis and Ligand and has received consulting fees from Kalypsys, Exelixis, Daiichi-Sankyo, and Bay City Capital and lecture fees from Bristol-Myers Squibb, Gene Logic, Wyeth, and ISIS. S.A.K. owns equity in Intercept and Intekrin and has received consulting fees from Intekrin, GlaxoSmithKline, and Daiichi-Sankyo and lecture fees from Daiichi-Sankyo and Merck.
First Published Online March 6, 2008
Abbreviations: ABC, ATP-binding cassette; ASBT, apical sodium-dependent bile acid transporter; BrdU, bromodeoxyuridine; BSEP, bile salt export pump; CYP7A1, cholesterol 7α-hydroxylase; CYP8B1, sterol 12α-hydroxylase; ES, embryonic stem; FGF, fibroblast growth factor; FXR, farnesoid X receptor; hepKO, hepatocyte knockout; HNF4α, hepatocyte nuclear factor 4α; IBABP, ileal bile acid-binding protein; ieKO, intestinal epithelium knockout; LRH-1, liver receptor homolog-1; MCA, muricholic acid; MRP, multidrug resistance protein; OST, organic solute transporter; RT-qPCR, real-time quantitative PCR; SCARB1, scavenger receptor B1; SHP, small heterodimer partner.
References
- Galarneau L, Pare JF, Allard D, Hamel D, Levesque L, Tugwood JD, Green S, Belanger L 1996 The α1-fetoprotein locus is activated by a nuclear receptor of the Drosophila FTZ-F1 family. Mol Cell Biol 16:3853–3865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta M, Ku S, Brown C, Okamoto AY, Shan B 1999 CPF: an orphan nuclear receptor that regulates liver-specific expression of the human cholesterol 7α-hydroxylase gene. Proc Natl Acad Sci USA 96:6660–6665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinger-Ziegelbauer H, Hihi AK, Laudet V, Keller H, Wahli W, Dreyer C 1994 FTZ-F1-related orphan receptors in Xenopus laevis: transcriptional regulators differentially expressed during early embryogenesis. Mol Cell Biol 14:2786–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayard E, Auwerx J, Schoonjans K 2004 LRH-1: an orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends Cell Biol 14:250–260 [DOI] [PubMed] [Google Scholar]
- Lee YK, Choi YH, Chua S, Park YJ, Moore DD 2006 Phosphorylation of the hinge domain of the nuclear hormone receptor LRH-1 stimulates transactivation. J Biol Chem 281:7850–7855 [DOI] [PubMed] [Google Scholar]
- Krylova IN, Sablin EP, Moore J, Xu RX, Waitt GM, MacKay JA, Juzumiene D, Bynum JM, Madauss K, Montana V, Lebedeva L, Suzawa M, Williams JD, Williams SP, Guy RK, Thornton JW, Fletterick RJ, Willson TM, Ingraham HA 2005 Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell 120:343–355 [DOI] [PubMed] [Google Scholar]
- Ortlund EA, Lee Y, Solomon IH, Hager JM, Safi R, Choi Y, Guan Z, Tripathy A, Raetz CR, McDonnell DP, Moore DD, Redinbo MR 2005 Modulation of human nuclear receptor LRH-1 activity by phospholipids and SHP. Nat Struct Mol Biol 12:357–363 [DOI] [PubMed] [Google Scholar]
- Wang W, Zhang C, Marimuthu A, Krupka HI, Tabrizizad M, Shelloe R, Mehra U, Eng K, Nguyen H, Settachatgul C, Powell B, Milburn MV, West BL 2005 The crystal structures of human steroidogenic factor-1 and liver receptor homologue-1. Proc Natl Acad Sci USA 102:7505–7510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman LA, Kennedy A, Wu J, Bark S, Remaley AT, Santamarina-Fojo S, Brewer Jr HB 2004 The orphan nuclear receptor LRH-1 activates the ABCG5/ABCG8 intergenic promoter. J Lipid Res 45:1197–1206 [DOI] [PubMed] [Google Scholar]
- Frankenberg T, Rao A, Chen F, Haywood J, Shneider BL, Dawson PA 2006 Regulation of the mouse organic solute transporter α-β, Ostα-Ostβ, by bile acids. Am J Physiol Gastrointest Liver Physiol 290:G912–G922 [DOI] [PubMed] [Google Scholar]
- Chen F, Ma L, Dawson PA, Sinal CJ, Sehayek E, Gonzalez FJ, Breslow J, Ananthanarayanan M, Shneider BL 2003 Liver receptor homologue-1 mediates species- and cell line-specific bile acid-dependent negative feedback regulation of the apical sodium-dependent bile acid transporter. J Biol Chem 278:19909–19916 [DOI] [PubMed] [Google Scholar]
- Botrugno OA, Fayard E, Annicotte JS, Haby C, Brennan T, Wendling O, Tanaka T, Kodama T, Thomas W, Auwerx J, Schoonjans K 2004 Synergy between LRH-1 and β-catenin induces G1 cyclin-mediated cell proliferation. Mol Cell 15:499–509 [DOI] [PubMed] [Google Scholar]
- Mueller M, Cima I, Noti M, Fuhrer A, Jakob S, Dubuquoy L, Schoonjans K, Brunner T 2006 The nuclear receptor LRH-1 critically regulates extra-adrenal glucocorticoid synthesis in the intestine. J Exp Med 203:2057–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste A, Dubuquoy L, Barnouin R, Annicotte JS, Magnier B, Notti M, Corazza N, Antal MC, Metzger D, Desreumaux P, Brunner T, Auwerx J, Schoonjans K 2007 LRH-1-mediated glucocorticoid synthesis in enterocytes protects against inflammatory bowel disease. Proc Natl Acad Sci USA 104:13098–13103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DW 2003 The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem 72:137–174 [DOI] [PubMed] [Google Scholar]
- Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, Galardi C, Wilson JG, Lewis MC, Roth ME, Maloney PR, Willson TM, Kliewer SA 2000 A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell 6:517–526 [DOI] [PubMed] [Google Scholar]
- Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, Mangelsdorf DJ 2000 Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell 6:507–515 [DOI] [PubMed] [Google Scholar]
- Seol W, Choi HS, Moore DD 1996 An orphan nuclear hormone receptor that lacks a DNA binding domain and heterodimerizes with other receptors. Science 272:1336–1339 [DOI] [PubMed] [Google Scholar]
- Lee YS, Chanda D, Sim J, Park YY, Choi HS 2007 Structure and function of the atypical orphan nuclear receptor small heterodimer partner. Int Rev Cytol 261:117–158 [DOI] [PubMed] [Google Scholar]
- Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, Gonzalez FJ 2000 Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell 102:731–744 [DOI] [PubMed] [Google Scholar]
- Kerr TA, Saeki S, Schneider M, Schaefer K, Berdy S, Redder T, Shan B, Russell DW, Schwarz M 2002 Loss of nuclear receptor SHP impairs but does not eliminate negative feedback regulation of bile acid synthesis. Dev Cell 2:713–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lee YK, Bundman D, Han Y, Thevananther S, Kim CS, Chua SS, Wei P, Heyman RA, Karin M, Moore DD 2002 Redundant pathways for negative feedback regulation of bile acid production. Dev Cell 2:721–731 [DOI] [PubMed] [Google Scholar]
- Pare JF, Malenfant D, Courtemanche C, Jacob-Wagner M, Roy S, Allard D, Belanger L 2004 The fetoprotein transcription factor (FTF) gene is essential to embryogenesis and cholesterol homeostasis, and regulated by a DR4 element. J Biol Chem 279:21206–21216 [DOI] [PubMed] [Google Scholar]
- del Castillo-Olivares A, Campos JA, Pandak WM, Gil G 2004 The role of α1-fetoprotein transcription factor/LRH-1 in bile acid biosynthesis: a known nuclear receptor activator that can act as a suppressor of bile acid biosynthesis. J Biol Chem 279:16813–16821 [DOI] [PubMed] [Google Scholar]
- Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG, Luo G, Jones SA, Goodwin B, Richardson JA, Gerard RD, Repa JJ, Mangelsdorf DJ, Kliewer SA 2005 Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab 2:217–225 [DOI] [PubMed] [Google Scholar]
- Kim I, Ahn SH, Inagaki T, Choi M, Ito S, Guo GL, Kliewer SA, Gonzalez FJ 2007 Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine. J Lipid Res 48:2664–2672 [DOI] [PubMed] [Google Scholar]
- del Castillo-Olivares A, Gil G 2000 Role of FXR and FTF in bile acid-mediated suppression of cholesterol 7α-hydroxylase transcription. Nucleic Acids Res 28:3587–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulias K, Katrakili N, Bamberg K, Underhill P, Greenfield A, Talianidis I 2005 Regulation of hepatic metabolic pathways by the orphan nuclear receptor SHP. EMBO J 24:2624–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crestani M, Sadeghpour A, Stroup D, Galli G, Chiang JY 1998 Transcriptional activation of the cholesterol 7α-hydroxylase gene (CYP7A) by nuclear hormone receptors. J Lipid Res 39:2192–2200 [PubMed] [Google Scholar]
- Lee YK, Dell H, Dowhan DH, Hadzopoulou-Cladaras M, Moore DD 2000 The orphan nuclear receptor SHP inhibits hepatocyte nuclear factor 4 and retinoid X receptor transactivation: two mechanisms for repression. Mol Cell Biol 20:187–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Fabiani E, Mitro N, Anzulovich AC, Pinelli A, Galli G, Crestani M 2001 The negative effects of bile acids and tumor necrosis factor-α on the transcription of cholesterol 7α-hydroxylase gene (CYP7A1) converge to hepatic nuclear factor-4: a novel mechanism of feedback regulation of bile acid synthesis mediated by nuclear receptors. J Biol Chem 276:30708–30716 [DOI] [PubMed] [Google Scholar]
- Inoue Y, Yu AM, Yim SH, Ma X, Krausz KW, Inoue J, Xiang CC, Brownstein MJ, Eggertsen G, Bjorkhem I, Gonzalez FJ 2006 Regulation of bile acid biosynthesis by hepatocyte nuclear factor 4α. J Lipid Res 47:215–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agellon LB, Drover VA, Cheema SK, Gbaguidi GF, Walsh A 2002 Dietary cholesterol fails to stimulate the human cholesterol 7α-hydroxylase gene (CYP7A1) in transgenic mice. J Biol Chem 277:20131–20134 [DOI] [PubMed] [Google Scholar]
- Chiang JY, Kimmel R, Stroup D 2001 Regulation of cholesterol 7α-hydroxylase gene (CYP7A1) transcription by the liver orphan receptor (LXRα). Gene 262:257–265 [DOI] [PubMed] [Google Scholar]
- Goodwin B, Watson MA, Kim H, Miao J, Kemper JK, Kliewer SA 2003 Differential regulation of rat and human CYP7A1 by the nuclear oxysterol receptor liver X receptor-α. Mol Endocrinol 17:386–394 [DOI] [PubMed] [Google Scholar]
- Horton JD, Cuthbert JA, Spady DK 1995 Regulation of hepatic 7α-hydroxylase expression and response to dietary cholesterol in the rat and hamster. J Biol Chem 270:5381–5387 [DOI] [PubMed] [Google Scholar]
- Jelinek DF, Andersson S, Slaughter CA, Russell DW 1990 Cloning and regulation of cholesterol 7α-hydroxylase, the rate-limiting enzyme in bile acid biosynthesis. J Biol Chem 265:8190–8197 [PMC free article] [PubMed] [Google Scholar]
- Pandak WM, Li YC, Chiang JY, Studer EJ, Gurley EC, Heuman DM, Vlahcevic ZR, Hylemon PB 1991 Regulation of cholesterol 7α-hydroxylase mRNA and transcriptional activity by taurocholate and cholesterol in the chronic biliary diverted rat. J Biol Chem 266:3416–3421 [PubMed] [Google Scholar]
- Rudel L, Deckelman C, Wilson M, Scobey M, Anderson R 1994 Dietary cholesterol and downregulation of cholesterol 7α-hydroxylase and cholesterol absorption in African green monkeys. J Clin Invest 93:2463–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Salen G, Shefer S, Ness GC, Nguyen LB, Parker TS, Chen TS, Zhao Z, Donnelly TM, Tint GS 1995 Unexpected inhibition of cholesterol 7α-hydroxylase by cholesterol in New Zealand white and Watanabe heritable hyperlipidemic rabbits. J Clin Invest 95:1497–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inokuchi A, Hinoshita E, Iwamoto Y, Kohno K, Kuwano M, Uchiumi T 2001 Enhanced expression of the human multidrug resistance protein 3 by bile salt in human enterocytes. A transcriptional control of a plausible bile acid transporter. J Biol Chem 276:46822–46829 [DOI] [PubMed] [Google Scholar]
- Schoonjans K, Annicotte JS, Huby T, Botrugno OA, Fayard E, Ueda Y, Chapman J, Auwerx J 2002 Liver receptor homolog 1 controls the expression of the scavenger receptor class B type I. EMBO Rep 3:1181–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananthanarayanan M, Balasubramanian N, Makishima M, Mangelsdorf DJ, Suchy FJ 2001 Human bile salt export pump promoter is transactivated by the farnesoid X receptor/bile acid receptor. J Biol Chem 276:28857–28865 [DOI] [PubMed] [Google Scholar]
- Kast HR, Goodwin B, Tarr PT, Jones SA, Anisfeld AM, Stoltz CM, Tontonoz P, Kliewer S, Willson TM, Edwards PA 2002 Regulation of multidrug resistance-associated protein 2 (ABCC2) by the nuclear receptors pregnane X receptor, farnesoid X-activated receptor, and constitutive androstane receptor. J Biol Chem 277:2908–2915 [DOI] [PubMed] [Google Scholar]
- Li-Hawkins J, Gafvels M, Olin M, Lund EG, Andersson U, Schuster G, Bjorkhem I, Russell DW, Eggertsen G 2002 Cholic acid mediates negative feedback regulation of bile acid synthesis in mice. J Clin Invest 110:1191–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Pircher PC, Schulman IG, Westin SK 2005 Regulation of complement C3 expression by the bile acid receptor FXR. J Biol Chem 280:7427–7434 [DOI] [PubMed] [Google Scholar]
- Grober J, Zaghini I, Fujii H, Jones SA, Kliewer SA, Willson TM, Ono T, Besnard P 1999 Identification of a bile acid-responsive element in the human ileal bile acid-binding protein gene. Involvement of the farnesoid X receptor/9-cis-retinoic acid receptor heterodimer. J Biol Chem 274:29749–29754 [DOI] [PubMed] [Google Scholar]
- Mataki C, Magnier BC, Houten SM, Annicotte JS, Argmann C, Thomas C, Overmars H, Kulik W, Metzger D, Auwerx J, Schoonjans K 2007 Compromised intestinal lipid absorption in mice with a liver-specific deficiency of liver receptor homolog 1. Mol Cell Biol 27:8330–8339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley FW, Soriano P, Steffen LS, Dymecki SM 2000 Widespread recombinase expression using FLPeR (flipper) mice. Genesis 28:106–110 [PubMed] [Google Scholar]
- Postic C, Shiota M, Niswender KD, Jetton TL, Chen Y, Moates JM, Shelton KD, Lindner J, Cherrington AD, Magnuson MA 1999 Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic β cell-specific gene knock-outs using Cre recombinase. J Biol Chem 274:305–315 [DOI] [PubMed] [Google Scholar]
- Madison BB, Dunbar L, Qiao XT, Braunstein K, Braunstein E, Gumucio DL 2002 Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J Biol Chem 277:33275–33283 [DOI] [PubMed] [Google Scholar]
- Burkard I, von Eckardstein A, Rentsch KM 2005 Differentiated quantification of human bile acids in serum by high-performance liquid chromatography-tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 826:147–159 [DOI] [PubMed] [Google Scholar]
- Turley SD, Schwarz M, Spady DK, Dietschy JM 1998 Gender-related differences in bile acid and sterol metabolism in outbred CD-1 mice fed low- and high-cholesterol diets. Hepatology 28:1088–1094 [DOI] [PubMed] [Google Scholar]
- Bookout AL, Mangelsdorf DJ 2003 Quantitative real-time PCR protocol for analysis of nuclear receptor signaling pathways. Nucl Recept Signal 1:e012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Choi M, Suino K, Kovach A, Daugherty J, Kliewer SA, Xu HE 2005 Structural and biochemical basis for selective repression of the orphan nuclear receptor liver receptor homolog 1 by small heterodimer partner. Proc Natl Acad Sci USA 102:9505–9510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaany NY, Gauthier KC, Zavacki AM, Mammen PP, Kitazume T, Peterson JA, Horton JD, Garry DJ, Bianco AC, Mangelsdorf DJ 2005 LXRs regulate the balance between fat storage and oxidation. Cell Metab 1:231–244 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.