Abstract
Poliovirus (PV) causes a drastic inhibition of cellular cap-dependant protein synthesis due to the cleavage of translation factors eukaryotic initiation factor 4G (eIF4G) and poly (A) binding protein (PABP). Only about half of cellular PABP is cleaved by viral 2A and 3C proteinases during infection. We have investigated PABP cleavage determinants that regulate this partial cleavage. PABP cleavage kinetics analyses indicate that PABP exists in multiple conformations, some of which are resistant to 3Cpro or 2Apro cleavage and can be modulated by reducing potential. Cleavage reactions containing a panel of PABP-binding proteins revealed that eukaryotic release factor 3 (eRF3) and PABP-interacting protein 2 (Paip2) modulate and interfere with the cleavage susceptibility of PABP, whereas all other PABP-binding proteins tested do not. We show that PABP on cellular polysomes is cleaved only by 3Cpro and that Paip2 does not sediment with polysomes. Also, viral polysomes contained only full length PABP, however, cellular or viral ribosomes were equally susceptible to 3Cpro cleavage in vitro. Finally, we determined that precursor 3CD and mature 3Cpro have equivalent cleavage activity on purified PABP, but only 3Cpro cleavage activity was stimulated by PABP binding viral RNA. The results further elucidate complex mechanisms where multiple inherent PABP conformations and protein and RNA interactions both serve to differentially regulate PABP cleavage by 3CD, 3Cpro and 2Apro.
Keywords: poly(A)-binding protein, PABP, poliovirus, Paip2, eRF3, proteinase
Introduction
Poliovirus (PV) is the prototypic member of the Picornaviridae family. It contains a 7.5 kb positive single stranded RNA genome and translation of the poliovirus genome during viral infection generates two mature viral proteinases, 2A and 3C proteinases (2Apro and 3Cpro), and a proteinase-active polypeptide precursor (3CD). These viral proteinases target a wide variety of proteins in the infected cell with many of these targets being cleaved to completion. During poliovirus infection multiple cellular processes are disrupted; most notably inhibition of cap-dependent host protein synthesis through the cleavage of eukaryotic initiation factor 4G I and II (eIF4G -I and -II) by 2Apro and poly(A)-binding protein (PABP) by 2Apro and 3Cpro (Etchison et al., 1982; Joachims et al., 1999; Kuyumcu-Martinez et al., 2002; Kuyumcu-Martinez et al., 2004b; Lamphear et al., 1993). PABP and eIF4G both possess RNA-binding capabilities and act as scaffold proteins, supporting protein-protein interactions with multiple translation factors. While eIF4G is rapidly and completely cleaved by cellular- and poliovirus 2A proteinases (Bovee et al., 1998; Krausslich et al., 1987; Zamora et al., 2002), only about half of the total PABP in the cell is by cleaved late infection, though this is preferentially PABP associated with the translational machinery (Kuyumcu-Martinez et al. 2002). The molecular determinants that regulated PABP cleavage are unknown.
PABP is a highly abundant cytoplasmic protein, ~4μM in HeLa cells, suggesting a threefold excess of PABP protein over potential mRNA binding sites (Gorlach et al., 1994). Besides now being recognized as a translation initiation factor (Kahvejian et al., 2005), PABP has also been implicated in mRNA maturation, export and mRNA stability (Dehlin et al., 2000; Gorlach et al., 1994; Wormington et al., 1996). PABP is broadly comprised of two functional domains, an N-terminal domain with four RNA Recognition Motifs (RRMs) and a C-terminal domain (PABP-CTD) containing a proline-rich linker region tied to a globular protein-binding domain (PABC). The four RRMs at the N-terminus each display different RNA binding affinities and specificities. RRM1 and RRM2 play a pivotal role in poly(A) binding, while RRM3 and RRM4 have relatively higher binding affinity to A-rich sequences interspersed with other nucleotides (Deo et al., 1999; Khanam et al., 2006; Sladic et al., 2004). The PABP-CTD contains an undefined homo-oligomerization domain and the ~70 amino acid conserved C-terminal PABC motif bears binding motifs for multiple proteins.
PABP associates with a wide variety of proteins. Several viral proteins, including turnip mosaic virus Vpg-Pro polypeptide (Leonard et al., 2004), herpes simplex virus ICP27 (Fontaine-Rodriguez et al., 2004), Kaposi’s sarcoma-associated herpesvirus K10/10.1 protein (Kanno et al., 2006) and poliovirus 3CD polypeptide (Herold and Andino, 2001) interact with PABP to possibly regulate viral or cellular processes. Among cellular proteins, BRCA1, eIF4G and Upstream of N-Ras (unr) associate with different regions within the N-terminus of PABP (Chang et al., 2004; Dizin et al., 2006; Imataka et al., 1998). Two different PABP Associating Motifs, termed PAM1 and PAM2 (Roy et al., 2002), have been identified in a wide array of eukaryotic proteins (Albrecht and Lengauer, 2004), mediating their interaction with the PABC domain. Proteins containing PAM motifs include eIF4B, eukaryotic Release Factor 3 (eRF3), Poly r(C) Binding Protein 2 (PCBP2), Apc5, Transducer of erbB2 (Tob) and ataxin-2 (Herold and Andino, 2001; Hoshino et al., 1999; Koloteva-Levine et al., 2004; Le et al., 1997; Okochi et al., 2005; Satterfield and Pallanck, 2006). Additionally, PABP has two binding motifs for the PABP-Interacting Proteins 1 and 2 (Paip1/2), one near the N-terminus and one within the PABC (Craig et al., 1998; Khaleghpour et al., 2001a; Khaleghpour et al., 2001b; Roy et al., 2002). With the extensive list of known and predicted PABP binding partners (Albrecht and Lengauer, 2004), it can only be expected that many of these interactions are highly dynamic and dependent on the state of the cell and that PABP pools participate in several distinct types of protein/RNA complexes at any time.
The PABP-CTD is targeted for proteolytic cleavage by enteroviral proteinases, and contains one cleavage site for 2A proteinase and two main cleavage sites for 3C proteinase, all within the proline-rich linker domain. A third poliovirus proteinase, 3CD, the precursor for the mature form of 3C proteinase, shares substantial substrate specificity with 3C proteinase, yet, it is unknown if 3CD can also target PABP for cleavage. While it has been shown that polioviral proteinases preferentially target PABP molecules associated with crude cell fractions containing translation components and PABP bound to poly(A) RNA (Kuyumcu-Martinez et al., 2002), it is unclear why the majority of PABP in cells is not cleaved. Moreover, the molecular mechanisms and determinants that regulate PABP substrate recognition by the viral proteinases are unknown. Since most PABP-protein interactions involve the PABP-CTD, it is of interest to determine if these interactions modulate the susceptibility of PABP to viral proteinases. Further, it is unknown if viral proteinases differentially target PABP bound to cellular versus viral mRNA. Here we show kinetics analyses of PABP cleavage reactions that suggest PABP exists in multiple conformations, one of which is resistant to 3Cpro or 2Apro. We determined that Paip2 and eRF3 negatively modulate the viral proteinase susceptibility of PABP when using different sources of PABP and that Paip2 strongly inhibits 2Apro cleavage of non-ribosome associated PABP pools, while it only partly inhibited 3Cpro cleavage on PABP associated with ribosomes. PABP pools associated with viral and cellular polysomes were equally susceptible to 3Cpro cleavage, yet active viral polysomes contained only intact PABP. Additionally, we show that 3Cpro and 3CD have equivalent proteolytic activity versus several PABP substrates. These results further define the PABP complexes targeted for proteolytic cleavage during infection.
Results
Biphasic PABP cleavage kinetics by 3C
Figure 1A depicts a schematic of PABP showing protein motifs and known viral proteinase cleavage sites. PABP interacts with a varied array of cellular and viral proteins and some of the mapped interaction domains on PABP are shown. Since it has been established that viral infection does not result in complete PABP cleavage, we sought to investigate mechanisms that may limit or regulate PABP cleavage. Figure 1B shows kinetics analysis of cleavage of purified recombinant PABP with 3Cpro and reveals a typical biphasic pattern where about 30% of total PABP was rapidly cleaved in 10 min, followed by a much slower cleavage rate that did not reach completion by 60 min or extended incubation periods (data not shown). The slower cleavage rate was not significantly enhanced by addition of fresh proteinase after the first hour incubation (data not shown). 3Cpro cleavage of endogenous PABP in HeLa S10 lysates also displayed a rapid initial cleavage rate where 70% cleavage required only 10 min incubation but very little additional substrate was cleaved upon extended incubation periods. In contrast, the Ras-GTPase activating protein SH3 domain-Binding Protein 1 (G3BP1), a novel substrate targeted for proteolytic cleavage by 3C proteinase (White et al., 2007), was rapidly cleaved to completion when supplied as purified recombinant protein or as endogenous protein in HeLa S10 lysates (Fig 1C), indicating that the 3Cpro was highly active. Therefore, the lack of complete cleavage of purified recombinant PABP by this proteinase suggests PABP exists in multiple conformations, some of which are not proteinase-susceptible and that PABP does not exchange conformations rapidly. In addition, as previously reported, cleavage of purified recombinant PABP with 3C proteinase heavily favored the 3Cpro primary cleavage site (Q537/G538) over the 3CAlt cleavage site (Q413/T414); very little or no cleavage product resulting from the latter was observed in most reactions (data not shown). In contrast, in vitro cleavage of PABP within HeLa S10 cell lysates or during poliovirus infection in vivo consistently resulted in significant cleavage at both the primary 3C and 3CAlt cleavage sites (Fig 1B) (Kuyumcu-Martinez et al., 2002). This suggests that recombinant and endogenous PABP may exist in different conformations, some inherently resistant to cleavage, or that different protein-protein interactions within the S10 lysate may modulate cleavage or expose different cleavage sites.
Figure 1. Biphasic 3C cleavage kinetics on PABP.
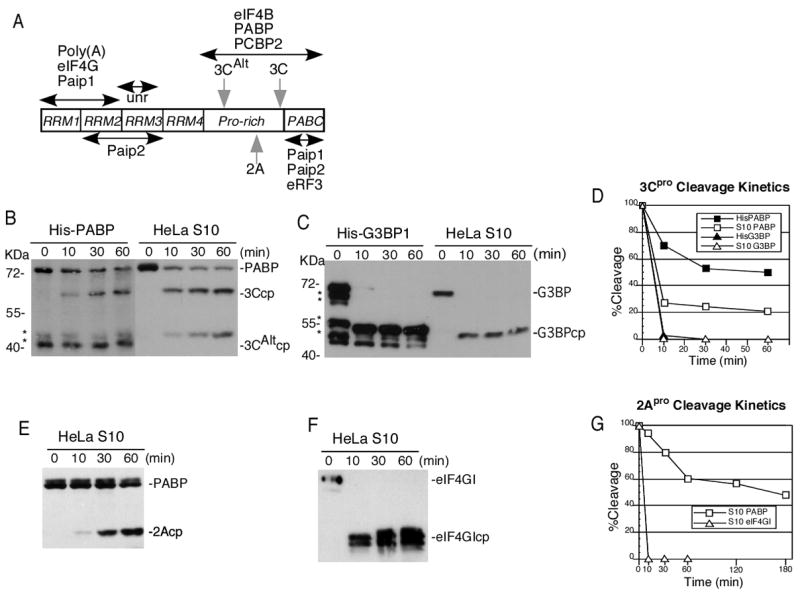
A. Schematic of PABP, depicting the location of RRMs, the primary sites for cleavage of the enteroviral proteinases (gray arrows) and some of the mapped recognition sites for the different RNA and protein-protein interactions (black arrows). B. PABP in vitro cleavage with 3Cpro. Purified His-PABP or HeLa S10 cell lysates were used as substrate for cleavage with 3C proteinase (1μg) for 0, 10, 30 or 60 minutes. C. G3BP in vitro cleavage with 3Cpro. Recombinant His-G3BP or S10 lysates were treated with proteinase as in panel B. D. Graph of cleavage of PABP and G3BP determined from densitometric analysis of immunoblots. E. PABP in vitro cleavage with 2Apro (0.5 μg) incubated with HeLa S10 lysate. F. Cleavage of eIF4GI in HeLa lysates by 2Apro. G. Graph of cleavage of PABP or eIF4GI in HeLa lysates by 2Apro, determined from densitometric analysis of immunoblots. The reactions in B, C, E and F were resolved by SDS-PAGE followed by immunoblot analysis with antibodies against the indicated proteins. Asterisks indicate PABP or G3BP degradation products produced in bacteria. G3BP1 cleavage generates a 52KDa fragment (White et al., 2007), while cleavage of eIF4GI isoforms generates multiple cleavage products (2Acps).
Similar cleavage assays with 2Apro and PABP revealed that PABP in a HeLa S10 lysate is also highly resistant to cleavage by this proteinase despite high 2Apro activity versus eIF4GI (Fig 1E, F). In this case the initial cleavage rate was very slow and remained constant for 60 min before slowing down further upon extended incubation (Fig. 1G). Extended incubation also revealed a biphasic cleavage profile. Incubation of 2Apro with recombinant His-PABP under typical assay conditions resulted in relatively poor and variable cleavage ranging from 0–20% (Fig 2, lanes 1–4). This suggested most purified His-PABP usually exists in a conformation not suitable for 2Apro recognition and binding.
Figure 2. Differential activation of PABP cleavage by reducing potential.
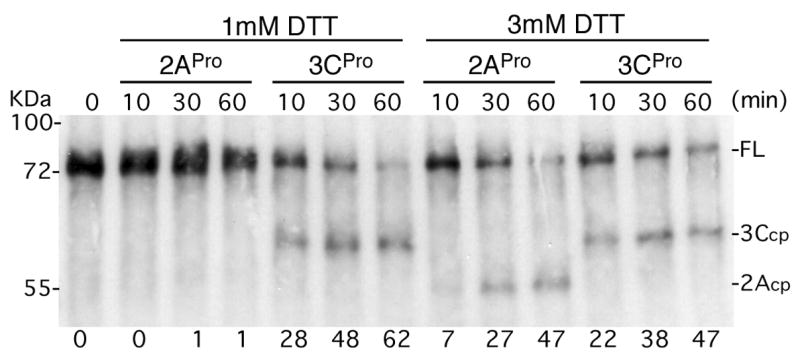
Recombinant His-PABP (500 ng) was incubated with 2Apro (0.5 μg) or 3Cpro (1μg) for various times in buffer containing the indicated concentrations of DTT. PABP and cleavage products were analyzed by immunoblot and full length (FL) and cleavage products are indicated on the right. Migration of molecular weight standards is shown on the left. Percent PABP cleavage determined by densitometric analysis is shown below each lane.
A recent report suggested PABP exists in several configurations via multimerization or formation of a looped structure stabilized by disulfide bridging (Yao et al., 2007). Our assay conditions and protein buffers normally contain 1 mM DTT, however we tested the effect of increased DTT on cleavage reactions using His-PABP. These results indicated that partial cleavage of PABP by 3Cpro not be significantly altered by additional DTT (Fig. 2). In contrast, higher reducing potential in the buffer did activate partial cleavage by 2Apro but had no effect on 2Apro-mediated cleavage of eIF4G (data not shown). Thus, substrate conformational changes imposed by higher DTT differentially affected only one protease-PABP interaction. Taken together, the results suggest PABP exists in various molecular conformations that exhibit differing inherent cleavage sensitivity by viral proteinases.
Paip2 and eRF3 modulate the cleavage susceptibility of recombinant PABP by viral proteinases
Since PABP interacts with a large number of cellular proteins we wished to determine if protein-protein interactions alter the susceptibility of PABP to viral proteinases. Figure 3 shows in vitro cleavage assays using a panel of paired recombinant proteins. Similar to Fig. 1 and Fig. 2, addition of 3Cpro resulted in cleavage of only about 40% of PABP in repeat assays. Likewise, addition of 2Apro under suitable buffer conditions to measure cleavage also resulted in only about 18% PABP cleavage, reinforcing the existence of proteinase-resistant PABP conformations within the population. When several PABP-binding proteins were added to these reactions they had no significant effect on proteinase cleavage catalyzed by either 2Apro or 3Cpro. These proteins included GST- Paip1, a GST-fused fragment of the eukaryotic initiation factor 4G-I (eIF4GI) containing the binding motif for PABP (amino acids 41–220) (GST- ΔeIF4GI), maltose-binding protein-unr fusion (MBP-unr), His-PCBP2 or His-tagged eIF4B. The cleavage of PABP by viral proteinases also remained unaffected when GST alone or a GST-fused truncated form of Tob1 (GST-Tob 1–170) were added to the cleavage reaction (data not shown). In contrast, GST-Paip2 partly inhibited the cleavage of recombinant His-PABP with 3Cpro or 2Apro proteinases (Fig. 3). Similarly, addition of His-eRF3 led to significant inhibition of PABP cleavage with 2A and 3C proteinases (Fig. 3B, C).
Figure 3. PABP-binding proteins modulate cleavage by viral proteinases.
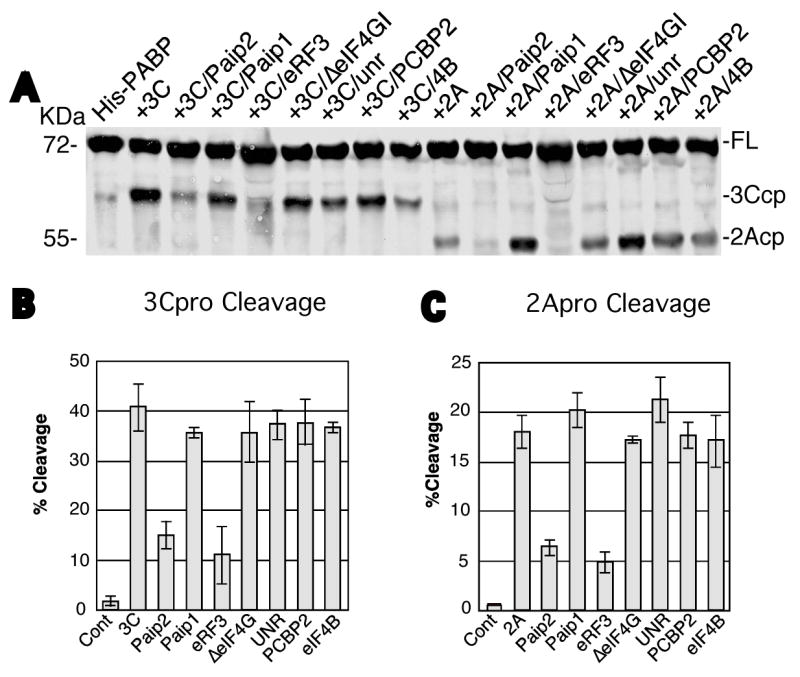
A. Recombinant His-PABP (500 ng) was incubated with buffer control (His-PABP) or with 3Cpro (1μg) or 2Apro (0.5 μg). Test cleavage reactions contained 500 ng of recombinant GST-Paip2, GST-Paip1, His-eRF3, GST- eIF4GI, MBP-unr, His-PCBP2 or His-eIF4B as indicated plus 3 mM DTT. Cleavage of PABP was performed for 1hr at 37°C, followed by immunoblot analysis with PABP antibody. The migration of the molecular weight markers is shown on the left, and on the right, the location of full-length PABP (FL) and its cleavage products (cp) for the 2A- and 3C-proteinases. B and C. Percent cleavage by 3Cpro or 2Apro, respectively, determined by densitometric analysis from 2–3 separate experiments. Values depict the mean and standard error of the mean.
Dose dependence of PABP cleavage inhibition by Paip2 and eRF3
To investigate the stoichiometry of the modulation of PABP cleavage by Paip2 and eRF3, we conducted cleavage assays examining dose responses of proteinase and proteins in these assays. Figure 4A depicts a cleavage assay utilizing His-PABP as a substrate for cleavage by 3C (lanes 1–8) or 2A proteinases (lanes 9–16). Addition of increasing concentrations of Paip1 (lanes 2–4 and 10–12) did not affect 3Cpro- or 2Apro- mediated cleavage of PABP (lanes 1 and 9, respectively). At the highest concentration of Paip1 tested, increasing concentrations of proteinases led to a dose-dependant increase in cleavage of PABP by 3Cpro and 2Apro proteinases (lanes 6–8 and 14–16, respectively). The same assay was conducted utilizing increasing concentrations of Paip2 (Fig 4B). Unlike Paip1, increasing concentrations of Paip2 inhibited cleavage of PABP by 3Cpro (lanes 2–4) in a dose dependent manner. The inhibitory effect exerted by Paip2 was more pronounced on 2Apro, as the lesser concentrations of Paip2 that led to partial inhibition of PABP cleavage by 3Cpro resulted in complete inhibition of 2Apro-directed cleavage of PABP (compare lanes 2–4 and 10–12). Paip2 inhibited proteolytic cleavage of PABP by 3Cpro at low proteinase concentrations, but the inhibition was abrogated with higher concentrations of 3Cpro (lanes 6–8). On the other hand, higher concentrations of 2Apro did not overcome Paip2 inhibition.
Figure 4. Paip2 and eRF3 inhibit cleavage of recombinant His-PABP in a dose dependent manner.
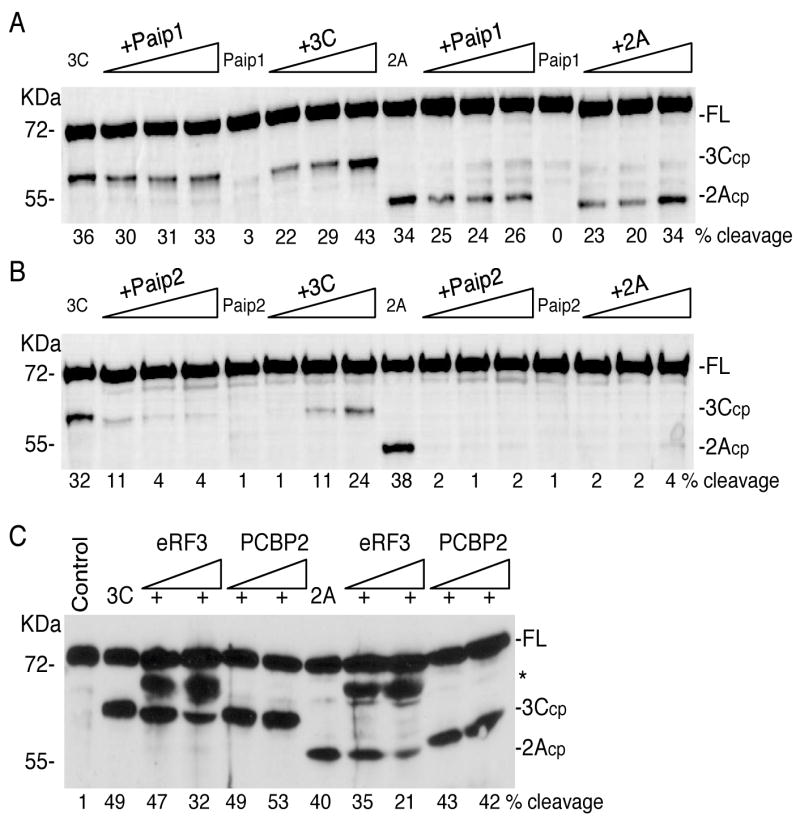
Recombinant His-PABP (~500ng) was subjected to cleavage by viral proteinases in the presence or absence of increasing concentrations of GST-Paip1 (panel A) or GST-Paip2 (panel B). His-PABP was cleaved with 3Cpro (3μg) in the absence (lane 1) or presence of increasing concentrations of recombinant GST-Paip proteins (1, 2 or 3μg (3–9-fold molar excess of Paip1or Paip2 over PABP), lanes 2–4). Alternatively, His-PABP was incubated with GST-Paip(1 or 2) (3μg) alone (lane 5) or with increasing concentrations of 3Cpro (1, 2 or 3μg – lanes 6–8). His-PABP was also cleaved with 1.5μg of 2A proteinase alone (lanes 9) or in the presence of increasing concentrations of GST-Paip(1 or 2) proteins (1, 2 or 3μg lanes 10–12) or incubated with the GST-Paip alone (lane 13) or with increasing concentrations of 2Apro (0.5, 1 or 1.5μg of 2A, lanes 14–16). C. Recombinant His-PABP (1μg) was incubated alone (lane 1) or with 1μg of 3Cpro or 0.5μg of 2Apro in the absence (lanes 2 and 7, respectively) or presence of increasing concentrations of of His-eRF3 (1 or 2μg, 1–2-fold molar excess of eRF3 over PABP, lanes 3–4 and 8–9) or His-PCBP2 (1 or 2μg, 2–4-fold molar excess of PCBP2 over PABP, lanes 5–6 and 10–11). All cleavage reactions contained 3 mM DTT and were analyzed by immunoblot with an anti-PABP polyclonal antibody. Percent cleavage in all panels was determined by densitometry. The migration of the molecular weight markers is shown on the left, and on the right, the location of full-length PABP (FL) and its cleavage products (cp) for the 2A- and 3C- proteinases. The asterisk represents a crossreactive protein in the His-eRF3 preparation.
We also examined if eRF3 inhibition of PABP cleavage by viral proteinases was dose dependent (Figure 4C). Increasing concentrations of recombinant His-tagged eRF3 led to partial inhibition of 3Cpro- and 2Apro- proteolytic cleavage of PABP that increased modestly (lanes 2–4 and 7–9). This inhibitory effect was not seen when similar amounts of His-tagged PCBP2 were added in the same assay as a negative control.
Paip2 and eRF3 inhibit cleavage of PABP in cell lysates
We were interested in determining if the observed inhibition of PABP cleavage by Paip2 and eRF3 occurred with native PABP in the context of a HeLa cell lysate. As shown previously, addition of 3Cpro cleaved only 60–70% of the PABP in the lysate (Fig. 5A). However, addition of either GST-Paip2 or His-eRF3 inhibited cleavage of endogenous PABP by 3Cpro by about 3 fold (Fig. 5A). When 2Apro was incubated with lysate, a smaller portion of PABP was cleaved and addition of Paip2 and eRF3 significantly inhibited this cleavage.
Figure 5. Paip2 and eRF3 inhibit cleavage of PABP in cell lysates.
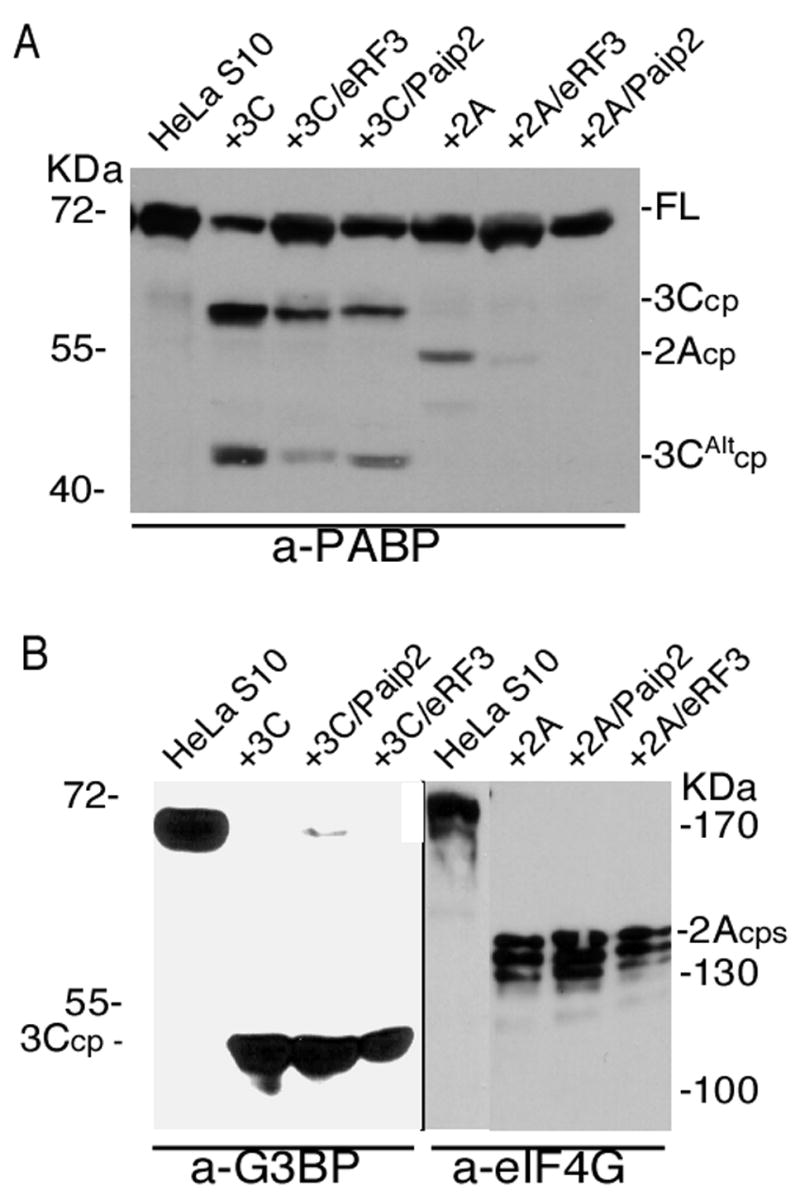
A. HeLa S10 lysate (200 μg) was incubated with buffer control or subjected to cleavage with 3Cpro (1 μg) or 2Apro (0.5 μg) in the presence or absence of eRF3 (500 ng) or Paip2 (500 ng). B. Control cleavage assays with 3Cpro or 2Apro of HeLa S10 lysates in the presence or absence of eRF3 or Paip2 were analyzed by immunoblot with polyclonal anti-G3BP1 or anti-eIF4GI antibodies.
In order to show that recombinant Paip2 or eRF3 did not inhibit 3Cpro or 2Apro activity per se, we conducted in vitro cleavage assays of endogenous G3BP1 or eIF4GI in HeLa S10 lysates. G3BP1 and eIF4G were efficiently cleaved by 3C- and 2A- proteinases, respectively (Fig 5B) and cleavage was not significantly affected by the addition of GST-Paip2 or His-eRF3. When other PABP-binding proteins were tested in similar cleavage assays with fractionated HeLa lysates (His-PCBP2, His-eIF4B, MBP-unr, GST, GST-Tob, GST-4G or GST-Paip1) no effect on the cleavage of PABP was observed with either 3Cpro or 2Apro (data not shown). Thus, eRF3 and Paip2 were found to inhibit cleavage of both recombinant and endogenous PABP, and several other PABP-binding proteins had no effect in either context.
Previously we have shown that cytoplasmic PABP that did not sediment with ribosomes (S200 fractions) was relatively resistant to cleavage with 3Cpro, whereas the fraction of PABP that did sediment with ribosomes (P200) or salt-washed ribosomes (SWRibo) was more susceptible to cleavage. The basis for the cleavage resistance in certain fractions may be due to the presence of endogenous Paip2 in these samples. We examined the distribution of PABP and Paip2 on sucrose gradients and found nearly all Paip2 to sediment slowly in gradients and not stably associated with 80S ribosomes or polysomes (Fig. 7B). This was consistent with Paip2 being present in S200 fractions and partly inhibiting cleavage. Similar analysis of eRF3 sedimentation was not performed due to unavailability of antisera. In direct assays, 3Cpro cleaved PABP in S200 fractions relatively poorly, however, addition of exogenous Paip2 only slightly inhibited this cleavage (Fig 6A, lanes 8,9). In contrast, 2Apro cleaved PABP in S200 fractions more efficiently and this cleavage was effectively inhibited by Paip2 addition (Fig 6B, lanes 5,6). This suggests that Paip2 may interact with a portion of the PABP susceptible to 2Apro in this fraction, but not PABP in other conformations or complexes that can be cleaved with 3Cpro. It also suggests that within the S200 fraction, a portion of PABP exists in two exclusive conformations or complexes, one that allows access to the 2Apro site and Paip2 but largely blocks access by 3Cpro, and vice versa.
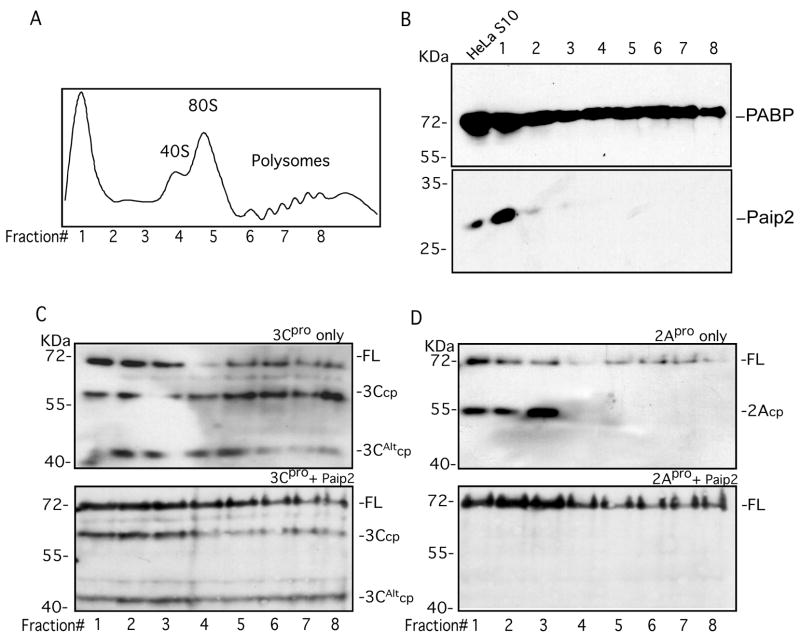
Figure 7. Paip2 differentially enhances the stability of PABP associated with polysomes.
A. HeLa polysome gradient fractions analyzed by UV trace. Location of ribosome and polysome peaks with the collected fractions is indicated in the graph. B. Immunoblot analysis of the distribution of PABP (top panel) or Paip2 (lower panel) in polysome gradient fractions. C. Sucrose gradient fractions (50 μl) were incubated with 3Cpro (1.5 μg) (upper panel) or 3Cpro plus GST-Paip2 (1 μg) (lower panel) and extent of cleavage determined by immunoblot analysis. D. Sucrose gradient fractions were incubated with 2Apro (upper panel) or 2Apro plus GST-Paip2 (lower panel) and the extent of cleavage determined by immunoblot analysis.
Figure 6. Paip2 inhibits cleavage of cleavage-susceptible or cleavage-resistant PABP fractions.
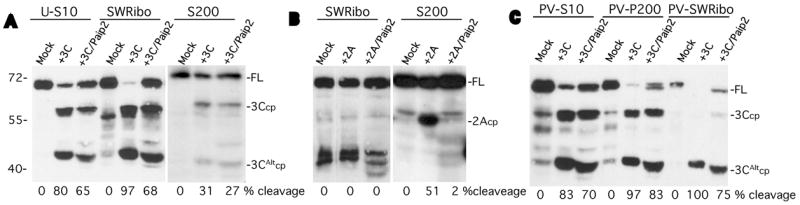
A. 3Cpro was incubated with HeLa S10 lysate, salt-washed ribosomes (SWRibo) or non-polysome fractions containing PABP (S200). Some reactions contained 3Cpro and GST-Paip2 (500 ng) as indicated or were incubated with buffer (mock). B. HeLa fractions were incubated with buffer (mock), with 2Apro alone or 2Apro plus GST-Paip2. C. Similar HeLa cell fractions prepared from PV-infected cells at 4 hrs post-infection were incubated with 3C and/or GST-Paip2 as indicated. PV-P200 indicates crude pelleted ribosome fraction before initiation factors are stripped off with high salt. All fractions were analyzed for PABP cleavage by immunoblot and densitometry. The migration of the molecular weight markers is shown on the left, and on the right, the location of full-length PABP (FL) and its cleavage products (cp) for the 2A- and 3C- proteinases.
In contrast to the partial cleavage-resistance of PABP in S200 fractions, ribosome-associated PABP is much more susceptible to cleavage. While only 40–75% of PABP in S10 lysates is cleaved in vitro by 3Cpro, the crude ribosome pellet fraction (SWRibo, Fig. 6A. lanes 5,6), which is deficient in endogenous Paip2, is cleaved by 95%. As reported previously, this fraction is very resistant to 2Apro cleavage (Fig. 6B, lanes 2,3). When we examined ribosome fractions taken from PV-infected cells at 4 hr p.i. we found the same high cleavage susceptibility to 3Cpro as observed in uninfected cells (Fig. 6C, lanes 5,8). At 4 hr p.i. 90% of translating ribosomes are associated with viral mRNA since host translation has been largely shutoff. Interestingly, the SWRibo fraction (which is stripped of most initiation factors) from infected cells contained no PABP cleavage products (Fig. 6C, lane7). Nonetheless, the intact PABP associated with viral mRNA or cellular mRNA was equally and highly susceptible to 3Cpro cleavage (compare Fig. 6A and 6C). One difference found was that cleavage site selection was altered; ribosome-associated PABP in uninfected cells was cleaved to produce both 3Ccp and 3CAltcp, whereas in mid-PV infection, there was a strong selection of 3CAltcp. This was not the case with crude ribosome/initiation factor fractions (PV-200) in which both cleavage sites were used (Fig. 6C, lanes 5,6). In agreement with these results, we found that polysome gradient fractions from mock infected cells, or cells infected with PV at 4 or 7hpi, were all equally susceptible to cleavage by 3Cpro (data not shown), again suggesting that PABP within cellular or viral polysomes is equally susceptible to 3Cpro cleavage. Paip2 was found to inhibit cleavage of uninfected and viral ribosome-associated PABPs equally, however, the inhibition of cleavage was incomplete (~50%) in each case.
Paip2 inhibition of 2A-mediated cleavage of non-ribosome associated PABP
Since Paip2 was found to inhibit PABP cleavage in crude ribosome fractions, we wanted to test potential inhibition of PABP cleavage within 40S, 80S or polyribosome fractions enriched by sucrose gradient sedimentation. Cleavage of PABP in polysomes has not been examined before. PABP sedimented with non-ribosome fractions, 40S and 80S ribosomes and polysome fractions (Fig. 7B). Addition of 3Cpro to each of these fractions led to partial cleavage in every case, and only slightly higher percent cleavage of PABP in polysome fractions (fraction 6–8) or 40S-80S fractions (fraction 4–5) was observed compared to crude SWRibo fractions. This suggests that factors that may enhance cleavage have been lost by the sucrose sedimentation procedure. When Paip2 was added to each cleavage reaction, a partial and variable inhibition of PABP cleavage was observed (Fig. 7C, compare panels). Production of 3Ccp from polysome fractions was inhibited more strongly by Paip2. When 2Apro was incubated with these sucrose fractions, only PABP in slow-sedimenting, non-ribosome-associated fractions (1–3) was cleaved, providing a quite different cleavage profile than 3C (Fig 7D). In contrast to 3Cpro cleavage, the 2Apro cleavage in these fractions was strongly inhibited by Paip2 (Fig 7D), in agreement with S200 cleavage data (Fig 6B). This is interesting since these fractions already contain endogenous Paip2. We endeavored to estimate endogenous Paip2 and PABP concentrations via immunoblot analysis of serial dilutions of fractions and recombinant Paip2 or PABP (data not shown). We estimate that PABP is present at a large molar excess in comparison to Paip2 (4 μM PABP versus 20 nM Paip2 in S10 lysate), even in fractions 1–3 of the gradient, thus providing an explanation for partial 2Apro cleavage of PABP in these fractions.
PABP associated with viral polysomes is cleaved late during poliovirus infection
Since crude viral ribosome fractions contained no detectable PABP cleavage products, and yet were equally susceptible to 3Cpro cleavage as uninfected ribosome fractions in vitro (Fig 6), we wondered if there was any temporal discrimination of cleavage of PABP associated with viral polysomes that may allow continued viral translation after host translation was shutoff. 35S-methionine pulse-label analysis on PV infected HeLa cells showed that as expected, a strong host translational shutoff was induced by the viral infection leading to the almost exclusive production of viral proteins by 4–5 hpi (Fig. 8A). Viral translation sharply declined by 7hpi. As seen before, PABP from mock-infected cells was distributed throughout the entire polysome gradient (Fig 8B, top panel). By 4 hpi, during the phase of viral infection when host translation is shutoff (host polysomes are disassembled) and viral polysomes become predominant, there was a large, but incomplete loss of PABP in the polysome region (Fig 8, middle panel, fraction 6,8) and a shift toward non-ribosome associated (fraction 1) and 40–80S ribosome fractions (fractions 3,5). PABP cleavage products mostly remained associated with 40–80S ribosome fractions and did not release into the slowest sedimenting non-ribosome associated fractions. Interestingly, PABP cleavage products were not associated with active viral polysomes, suggesting intact PABP is required for viral translation (Kuyumcu-Martinez et al., 2004b). Interestingly, by 7 hpi when viral translation was steeply declining there was still intact PABP associated with polysomes, however, PABP cleavage products now remained associated with viral polysome fractions (Fig 8B, bottom panel, fractions 6,8). These PABP cps may contribute to inhibition of viral translation.
Figure 8. Viral polysomes contain exclusively intact PABP.
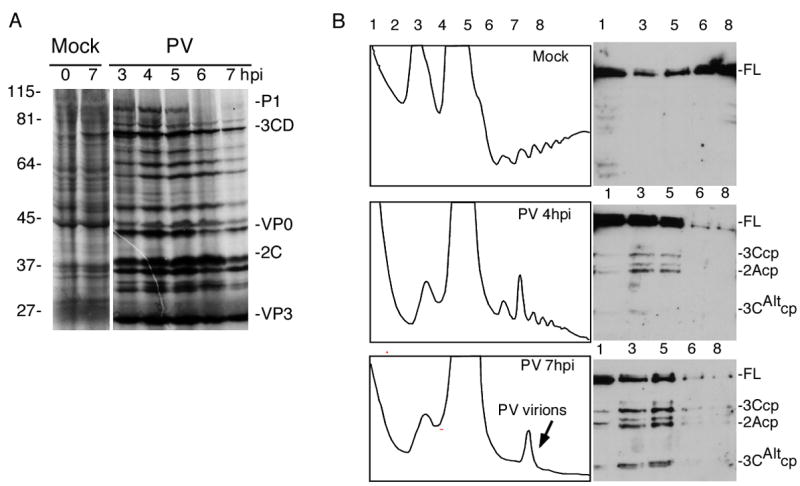
A. Autoradiograph showing 35S-met/cys pulse-label analysis of host and viral translation in mock-infected or PV-infected HeLa cells. B. Analysis of PABP cleavage in polysomes in Mock- (top panel) or PV-infected cells at 4hpi (middle panel) or 7hpi (bottom panel). UV absorbance profiles are depicted in panels on the left. Immunoblots show the presence of PABP or its 2A- or 3C- cleavage products (cp) (right panels).
3CD and 3Cpro cleave PABP with equal efficiency
3CD is the precursor of mature 3Cpro and is present in infected cells at much higher concentrations than fully processed 3Cpro. 3CD has no polymerase activity but serves as an active proteinase with cleavage specificity that overlaps, but is distinct from 3Cpro, especially in its ability to cleave PV capsid precursors with much higher efficiency (Harris et al., 1992; Jore et al., 1988; Parsley et al., 1999; Ypma-Wong et al., 1988). 3CD has not been tested versus PABP substrates in cleavage assays, thus the relevant viral proteinase that cleaves PABP in cells remains undetermined. Active 3CD (containing mutations at the 3Cpro-3Dpol junction that inhibit processing to 3Cpro and 3Dpol) and 3Cpro were standardized to equal proteinase units using radiolabeled polypeptide substrate containing the P2-P3 junction of the viral polyprotein as reported previously (Fig 9A) (Parsley et al., 1999). The recombinant 3CD was shown to have high cleavage activity against a radiolabeled P1 portion of the viral polyprotein, whereas equimolar amounts of 3C proteinase failed to cleave the P1 peptide, as reported previously (Parsley et al., 1999) (data not shown). When equivalent proteinase units of 3CD and 3Cpro were used to cleave endogenous PABP in HeLa S10 lysates, SWRibo fractions, as well as recombinant His-PABP substrates, we found that both proteinases cleaved all PABP substrates with near equivalent efficiency (Fig. 9B). Another recombinant HA-tagged PABP was also equally efficiently cleaved by both proteinases (data not shown). This suggested that in the context of a viral infection, more PABP processing may be catalyzed by 3CD rather than 3Cpro, due to large molar excess of the former.
Figure 9. Poliovirus 3CD proteinase cleaves PABP.
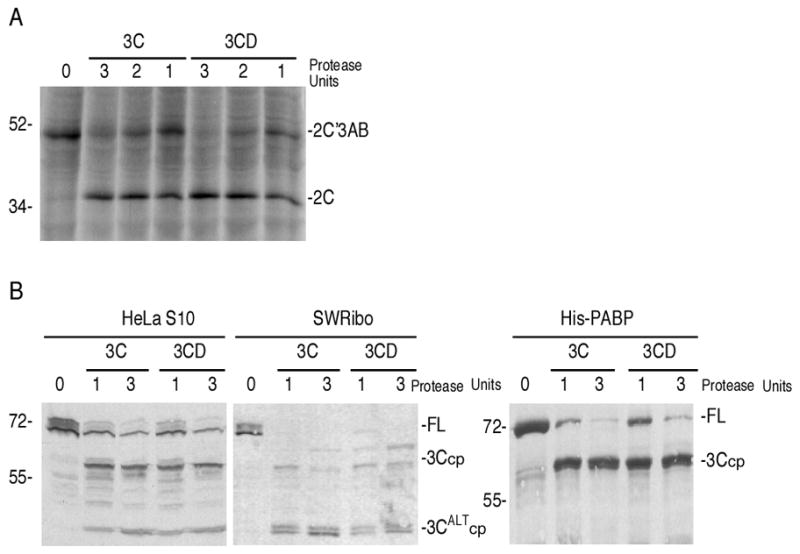
A. Standardization of 3Cpro and 3CD proteinase activity based on the ability of each proteinase to cleave at the P2-P3 junction of the viral polyprotein (previously shown to have equal proteinase susceptibility against 3Cpro and 3CD proteinases (Parsley et al., 1999)). One proteinase unit was defined as the amount of proteinase required to cleave 50% of the 2C’3AB substrate in 30 mins at 30°C. B. HeLa S10 lysates (100μg), SWRibo fraction (100 μg), or recombinant His-PABP (200ng) was incubated with 1 or 3 proteinase units of either 3Cpro or 3CD proteinase. Equivalent proteinase units of 3Cpro and 3CD contained approximately 1.2-fold molar excess of 3CD over 3Cpro. Cleavage was performed at 37°C for 1hr, followed by immunoblot analysis.
We examined PABP cleavage in the context of viral RNA sequences and PCBP2 in order to determine if formation of an RNP complex containing PCBP2 and PABP bound to the 5′ and 3′ regions of a viral minigenome RNA would activate a higher degree of PABP cleavage. The relative cleavage activity of 3Cpro and 3CD were also measured separately and together with RNA and PCBP2 in this context. 3Cpro and 3CD were used at equivalent protein concentration (0.1 μg/μl), resulting in higher cleavage with 3Cpro than 3CD in these assays due to higher molar concentration (Fig. 10). When viral minigenome RNA containing a poly(A) segment was added, the PABP was cleaved more efficiently by 3Cpro (76% to 88%), as has been described before (Kuyumcu-Martinez et al., 2002), but PABP cleavage by 3CD did not change (31% vs 32%). Addition of PCBP2 alone caused a slight decrease in PABP cleavage by both proteinases which also mitigated the RNA enhancement of 3Cpro cleavage activity. With either protease, addition of RNA plus PCBP2 did not activate higher levels of PABP cleavage than with PABP alone. When both 3Cpro and 3CD were incubated together (retaining the same concentration of total protease) no further increase in PABP cleavage was observed, suggesting that any potential 3Cpro-3CD complexes did not have increased cleavage activity versus this substrate. When the proteases were used in combination, the addition of RNA did not increase PABP cleavage, nor did PCBP2. We verified that PCBP2 and PABP interacted with the minogenome RNA by performing electrophoretic mobility assays and observed a supershift when both proteins were used together (data not shown). Taken together, since 3Cpro cleavage activity, but not 3CD cleavage activity was enhanced by viral poly(A) RNA, this suggests that 3Cpro may be the more active protease versus PABP in cells.
Figure 10. Cleavage of PABP in mRNP complexes.
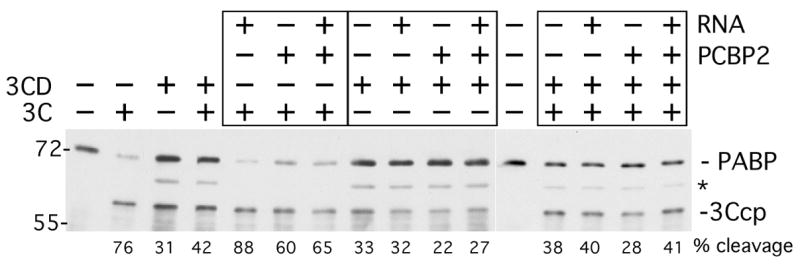
Purified PABP and PCBP2 were incubated with minigenome viral RNA containing a complete 5′ UTR and 3′ UTR-A71 bracketing a FLuc open reading frame. RNA, PABP and PCBP2 were used in a 1:3:8 molar ratio (0.24 μg, 0.15 μg, 0.2 μg). Combinations of mRNPs, or PABP alone or PABP plus PCBP2 were incubated with 3Cpro, 3CD or a combination of 3Cpro and 3CD. 3Cpro (0.1μg) and 3CD (0.15μg) were used at a molar ratio of 2:1 and when combined, the amount of each was halved in reactions. Reactions were incubated for 3 hrs and examined by immunoblot and densitometry. PABP and PABPcp are indicated, the asterisk indicates a cross reactive protein. Calculated percent cleavage is shown below each lane.
Discussion
One of the hallmarks of enterovirus infection is the inhibition of host cell protein synthesis through cleavage of eIF4G and PABP. A growing number of viruses are now known to cleave PABP during infection, including caliciviruses, hepatitis A virus and HIV, thus its evolutionary importance in virus replication is growing (Alvarez et al., 2006; Kuyumcu-Martinez et al., 2004a; Zhang et al, 2007). While eIF4G is efficiently cleaved to completion early during PV infection, only a third of cytoplasmic PABP is cleaved at this time, and only 50–60% by late infection. Though partial PABP cleavage during viral infection may result from its huge abundance in cells, eluciation of the precise and restricted PABP-RNP complex(es) that is preferentially targeted by viral proteases is important to determine. We have shown that PABP is cleaved with biphasic kinetics by both viral proteinases, revealing a cleavage-resistant PABP population and we have shown that two PABP-binding proteins, eRF3 and Paip2, block PABP cleavage by two different proteinases.
Our results indicate that purified PABP inherently adopts conformations that are proteinase resistant. Because the PABP 3Cpro cleavage sites do not perfectly match the 3Cpro consensus cleavage site, incomplete PABP cleavage in cells could have resulted from reduced binding and catalytic rates. The results in Fig. 1 show that 3Cpro can easily cleave PABP with fast initial cleavage kinetics, indicating that 3Cpro was amply active and that the non-consensus cleavage site sequence only minimally impedes cleavage. However, the abrupt reduction in PABP cleavage rate after 10–30 min suggests that at least two PABP pools exist in the population, one highly susceptible to cleavage and another configuration(s) that is refractory to cleavage. Slow secondary rate of cleavage by 3Cpro suggests that interconversion between PABP conformations is inefficient. PABP cleavage by 2Apro showed slower initial cleavage rates than 3Cpro, but extended incubation time also revealed a biphasic cleavage profile. In these reactions, PABP likely interacts with itself through a well known, but poorly characterized oligomerization property (Kuhn and Pieler, 1996). We have examined our PABP preparations by gel filtration and found anomalous migration (data not shown). Additionally, a recent report shows yeast PABP migrates in native gels as clear oligomers. This report also suggested that cysteine residues (conserved in human PABP) within the N-terminal RNA binding component of yeast PABP allow for a circular conformation to form in some PABP molecules through the formation of a disulfide bond. (Yao et al., 2007). Such a configuration may block cleavage with 2Apro, which cleaved more readily with high DTT concentration (Fig. 2), however 3Cpro cleavage was not influenced by this parameter. This suggests that 2Apro and 3Cpro may recognize two different PABP pools. Importantly, biphasic PABP cleavage kinetics was also observed in HeLa extracts as well, however cleavage was consistently more efficient than with purified PABP. This suggests that factors present in lysates (e.g. polyA RNA) enhance PABP cleavage.
We previously reported that 3Cpro primarily targets PABP molecules associated with the translational machinery, but molecular details were undetermined. Here we show that cleavage of PABP by viral proteinases is inhibited by Paip2 and eRF3, both of which function as translational repressors, but not by other the PABP-associated factors Tob1, Paip1, eIF4B, eIF4G, PCBP2 or unr (Fig. 3 and data not shown). eRF3 is proposed to transiently interact with polysome-bound PABP when ribosomes pause at stop codons, thus, eRF3 may only exert a minor inhibitory effect on overall PABP cleavage in a cell. However, other unknown interactions of eRF3 and PABP away from the context of translating polysomes remain possible. Paip2 is a general inhibitor of PABP function that can strip PABP off poly(A) RNA. This interaction is relatively stable and could interfere with PABP cleavage by steric effects and by reducing pools of poly(A)-bound PABP, which is more susceptible to 3Cpro than free PABP (Khaleghpour et al., 2001b; Kuyumcu-Martinez et al., 2002). A new Paip2 homolog, termed Paip2B, has similar functions in regulating PABP and releasing it from poly(A) RNA (Berlanga et al., 2006), thus it is likely that Paip2B can also inhibit the cleavage of PABP by viral proteinases.
Interestingly, Paip1 did not inhibit PABP cleavage, despite the fact it binds PABP at two sites that partly overlap the Paip2 binding sites. This difference could be attributed to the fact that the binding of Paip1 to PABP occurs with a 1:1 stoichiometry and with an apparent Kd of 1.9nM, whereas Paip2 binds PABP with a 2:1 stoichiometry and Kd values of 0.66 and 74nM (Khaleghpour et al. 2001). Indeed, structural and thermodynamic studies of the binding of eRF3 and Paip1 or 2 peptides to the PABC domain of PABP indicated that binding of Paip1 causes a more drastic conformational change than Paip2 (Kozlov et al., 2004; Kozlov et al., 2001). Moreover, binding of Paip2 and eRF3 peptides (more than Paip1) is highly dependent on hydrophobic interactions or involves a larger protein/peptide contact surface (Kozlov et al., 2004). Since proteinases probe subtrate structure, the differential cleavage results obtained with Paip1 and Paip2 suggest that PABP associated with these proteins adopts different conformations or that steric hindrance occurs only with Paip2.
Despite the finding that Paip2 can inhibit PABP cleavage by both proteinases and in all HeLa fractions tested, it probably plays a lesser role in modulating overall PABP cleavage in cells. Examination of actual protein concentrations in lysates indicated that Paip2 exists at levels approximately 50-fold lower than PABP, seemingly too low to account for the differential and incomplete PABP cleavage by 2Apro or 3Cpro in lysates and fractions thereof. 2Apro only cleaved PABP within slowly sedimenting complexes in polysome gradients, however, the bulk of the cellular Paip2 was already present in these same polysome gradient fractions, demonstrating insufficient endogenous Paip2 was present to block cleavage. Further, the PABP in these fractions could interact with Paip2 since addition of exogenous Paip2 strongly inhibited its cleavage with 2Apro (Fig. 7).
We also found an interesting inverse relationship between 2Apro and 3Cpro cleavage of PABP in HeLa cell fractions where PABP molecules within non-ribosome complexes (S200) were cleaved more efficiently by 2Apro than by 3Cpro. Conversely, PABP found within crude salt-washed ribosome extracts (SWRibo) was efficiently cleaved by 3Cpro, but not 2Apro (Fig 6). This relationship partly extended to polysome fractions where 2A only cleaved non-ribosome associated PABP. The 2Apro cleavage site on PABP lies between the two 3Cpro sites in the linear sequence, yet no knowledge of true structure in this region is available, and the cleavage sites may actually lie in different surface regions. Our data show that each PV proteinase targets one of at least two distinct PABP populations, which present different conformational constraints. These unique conformations are likely modulated by differing host factors in complexes with PABP, which promote or inhibit cleavage of one or the other proteinase.
Why would 2Apro evolve to target non-ribosome-associated PABP but not 3Cpro? 3Cpro is the default picornavirus proteinase that has evolutionary homologs in other virus families such as norovirus, whereas 2Apro homologs are absent in many other virus families. Thus 3Cpro may have evolved early on to regulate viral and cellular translation and thus targets PABP in active polysomes. One possible function of the pool of PABP that is not associated with mRNA is to provide PABP for nascent RNA transcripts. Cleavage of this PABP population by 2Apro, the accessory proteinase, may serve to prevent nascent viral RNAs from acquiring intact PABP and being translated later in infection, thus aiding the process leading to packaging of vRNA. We have determined that PV virion RNA is completely devoid of PABP (data not shown).
PABP cleavage inhibits not only cap-dependent translation, but also PV translation (Bonderoff et al., 2008) and HAV translation (Zhang et al., 2007). Since viral translation persists for several hours longer than cellular translation in cells, we also investigated if viral proteinases preferentially target PABP cleavage on cellular versus viral polysomes as part of a mechanism that promotes viral translation. Surprisingly, we determined that PABP on either type of polysome pool was equally and highly susceptible to cleavage with 3Cpro (Fig. 6). This suggests that viral polysomes do not stably bind factors that inhibit PABP cleavage. However, examination of PABP on active viral polysomes purified from infected cells shows that exclusively intact PABP was present; all PABP cleavage products migrated with 40–80S ribosome subunits (Fig. 6 and 8). Thus, some form of PABP cleavage discrimination may exist in cells to restrict PABP cleavage on viral polysomes until after cleavage on cellular polysomes occurs. Also, viral proteinases must retain a population of intact PABP during the exponential phase of the viral growth cycle to support translation of the expanding pool of nascent viral RNAs. One hypothesis is that the non-poly(A) associated PABP which is resistant to 3Cpro cleavage (S200 pool) may represent this population of intact PABP for nascent viral mRNA. Further, there was a temporal correlation between the downregulation of viral translation during late PV infection and appearance of PABP cps associating with viral polysomes (Fig. 8B). This supports the hypothesis that intact PABP is required for viral translation.
We also investigated if 3Cpro or 3CD is the major proteinase that cleaves PABP. Since 3Cpro and 3CD had nearly equivalent cleavage activity on purified PABP, yet 3CD is more abundant in infected cells, 3CD may be dominant. However, RNA stimulated 3Cpro cleavage of PABP but not 3CD cleavage of PABP (Fig 10), suggesting that 3Cpro is more effective versus poly(A)-bound PABP, which is presumably the more important target. These results make it unlikely that any preferential cleavage of cellular versus viral polysome-bound PABP could stem from regulation of 3CD processing into mature 3Cpro during the course of infection. However, since we have shown 3CD is an active PABP-specific proteinase, and was not inhibited by RNA, it likely plays a role in the cleavage of PABP on viral mRNA.
3CD may play an important role in the switch from viral translation to RNA replication, as it associates with the cloverleaf structure of the viral 5′ UTR and may mediate the circularization of the viral genome by interacting with PABP (Herold and Andino, 2001). Since PABP cleavage, along with cleavage of PTB and PCBP2 (Back et al., 2002; Perera et al., 2007) all regulate the end of viral translation, 3CD and PCBP2 coordinately binding the viral cloverleaf and PABP may strongly stimulate PABP cleavage as well as cleavage of PCBP2 or PTB. We tested this hypothesis in vitro and did not observe a stimulation of cleavage activity when complexes of 3CD, PCBP2, PABP and viral minigenome RNA could form. It would be interesting to examine if addition of other viral replicase components 3AB, VPg-pUpU or 2C to this system can stimulate PABP or even PCBP2 cleavage.
In summary, this study provides insights about the mechanisms that modulate PABP cleavage during PV infection. PABP inherently exhibits a dual proteinase sensitive/resistant phenotype due to formation of multiple conformations, partly via self-oligomerization. Further, several types of PABP/protein/mRNP complexes influence these cleavage reactions but remain undefined at a molecular level. Interactions of PABP with eRF3 and Paip2 inhibit cleavage reactions with both viral proteinases, however, likely play a minor role in regulation of PABP cleavage during virus infection. More work remains to determine the primary PABP substrate complexes that 3CPro and 3CD have evolved to cleave most efficiently. Such complexes may exist only in a transient conformation that PABP adopts at a particular stage of the translation initiation, elongation or termination phases, or at a particular PABP moiety bound to poly(A).
Materials and Methods
Cells and Virus
HeLa S3 cells were grown in suspension in defined SMEM-Joklik media supplemented with 9% Bovine Calf Serum, 1% Fetal Bovine Serum and Penicillin/Streptomycin (Invitrogen). Poliovirus type 1 (Mahoney) was grown and purified as previously described (Brown and Ehrenfeld, 1979). HeLa S3 cells were infected with poliovirus at a multiplicity of infection of 10 pfu/cell. Serum was added to the cells 30 min after infection to a final concentration of 3%. Aliquots of cells were taken at different time points throughout the infection and subjected to fractionation or used to examine translation levels during infection. Translational shutoff during viral infection was assessed by pulse labeling 1×106 cells at different time points for 30 min, for which the cells were resuspended in 100μl of (-Met/-Cys) DMEM, supplemented with 30μCi of 35S-methionine. After the 30-minute pulse, the cells were harvested and lysed in CHAPS buffer on ice for 30 min. The supernatant containing the soluble proteins was then loaded onto a 12% SDS-PAGE gel and analyzed by autoradiography.
Plasmids
The PABP expression plasmids pET28a-His-PABP has been described elsewhere (Kuyumcu-Martinez et al., 2004b). The plasmid pET-His-PCBP2 was constructed by insertion of the PCBP2 coding sequence from pQE30-PCBP2 (Blyn et al., 1996) into pET28a(+). Plasmids encoding GST-Paip1 and GST-Paip2 (pGEX6P-Paip1/2) were a kind gift from Dr. Nahum Sonenberg. The plasmid pET-His-4B, encoding human eIF4B was obtained from Dr. Nadia Korneeva. The plasmid pheRF3, encoding human eRF3 was a kind gift of Dr. Bertrand Cosson. The pMBP-unr construct was a gift from Dr. Ann Bin Shyu. The plasmid pGEX5X-1-TobN(1–170) was a kind gift from Dr Tadashi Yamamoto. The construct pGST-4G (41–220) was made by PCR-amplification of the PABP binding motif from pSPORT-4GI and the PCR product corresponding to residues 41–220 of eIF4G flanked by XhoI sites was then inserted into the XhoI site of pGEX-4T2. The resulting construct, GST-4G (41–220) was confirmed by sequencing and the GST-4G peptide expressed in DH5α cells and purified according to manufacturer’s protocols.
Production of Recombinant Proteins
Purified Coxsackievirus B3 (CVB3) 2A- and PV 3C- proteinases were expressed and purified as described previously (Joachims et al., 1999; Liebig et al., 1993). Active 3CD bearing a Ser insertion mutation near the 3C-3D junction to block autocleavage was expressed and purified as previously described (Parsley et al., 1999). Recombinant His-PABP was expressed from the plasmid pET28-His-PABP and purified by metal affinity chromatography as described elsewhere (Kuyumcu-Martinez et al., 2004b). Recombinant His-eIF4B, His-PCBP2 and His-eRF3 were expressed and purified using the same protocol as for the His-PABP. The MBP-unr protein was expressed and purified according to published protocols (Chang et al., 2004). GST-Paip1 and GST-Paip2 were expressed and purified from bacteria as previously described (Khaleghpour et al., 2001a; Roy et al., 2002). GST and GST-Tob 1–170 were purified similarly to the GST-Paip proteins. Following purification, all proteins were dialyzed in standard dialysis buffer containing 100mM NaCl, 25 mM Tris (pH 8.0) and 10% glycerol).
Proteinase activity standardization
The plasmid pTM1–2C3AB, kindly provided by Dr. Bert Semler, was linearized with XhoI and used for in vitro transcription with T7 polymerase. The resulting RNA was used for in vitro translation in rabbit reticulocyte lysate (Promega), generating a 52 KDa polypeptide comprised of the 2C and the 3AB polypeptides of the viral polyprotein. Serial dilutions of equimolar concentrations of the purified 3C and 3CD proteinases were used to cleave the radiolabeled substrate for 30 min at 30°C. One proteinase unit was defined as the amount of proteinase required for cleavage of 50% of the substrate in this assay.
In Vitro Cleavage Assays
Purified PABP-HA, His-PABP or HeLa cell lysates were incubated with the corresponding amount of proteinase and exogenous protein or standard dialysis buffer control at 37°C for 1hr, or for extended times as indicated. Typical cleavage reaction conditions contained 20mM Hepes (pH 7.4), 150mM KOAc and 1 mM DTT or 3 mM DTT as indicated. Proteins were then resolved on a 9% SDS-PAGE gel and analyzed by Western Blot with an anti-PABP rabbit polyclonal antibody using enhanced chemiluminescence reagents (Pierce).
HeLa Cell Fractionation
Mock- or PV-infected HeLa cells were harvested at different times during infection and fractionated into further compartments. The cell pellet was washed with PBS plus cycloheximide (160μg/ml). The cell pellet was then resuspended in two volumes of passive lysis buffer (10mM potassium chloride (KCl), 2.5mM dithiothreitol, 1.2mM magnesium acetate and 25mM Hepes, pH 7.4), incubated on ice for 10 mins, then lysed with 25 strokes in a Dounce homogenizer. The total lysate was clarified by centrifugation at 4°C at 10,000xg for 15 min to obtain the total cytoplasmic extract (S10 lysate). Further fractionation into S200/P200, RSW and SWRibo fractions was performed as described before (Kuyumcu-Martinez et al., 2002).
Polysome gradients
The HeLa S10 cell lysates were supplemented with cycloheximide and layered on top of 10–50% sucrose gradients containing 10mM Tris (pH 7.2), 140mM sodium chloride and 1.5mM magnesium chloride. The gradients were subjected to ultracentrifugation in a SW41 rotor for 3 hours at 35,000 RPM at 4°C. The gradients were collected with continuous monitoring at 260nm using an ISCO UA-6 UV detector and Retriever 500 fraction collector. Each fraction was then analyzed by immunoblot for the presence of PABP or Paip2 protein.
Immunoblot Analysis
Western Blot analysis was conducted using rabbit polyclonal antisera raised against the synthetic peptide (GIDDERLRKEFSPFGT) within the RRM4 of PABP as shown before (Kuyumcu-Martinez et al., 2004a). Rabbit polyclonal antisera against eIF4GI and G3BP1 have been described elsewhere (Byrd et al., 2005; White et al., 2007). Anti-Paip2 immunoblots were performed using a rabbit polyclonal antibody against Paip 2, kindly supplied by Dr. Nahum Sonenberg. Densitometric analysis of protein cleavage in immunoblots was determined using ImageJ software. Percent cleavage was computed from densitometry data as the combined average of percent decline in PABP signal and the ratio of cleavage product/(cleavage product + PABP) in each lane.
Acknowledgments
The authors would like to thank Dr. Nahum Sonenberg for antisera and plasmids, Dr. Ann-Bin Shyu, Dr. Bert Semler, Dr. Nadia Korneeva, Dr. Bertrand Cosson and Dr. Tadashi Yamamoto for expression constructs for some of the proteins utilized in this study. We would also like to acknowledge Jennifer M. Bonderoff and James P. White for help with reagents and for their feedback in reviewing this manuscript. This work was supported by NIH grants AI50237 and GM59803.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvarez E, Castello A, Menendez-Arias L, Carrasco L. HIV protease cleaves poly(A)-binding protein. Biochem J. 2006;396:219–26. doi: 10.1042/BJ20060108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht M, Lengauer T. Survey on the PABC recognition motif PAM2. Biochemical & Biophysical Research Communications. 2004;316:129–138. doi: 10.1016/j.bbrc.2004.02.024. [DOI] [PubMed] [Google Scholar]
- Back SH, Kim YK, Kim WJ, Cho S, Oh HR, Kim JE, Jang SK. Translation of polioviral mRNA is inhibited by cleavage of polypyrimidine tract-binding proteins executed by polioviral 3C(pro) J Virol. 2002;76:2529–2542. doi: 10.1128/jvi.76.5.2529-2542.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlanga JJ, Baass A, Sonenberg N. Regulation of poly (A) binding protein function in translation: Characterization of the Paip2 homolog, Paip2B. RNA. 2006;12:1556–1568. doi: 10.1261/rna.106506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blyn LB, Swiderek KM, Richards O, Stahl DC, Semler BL, Ehrenfeld E. Poly(rC) binding protein 2 binds to stem-loop IV of the poliovirus RNA 5′ noncoding region: identification by automated liquid chromatography-tandem mass spectrometry. Proc Natl Acad Sci USA. 1996;93:11115–11120. doi: 10.1073/pnas.93.20.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonderoff JM, LaRey JL, Lloyd RE. Cleavage of poly(A)-binding protein by poliovirus 3C proteinase inhibits viral IRES-mediated translation. 2008 doi: 10.1128/JVI.00006-08. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovee ML, Marissen WE, Zamora M, Lloyd RE. The predominant eIF4G-specific cleavage activity in poliovirus-infected HeLa cells is distinct from 2A proteinase. Virology. 1998;245:229–240. doi: 10.1006/viro.1998.9171. [DOI] [PubMed] [Google Scholar]
- Brown BA, Ehrenfeld E. Translation of poliovirus RNA in vitro: changes in cleavage pattern and initiation sites by ribosomal salt wash. Virology. 1979;97:396–405. doi: 10.1016/0042-6822(79)90350-7. [DOI] [PubMed] [Google Scholar]
- Byrd MP, Zamora M, Lloyd RE. Translation of eIF4GI proceeds from multiple mRNAs containing a novel cap-dependent IRES that is active during poliovirus infection. J Biol Chem. 2005;280:18610–18622. doi: 10.1074/jbc.M414014200. [DOI] [PubMed] [Google Scholar]
- Chang TC, Yamashita A, Chen CY, Yamashita Y, Zhu W, Durdan S, Kahvejian A, Sonenberg N, Shyu AB. UNR, a new partner of poly(A)-binding protein, plays a key role in translationally coupled mRNA turnover mediated by the c-fos major coding-region determinant. Genes Dev. 2004;18:2010–2023. doi: 10.1101/gad.1219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig AWB, Haghighat A, Yu ATK, Sonenberg N. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature. 1998;392:520–523. doi: 10.1038/33198. [DOI] [PubMed] [Google Scholar]
- Dehlin E, Wormington M, Korner CG, Wahle E. Cap-dependent deadenylation of mRNA. EMBO J. 2000;19:1079–1086. doi: 10.1093/emboj/19.5.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deo RC, Bonanno JB, Sonenberg N, Burley SK. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999;98:835–845. doi: 10.1016/s0092-8674(00)81517-2. [DOI] [PubMed] [Google Scholar]
- Dizin E, Gressier C, Magnard C, Ray H, Decimo D, Ohlmann T, Dalla Venezia N. BRCA1 interacts with poly(A)-binding protein: implication of BRCA1 in translation regulation. J Biol Chem. 2006;281:2436–46. doi: 10.1074/jbc.M602176200. [DOI] [PubMed] [Google Scholar]
- Etchison D, Milburn SC, Edery I, Sonenberg N, Hershey JWB. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eukaryotic initiation factor 3 and a cap binding protein complex. J Biol Chem. 1982;257:14806–14810. [PubMed] [Google Scholar]
- Fontaine-Rodriguez E, Taylor T, Olesky M, Knipe D. Proteomics of herpes simplex virus infected cell protein 27: association with translation initiation factors. Virology. 2004;330:487–92. doi: 10.1016/j.virol.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Gorlach M, Burd CG, Dreyfuss G. The mRNA poly(A)-binding protein: Localization, abundance, and RNA-binding specificity. Exp Cell Res. 1994;211:400–407. doi: 10.1006/excr.1994.1104. [DOI] [PubMed] [Google Scholar]
- Harris KS, Reddigari SR, Nicklin MJH, Hammerle T, Wimmer E. Purification and characterization of poliovirus polypeptide 3CD, a proteinase and a precursor for RNA polymerase. J Virol. 1992;66:7481–7489. doi: 10.1128/jvi.66.12.7481-7489.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold J, Andino R. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol Cell. 2001;7:581–591. doi: 10.1016/S1097-2765(01)00205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino S, Imai M, Kobayashi T, Uchida N, Katada T. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-Poly(A) tail of mRNA. Direct association of erf3/GSPT with polyadenylate-binding protein. J Biol Chem. 1999;274:16677–16680. doi: 10.1074/jbc.274.24.16677. [DOI] [PubMed] [Google Scholar]
- Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joachims M, van Breugel PC, Lloyd RE. Cleavage of poly(A)-binding protein by enterovirus proteinases concurrent with inhibition of translation in vitro. J Virol. 1999;73:718–727. doi: 10.1128/jvi.73.1.718-727.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jore J, DeGeus B, Jackson RJ, Pouwels PH, Enger-Valk B. Poliovirus protein 3CD is the active proteinase for processing of the precursor protein P1 in vitro. J Gen Virol. 1988;69:1627–1636. doi: 10.1099/0022-1317-69-7-1627. [DOI] [PubMed] [Google Scholar]
- Kahvejian A, Svitkin YV, Sukarieh R, M’Boutchou MN, Sonenberg N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 2005;19:104–113. doi: 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T, Sato Y, Sata T, Katano H. Expression of Kaposi’s sarcoma-associated herpesvirus-encoded K10/10.1 protein in tissues and its interaction with poly(A)-binding protein. Virology. 2006;352:100–109. doi: 10.1016/j.virol.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Khaleghpour K, Kahvejian A, De Crescenzo G, Roy G, Svitkin Y, Imataka H, O’connor-McCourt M, Sonenberg N. Dual interactions of the translational repressor Paip2 with poly (A) binding protein. Mol Cell Biol. 2001a;21:5200–5213. doi: 10.1128/MCB.21.15.5200-5213.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaleghpour K, Svitkin YV, Craig AW, DeMaria CT, Deo RC, Burley SK, Sonenberg N. Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol Cell. 2001b;7:205–216. doi: 10.1016/s1097-2765(01)00168-x. [DOI] [PubMed] [Google Scholar]
- Khanam T, Muddashatty RS, Kahvejian A, Sonenberg N, Brosius J. Poly(A)-binding protein binds to A-rich sequences via RNA-binding domains 1+2 and 3+4. RNA Biol. 2006;3:170–177. doi: 10.4161/rna.3.4.4075. [DOI] [PubMed] [Google Scholar]
- Koloteva-Levine N, Pinchasi D, Pereman I, Zur A, Brandeis M, Elroy-Stein O. The Apc5 subunit of the Anaphase-Promoting Complex/Cyclosome interacts with poly (A) binding protein and represses internal ribosome entry site-mediated translation. Mol Cell Biol. 2004;24(9):3577–87. doi: 10.1128/MCB.24.9.3577-3587.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov G, De Crescenzo G, Lim NS, Siddiqui N, Fantus D, Kahvejian A, Trempe JF, Elias D, Ekiel I, Sonenberg N, O’Connor-McCourt M, Gehring K. Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase. EMBO J. 2004;23:272–281. doi: 10.1038/sj.emboj.7600048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov G, Trempe JF, Khaleghpour K, Kahvejian A, Ekiel I, Gehring K. Structure and function of the C-terminal PABC domain of human poly(A)-binding protein. Proc Natl Acad Sci USA. 2001;98:4409–4413. doi: 10.1073/pnas.071024998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krausslich HG, Nicklin MJH, Toyoda H, Etchison D, Wimmer E. Poliovirus proteinase 2A induces cleavage of eukaryotic initiation factor 4F polypeptide p220. J Virol. 1987;61:2711–2718. doi: 10.1128/jvi.61.9.2711-2718.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn U, Pieler T. Xenopus poly(A) binding protein: functional domains in RNA binding and protein-protein interaction. J Mol Biol. 1996;256:20–30. doi: 10.1006/jmbi.1996.0065. [DOI] [PubMed] [Google Scholar]
- Kuyumcu-Martinez M, Belliot G, Sosnovtsev SV, Chang KO, Green KY, Lloyd RE. Calicivirus 3C-like proteinase inhibits cellular translation by cleavage of poly(A)-binding protein. J Virol. 2004a;78:8172–8182. doi: 10.1128/JVI.78.15.8172-8182.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuyumcu-Martinez NM, Joachims M, Lloyd RE. Efficient cleavage of ribosome-associated poly(A)-binding protein by enterovirus 3C proteinase. J Virol. 2002;76:2062–2074. doi: 10.1128/jvi.76.5.2062-2074.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuyumcu-Martinez NM, Van Eden ME, Younan P, Lloyd RE. Cleavage of Poly(A)-Binding Protein By Poliovirus 3C Proteinase Inhibits Host Cell Translation: A Novel Mechanism for Host Translation Shutoff. Mol Cell Biol. 2004b;24:1779–1790. doi: 10.1128/MCB.24.4.1779-1790.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamphear BJ, Yan RQ, Yang F, Waters D, Liebig HD, Klump H, Kuechler E, Skern T, Rhoads RE. Mapping the cleavage site in protein synthesis initiation factor-eIF-4g of the 2A proteinases from human coxsackievirus and rhinovirus. J Biol Chem. 1993;268:19200–19203. [PubMed] [Google Scholar]
- Le H, Tanguay RL, Balasta ML, Wei CC, Browning KS, Metz AM, Goss DJ, Gallie DR. Translation initiation factors eIF-iso4G and eIF-4B interact with the poly(A)-binding protein and increase its RNA binding activity. J Biol Chem. 1997;272:16247–16255. doi: 10.1074/jbc.272.26.16247. [DOI] [PubMed] [Google Scholar]
- Leonard S, Viel C, Beauchemin C, Daigneault N, Fortin M, Laliberte J. Interaction of VPg-Pro of turnip mosaic virus with the translation initiation factor 4E and the poly(A)-binding protein in planta. J Gen Virol. 2004;85:1055–1063. doi: 10.1099/vir.0.19706-0. [DOI] [PubMed] [Google Scholar]
- Liebig HD, Ziegler E, Yan R, Hartmuth K, Klump H, Kowalski H, Blaas D, Sommergruber W, Frasel L, Lamphear B, Rhoads R, Kuechler E, Skern T. Purification of two picornaviral 2A proteinases - interaction with eIF-4g and influence on in vitro translation. Biochemistry. 1993;32:7581–7588. doi: 10.1021/bi00080a033. [DOI] [PubMed] [Google Scholar]
- Okochi K, Suzuki T, Inoue J, Matsuda S, Yamamoto T. Interaction of anti-proliferative protein Tob with poly(A)-binding protein and inducible poly (A) binding protein: implication of Tob in translational control. Genes to Cells. 2005;10:151–163. doi: 10.1111/j.1365-2443.2005.00826.x. [DOI] [PubMed] [Google Scholar]
- Parsley TB, Cornell CT, Semler BL. Modulation of the RNA binding and protein processing activities of poliovirus polypeptide 3CD by the viral RNA polymerase domain. J Biol Chem. 1999;274:12867–12876. doi: 10.1074/jbc.274.18.12867. [DOI] [PubMed] [Google Scholar]
- Perera R, Daijogo S, Walter BL, Nguyen JH, Semler BL. Cellular protein modification by poliovirus: the two faces of poly (rC)-binding protein. J Virol. 2007;81:8919–8932. doi: 10.1128/JVI.01013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy G, De Crescenzo G, Khaleghpour K, Kahvejian A, O’Conner-McCourt M, Sonenberg N. Paip1 interacts with poly(A) binding protein through two independent binding motifs. Mol Cell Biol. 2002;22:3769–3782. doi: 10.1128/MCB.22.11.3769-3782.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterfield T, Pallanck L. Ataxin-2 and its Drosophila homolog, ATX2, physically assemble with polyribosomes. Hum Mol Genetics. 2006;15:2523–2532. doi: 10.1093/hmg/ddl173. [DOI] [PubMed] [Google Scholar]
- Sladic RT, Lagnado CA, Bagley CJ, Goodall GJ. Human PABP binds AU-rich RNA via RNA-binding domains 3 and 4. Eur J Biochem. 2004;271:450–457. doi: 10.1046/j.1432-1033.2003.03945.x. [DOI] [PubMed] [Google Scholar]
- White JP, Cardenas AM, Marissen WE, Lloyd RE. Inhibition of cytoplasmic stress granule formation by a viral proteinase. Cell Host & Microbe. 2007;2:295–305. doi: 10.1016/j.chom.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Wormington M, Searfoss AM, Hurney CA. Overexpression of poly(A) binding protein prevents maturation-specific deadenylation and translational inactivation in Xenopus oocytes. EMBO J. 1996;15:900–909. [PMC free article] [PubMed] [Google Scholar]
- Yao G, Chiang YC, Zhang C, Lee DJ, Laue TM, Denis CL. PAB1 Self-Association Precludes Its Binding to Poly (A), Thereby Accelerating CCR4 Deadenylation In Vivo. Mol Cell Biol. 2007;27:6243–6253. doi: 10.1128/MCB.00734-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ypma-Wong MF, Dewalt PG, Johnson VH, Lamb JG, Semler BL. Protein 3CD is the major poliovirus proteinase responsible for cleavage of the P1 capsid precursor. Virology. 1988;166:265–270. doi: 10.1016/0042-6822(88)90172-9. [DOI] [PubMed] [Google Scholar]
- Zamora M, Marissen WE, Lloyd RE. Multiple eIF4GI-specific proteinase activities present in uninfected and poliovirus-infeced cells. J Virol. 2002;76:165–177. doi: 10.1128/JVI.76.1.165-177.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Morace G, Gauss-Muller V, Kusov Y. Poly(A) binding protein, C-terminally truncated by the hepatitis A virus proteinase 3C, inhibits viral translation. Nucl Acid R. 2007;35:5975–5984. doi: 10.1093/nar/gkm645. [DOI] [PMC free article] [PubMed] [Google Scholar]