Abstract
Rotation of the sodium-driven polar flagellum of Vibrio alginolyticus requires four motor proteins: PomA, PomB, MotX, and MotY. PomA and PomB form a sodium-ion channel in the cytoplasmic membrane that functions as a stator complex to couple sodium-ion flux with torque generation. MotX and MotY are components of the T-ring, which is located beneath the P-ring of the polar flagellar basal body and is involved in incorporation of the PomA/PomB complex into the motor. Here, we describe the determination of the crystal structure of MotY at 2.9 Å resolution. The structure shows two distinct domains: an N-terminal domain (MotY-N) and a C-terminal domain (MotY-C). MotY-N has a unique structure. MotY-C contains a putative peptidoglycan-binding motif that is remarkably similar to those of peptidoglycan-binding proteins, such as Pal and RmpM, but this region is disordered in MotY. Motility assay of cells producing either of the MotY-N and MotY-C fragments and subsequent biochemical analyses indicate that MotY-N is essential for association of the stator units around the rotor, whereas MotY-C stabilizes the association by binding to the peptidoglycan layer. Based on these observations, we propose a model for the mechanism of stator assembly around the rotor.
Keywords: bacterial flagellum, peptidoglycan binding, sodium driven motor, T-ring
The bacterial flagellum is a rotating organelle that drives cell movement in response to environmental stimuli (1). The flagellum is driven by a reversible rotary motor embedded in the cell membrane at the base of the filament. The motor is powered by the transmembrane electrochemical gradient of a specific ion. Whereas the flagella of Escherichia coli and Salmonella enterica serovar Typhimurium use proton flux, the polar flagellum of Vibrio alginolyticus uses a flow of sodium ions (2, 3).
The motor consists of two parts, the rotor and the stator, and torque is generated between them. The stator is a transmembrane ion-channel composed of several different proteins. The stator units are produced and assembled around the rotor after completion of the hook–basal body structure. Even a single stator unit can drive rotation (4, 5). Although the stator unit of the Escherichia coli and Salmonella motors consists of two proteins, MotA and MotB, which form a proton-channel, the sodium-ion motor of V. alginolyticus requires four proteins, PomA, PomB, MotX, and MotY, to form a functional stator (6–8).
PomA and PomB are orthologs of MotA and MotB (6) and form a complex in the cytoplasmic membrane with the stoichiometry PomA4PomB2 (3, 9, 10). In their C-terminal periplasmic region, PomB and MotB both have a putative peptidoglycan-binding motif that is well conserved among outer-membrane proteins, such as outer membrane protein A (OmpA) from E. coli (11), peptidoglycan-associated lipoprotein (Pal) from E. coli (12), and reduction modifiable protein M (RmpM) from Neisseria meningitidis (13). Thus, PomB and MotB are thought to anchor the stator unit to the peptidoglycan layer (14, 15).
MotX and MotY are unique to the sodium-ion motors of Vibrio species. Null mutants of motX and motY exhibit nonmotile phenotypes (7, 8, 16), indicating that both proteins are essential for torque generation, but their actual roles remain unclear. MotX and MotY are translated with an N-terminal signal sequence, which is cleaved during their export to the periplasmic space, probably through the Vibrio equivalent of the E. coli Sec system (17). Mature MotY is a 32-kDa polypeptide with 272 amino acid residues. MotX and MotY form a complex in the periplasm, and MotX rapidly degrades in the absence of MotY, suggesting that complex formation stabilizes MotX (16, 18). MotX affects membrane localization of PomB, suggesting that MotX interacts with PomB (18).
The C-terminal region of MotY contains a putative peptidoglycan-binding motif similar to those of PomB and MotB, suggesting that MotY also binds to peptidoglycan (7). Recently, we discovered a ring-like structure (the T-ring) composed of MotX and MotY and localized beneath the P-ring of V. alginolyticus (19). In the absence of MotX or MotY, PomA and PomB do not localize to the flagellated cell pole, demonstrating that these two proteins are involved in the incorporation and/or stabilization of the PomA/PomB complex within the motor (19).
To understand the role of MotY in the assembly and function of the sodium-ion motor, we purified and crystallized (20) MotY from V. alginolyticus and determined its structure at 2.85 Å resolution. This structure and subsequent biochemical analyses clearly reveal distinct roles for the N- and C-terminal domains of MotY and provide insights into the process of stator complex assembly around the rotor.
Results
Overall Structure of MotY.
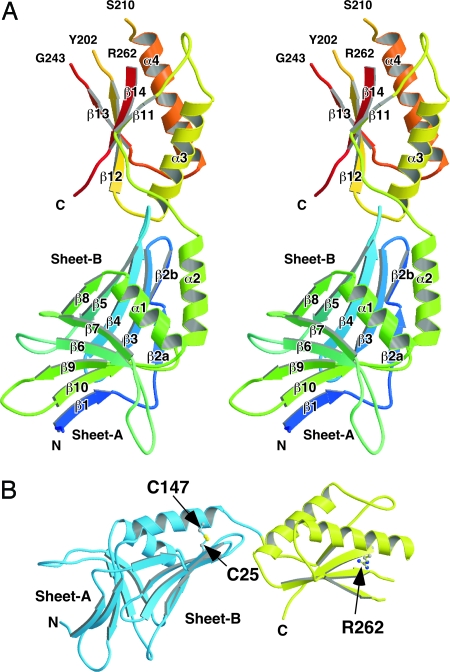
The asymmetric unit in the crystal contained a single MotY molecule. The atomic model of MotY was constructed from Val-1 to Gln-271 (Fig. 1). (This numbering does not include the 21 N-terminal residues that comprise the signal sequence). Residues Thr-203 through Lys-209, Lys-244 through Arg-261, Val-272, and the C-terminal hexahistidine tag were invisible in the electron density map, probably because of disorder, and are therefore missing in the model. Two distinct domains are present: an N-terminal domain (Val-1 through Tyr-154; MotY-N), and a C-terminal domain (Ser-155 through Val-272; MotY-C), with the approximate dimensions of 30 × 40 × 50 Å and 25 × 30 × 40 Å, respectively. The amino acid sequence and elements of secondary structure are shown in supporting information (SI) Fig. S1.
Fig. 1.
Overall structure of MotY. (A) Stereoview of the Cα ribbon drawing of MotY. (B) The N-terminal and C-terminal domains are shown in blue and yellow, respectively. The disulfide bond between Cys-25 and Cys-147 and the Arg-262 residue are displayed in ball-and-stick format.
Structure of MotY-N.
MotY-N is composed of two β-sheets, sheet-A and sheet-B, and two α-helices, α1 and α2. The two β-sheets form a distorted β-sandwich with a deep cleft, and the two α-helices cover the opening of the cleft. Each sheet contains five β-strands: β1, β6, β7, β9, and β10 in sheet-A; and β2, β3, β4, β5, and β8 in sheet-B (Fig. 1A). The strands are anti-parallel except for β1, which lies parallel to β10 in sheet-A. Sheet-B twists and extends toward MotY-C. Helix α1, which connects strands β8 and β9, is located just above the cleft and covers the hydrophobic core formed by the two β-sheets. Helix α2, which is followed immediately by MotY-C, lies on sheet-B along strand β3. The C-terminal end of helix α2 is anchored to sheet-B through a disulfide bridge between Cys-25 in β3 and Cys-147 in α2 (Fig. 1B). The two Cys residues are conserved in MotY from various bacteria with polar flagella (Fig. S1). This disulfide seems to be important for stabilizing MotY-N. MotY proteins containing the C25S, C147S, and C25S/C147S residues substitutions were susceptible to proteolysis and resulted in cells with reduced or lost motility (16).
Structure of MotY-C and Its Similarity to Other OmpA/MotB-Like Domains.
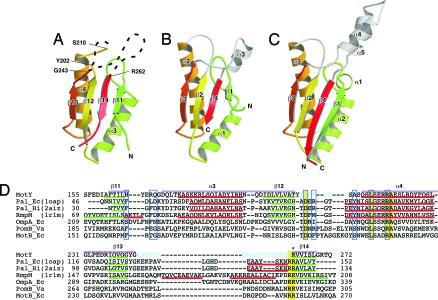
MotY-C forms an α/β sandwich composed of α3 and α4 and β11, β12, β13, and β14 (Fig. 1A and 2A). The strands form a mixed β-sheet; β11, β12, and β13 are parallel, and β14 is anti-parallel to them. The residues connecting β12 and α4, and β13 and β14, are invisible. The fold of MotY-C shows remarkable similarity to other OmpA/MotB-like proteins, such as the core domain of Pal (Fig. 2B) and the OmpA-like domain of RmpM (Fig. 2C) (13). MotY-C can be superimposed on RmpM [PDB ID code 1R1M (13)], E. coli Pal (PDB ID code 1OAP), and Haemophilus influenzae Pal [PDB ID code 2AIZ (21)], with rmsds for Cα atoms of 2.2 Å, 2.6 Å, and 1.9 Å, respectively. The corresponding sequence identities are 26%, 27%, and 21% (Fig. 2D). The major structural difference is that the residues connecting β13 and β14 are disordered in MotY, whereas they form helical structures in Pal and RmpM. The N terminus of helix α4 in MotY is also one turn shorter than in the corresponding helices of Pal and RmpM.
Fig. 2.
Comparison of OmpA/MotB-like domains. Cα ribbon representation of OmpA/MotB-like domain structures. (A) The C-terminal domain of MotY (MotY-C). (B) Pal of E. coli (1OAP). (C) The C-terminal domain of RmpM from N. meningitidis (1R1M). (D) Structure-based sequence alignment of the OmpA/MotB-like domains. The aligned sequences are: MotY, the C-terminal domain of V. alginolyticus MotY; Pal_Ec, E. coli Pal; Pal_Hi, H. influenzae Pal; RmpM, N. meningitidis RmpM; OmpA_Ec, E. coli OmpA; PomB_Va, V. alginolyticus PomB; and MotB_Ec, E. coli MotB. Residues highlighted in yellow are conserved in all seven proteins. Regions of secondary structure of MotY, Pal_Ec, Pal_Hi, and RmpM are indicated below the corresponding sequences as follows: Red line, α-helix; green line, β-strand. The secondary structure elements for MotY are labeled above the sequence. The residues contributing to the binding of the peptidoglycan precursor in Pal_Hi are indicated by blue boxes. The Arg-262 residue of MotY, which is important for motility and is conserved in OmpA/MotB-like proteins, is marked with an asterisk. Disordered residues in the MotY crystal structure are shown in cyan. The pink bars above the sequences represent the regions used to align the structures.
Disordering of the Putative Peptidoglycan-Binding Site.
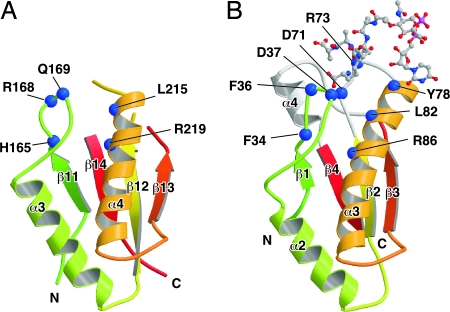
The NMR structure of H. influenzae Pal bound to the peptidoglycan precursor UDP-NAM-l-Ala-γ-d-Glu-l-Lys-d-Ala-d-Ala revealed that the peptidoglycan-binding pocket is formed by the loop between β1 and α2, the loop between β2 and α3, and α4 (21). Although the overall structure of the C-terminal domain of MotY is very similar to Pal (Fig. 2 A and B and Fig. 3), two of the three chains forming the binding pocket are disordered in the MotY crystal structure. The residues between β12 and α4 and between β13 and β14 in MotY, which correspond to the β2-α3 loop and helix α4 in Pal, are invisible in the electron density map. The C-terminal regions of β12 and β13 are slightly twisted and separate from α3; thus, the peptidoglycan-binding site is wide open in the MotY structure.
Fig. 3.
The peptidoglycan-binding regions of MotY (A) and Pal_Hi complexed with the peptidoglycan precursor (B). The residues contributing to binding of the peptidoglycan precursor and the corresponding residues in MotY (shown by the blue boxes in Fig. 2D) are indicated by blue balls. The polypeptide chains are drawn as Cα ribbon diagrams, with color coding going from green to red from the N terminus to the C terminus. The peptidoglycan precursor is shown in a ball-and-stick representation.
The conformation of the β11-α3 loop of MotY differs from the corresponding β1–α2 loop of Pal (Fig. 3), and the loops contain different numbers of residues (Fig. 2D). The residues in this loop are not conserved among the OmpA/MotB-like proteins, although Phe-36 and Asp-37 in Pal interact directly with the peptidoglycan precursor (Fig. 2D and 3B). The loop structure of RmpM deviates from the loops in both MotY and Pal. This loop may interact with peptidoglycan in different ways in these proteins. However, structure at the bottom of the peptidoglycan-binding site is well conserved. Leu-215 and Arg-219 in α4 of MotY are in the same relative positions as Leu-82 and Arg-86 in Pal; these residues interact directly with the peptidoglycan precursor (21). The orientation of Arg-219 in MotY is supported by His-165, just as Phe-34 supports Arg-86 in Pal.
MotY-N Is Essential for Motility.
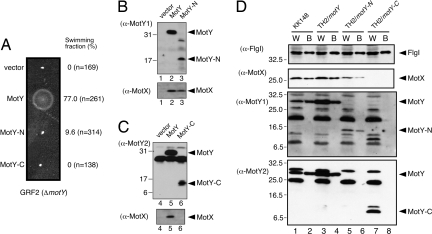
To elucidate the roles of the two domains of MotY, we analyzed the motility of ΔmotY (GRF2) cells expressing either MotY-N or MotY-C fused to a signal peptide at its N terminus. Expression of full-length MotY complemented the motility defect of GRF2 cells, whereas expression of MotY-N did not restore swarming in semisolid agar (Fig. 4A). However, ≈10% of the cells producing MotY-N swam to some extent when viewed by dark-field microscopy, indicating that the stator complex was installed into the motor even without MotY-C, albeit inefficiently. No motility was observed when MotY-C was expressed alone in GRF2 cells (Fig. 4A). Neither MotY-N nor MotY-C exhibited negative dominance when expressed from a multicopy plasmid in wild-type cells (data not shown).
Fig. 4.
Characterization of MotY-N and MotY-C. (A) Motility assays with GRF2 cells expressing MotY variants. Cells containing pSU41 (vector), pAS101 (MotY), pAS102 (MotY-N), and pAS103 (MotY-C) were inoculated into semisolid VPG agar and incubated at 30°C for 5 h. The fraction of cells swimming in liquid medium for each of the strains is shown at Right. (B and C) MotY-N prevents MotX degradation. Supernatants (Upper for MotY detection) and pellets (Lower for MotX detection) of lysates after ultracentrifugation were analyzed by SDS/PAGE followed by immunoblotting. Samples are for GRF2 (ΔmotY) cells containingthe pSU41 vector (lane 1 and 4), pAS101 (full-length MotY, lane 2 and 5), pAS102 (MotY-N, lane 3), and pAS103 (MotY-C, lane 6). Antibodies used for detection of proteins are shown on the left. Because we could not detect MotY-N with the affinity-purified MotY antibody, we used an MotY antiserum obtained from another rabbit for detection of MotY-N (α-MotY1). For detection of MotY and MotY-C, the affinity-purified antibody (α-MotY2) was used. Full-length MotY can be detected by both α-MotY1 and α-MotY2. (D) Association of MotY-N and MotY-C with the flagellar basal body. Samples of whole cells (W) and the basal bodies (B) of KK148 cells (lanes 1 and 2), TH2 cells expressing full-length MotY (lanes 3 and 4), TH2 cells expressing MotY-N (lanes 5 and 6), and TH2 cells expressing MotY-C (lanes 7 and 8) were analyzed by SDS/PAGE. The presence of the flagellar P-ring protein FlgI, MotX, and MotY (or its domains) were visualized by immunoblotting. Antibodies used for detection of proteins are shown on the left. As noted in C, MotY fragments were detected by α-MotY1 (for MotY-N) and α-MotY2 (for MotY-C). A number of extraneous bands appeared in the MotY blots are unidentified proteins that cross-reacted with the MotY antibodies.
MotY-N Is Responsible for Interaction with both MotX and the Basal-Body.
MotY interacts directly with MotX (18) and prevents degradation of MotX in the cell (16). Recently, we found that MotY, together with MotX, forms the T-ring structure beneath the P-ring (19). The ΔmotX strain had a smaller T-ring than the wild-type, and no T-ring was observed in ΔmotY and ΔmotXΔmotY strains, suggesting that MotY interacts with the basal body. GRF2 cells expressing MotY-N and MotY-C were grown to exponential phase, harvested, disrupted by sonication, and subjected to ultracentrifugation to isolate the insoluble membrane fraction and the soluble fraction. Both domains were detected in the soluble fraction (Fig. 4 B and C). MotX was also detected when intact MotY or MotY-N was coexpressed but not when MotY-C was coexpressed. These observations suggest that MotY-N is responsible for stabilizing the MotX structure, presumably through direct interaction between MotX and MotY-N.
To investigate the association of each domain with the basal body, we isolated basal bodies from TH2 cells (a ΔmotY strain derived from the multiflagellated KK148 mutant) (19) expressing either MotY-N or MotY-C and analyzed the preparation by immunoblotting (Fig. 4D). Although both proteins were found in the whole-cell fraction (Fig. 4D, lanes 5 and 7), only MotY-N was detected in the basal body fraction (Fig. 4D, lanes 6 and 8). MotX was detected in both the whole-cell and basal body fractions when MotY-N was coexpressed (Fig. 4D, lanes 5 and 6), confirming that MotY-N prevents degradation of MotX. The P-ring protein FlgI was detected in all fractions, indicating that the basal body was properly isolated. These results suggest that MotY-N binds both to the flagellar basal body and MotX.
Discussion
The crystal structure of MotY revealed that the protein consists of two distinct domains: the N-terminal MotY-N and the C-terminal MotY-C. MotY-C, which contains a putative peptidoglycan-binding sequence motif, showed remarkable structural similarities to the peptidoglycan-binding domains of Pal (21) and RmpM (13), but the MotY-N structure was unique.
Several experiments indicated that MotY-N is essential for formation of a functional stator complex, whereas MotY-C is not (Fig. 4). A ΔmotY strain expressing MotY-N had a weak but significant ability to swim, and significant amounts of MotY-N and MotX were detected in a purified basal body fraction, although the T-ring structure of this strain was difficult to visualize by electron microscopy (data not shown). These results suggest that MotY-N connects the basal body to MotX and that the PomA/PomB complex associates with MotX to form the functional stator complex around the rotor. MotY-N alone does not associate strongly with the basal body, but the partial T-ring structure made of the MotY-N/MotX complex must be sufficient to allow at least a few PomA/PomB stator complexes to be incorporated into the motor.
The similarity between MotY-C and other peptidoglycan-binding proteins suggests that MotY interacts with the peptidoglycan layer much like those proteins. In the crystal structure, however, the putative peptidoglycan-binding site of MotY-C was more widely open than in the structures determined for Pal and RmpM (Fig. 2), and most of the loops forming the peptidoglycan-binding pocket were disordered (Fig. 3). The disordered loops appear to be intrinsic to monomeric MotY and not an artifact of the crystallization conditions, because the native crystal obtained at pH 4.5, for which the model was refined, and the Se-Met derivative crystal obtained at pH 8.5 both show the disordered conformation (data not shown). A disordered peptidoglycan-binding pocket may be an important property for free MotY, because, after secretion into the periplasm, MotX and MotY presumably must diffuse around until they collide with the basal body and form the T-ring structure. Therefore, MotY should not bind tightly to the peptidoglycan layer before it encounters the basal body. Interaction with the basal body may induce folding of the disordered chains into a functional peptidoglycan-binding pocket.
A motY mutant containing the R262Q residue substitution exhibits greatly decreased swarming motility in semisolid agar (16). Arg-262 is near the N terminus of the β14 strand in MotY-C (Fig. 1B), and this residue is conserved in various proteins of the OmpA/MotB family (Fig. 2D). Although Arg-262 is near the putative peptidoglycan-binding site, its side chain extends away from the binding pocket. Thus, it is unclear why the R262Q substitution affects motility so strongly. One possible explanation is that Arg-262 may interact with a peptide chain of another peptidoglycan strand cross-linked to the peptidoglycan chain bound to the binding site. Another possibility is that Arg-262 is involved in association of MotY with the basal body to induce proper folding of the peptidoglycan-binding pocket.
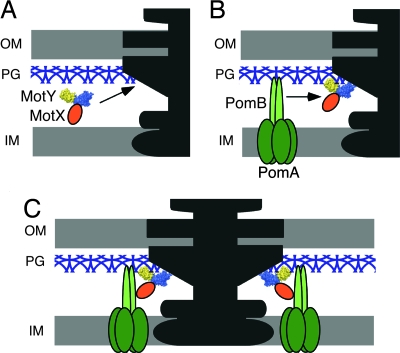
MotX and MotY are essential for torque generation by the sodium ion-driven polar flagellar motor of V. alginolyticus. Based on the results presented here, we propose a model for stator assembly (Fig. 5). MotY may form a complex with MotX and diffuse around the periplasm with little or no affinity for the peptidoglycan layer (Fig. 5A). When the complex associates with the basal body via MotY-N, conformational changes are induced that greatly increase the affinity of MotY-C for peptidoglycan. The PomA/PomB complex also forms before association with the motor and diffuses around in the cytoplasmic membrane (Fig. 5B). When the PomA/PomB complex encounters the basal body, association of the periplasmic C-terminal domain of PomB with MotX positions PomB to bind to the peptidoglycan layer in the proper orientation to interact productively with the rotor (Fig. 5C).
Fig. 5.
Model of stator assembly in the sodium ion-driven polar flagellar motor of Vibrio. MotY is shown as a space-filling model colored blue for MotY-N and yellow for MotY-C. MotX, PomA, and PomB are shown in orange, dark green, and light green, respectively. (A) MotX and MotY form a complex and diffuse around in the periplasm without binding to the peptidoglycan layer. (B) Interaction of the MotX/MotY complex with the flagellar basal body triggers a conformational change in MotY-C that increases its affinity for the peptidoglycan layer, and the complex associates tightly with the basal body via MotY-N to form the T-ring. The PomA/PomB stator complex diffuses through the membrane. (C) The PomA/PomB complexes assemble around the rotor through association with MotX in the T-ring and binding to the peptidoglycan layer via the C-terminal domain of PomB.
The proton-driven motors of E. coli and Salmonella do not need MotX or MotY to function, and a ΔpomABΔmotXΔmotY strain of Vibrio cholerae can swim by using proton motive force if it contains a plasmid that encodes the E. coli MotA/MotB complex (22). Furthermore, a stator complex composed of PomA and PotB, which is a chimeric protein containing the N-terminal transmembrane domain of PomB and the C-terminal periplasmic domain of E. coli MotB, is functional in V. alginolyticus without MotX or MotY (23). The PomA/PotB complex, which forms a sodium-ion channel, also allows the E. coli flagellar motor to rotate by using a sodium-ion current (23). These observations indicate that the periplasmic region of MotB can carry out the combined functions of the periplasmic domain of PomB, MotX, and MotY. Pseudomonas aeruginosa, which also has a polar flagellum, has a MotY homolog but no MotX (24). Comparative studies of the Vibrio, P. aeruginosa, E. coli, and Salmonella flagellar motors should elucidate the variety of mechanisms used to assemble stator complexes around the flagellar motors of γ Proteobacteria.
Materials and Methods
Preparation and Crystallization of MotY.
Details of expression, purification, and crystallization of MotY with a hexahistidine tag are described in ref. 20. Briefly, hexagonal P6522 crystals of MotY with unit cell dimensions a = b = 104.1 Å and c = 133.1 Å were grown from a solution containing 1.12 M NaH2PO4/K2HPO4, 5% PEG 1000, and 0.1 M acetate (pH 4.5) by the sitting drop vapor-diffusion method. Se-Met labeled MotY was prepared and crystallized in a solution containing 24% polyethylene glycol monoethyl ether 2000, 0.36 M ammonium sulfate, and 0.1 M Tris·HCl (pH 8.5). Although the crystallization conditions for the Se-Met derivative were different from those for native MotY, the unit cell dimensions (a = b = 104.7 Å and c = 132.3 Å) were almost the same as those of the native crystals.
Data Collection and Structure Determination.
X-ray diffraction data were collected at the synchrotron beamline BL41XU of SPring-8 (Harima, Japan). The statistics of the data are described in ref. 20. The crystals were frozen in liquid nitrogen. Because the Se-Met derivative crystals were highly sensitive to X-ray, we used a helium cryocooling device to reduce the radiation damage. The data were processed with MOSFLM (25) and scaled with SCALA (26). Phase calculation was performed by using SOLVE (27). The best electron-density map was obtained from MAD phases from the Se-Met derivative data, followed by density modification with DM (26). The model was constructed with O (28) and was refined against the native data at 2.85 Å, using CNS software (29). During the refinement process, iterative manual modifications were performed by using “omit map.” The refinement converged to an R factor of 29.0% and a free R factor of 31.2%. The Ramachandran plot showed that 71.8% and 25.5% of the residues were located in the most favorable and allowed regions, respectively. Structural refinement statistics are summarized in Table S1.
Expression and Fractionation of MotY Fragments in Vibrio Cells.
The plasmid pSU41 (Krm, vector), pAS101 (encoding full-length motY in pSU41), pAS102 (encoding MotY-N, residues 1–154 of MotY in pSU41), and pAS103 (encoding MotY-C, residues 154–272 in pSU41), were introduced into the ΔmotY strain GRF2 (16) by electroporation (30). MotY fragments were constructed with the native signal sequence fused at their N termini so that they are secreted to the periplasmic space. Transformants were grown exponentially at 30°C in vibrio peptone/tryptone glycerol (VPG) medium (1% bactotryptone, 0.4% K2HPO4, 3% NaCl, and 0.5% glycerol) containing kanamycin 100 μg/ml. The cells were harvested and washed once with buffer [20 mM Tris·HCl (pH 8.0), 500 mM NaCl], then resuspended in the same buffer. The cells were disrupted by sonication (Branson Ultrasonics), unbroken cells were removed by centrifugation (12,000 × g for 5 min), and the solution was ultracentrifuged (148,781 × g for 30 min). The fractionated samples were precipitated by trichloroacetic acid if necessary, then analyzed by SDS/PAGE followed by immunoblotting. Because we could not detect MotY-N with the affinity-purified MotY antibody, we used an MotY antiserum obtained from another rabbit for detection of MotY-N. For detection of MotY and MotY-C, the affinity-purified antibody was used.
Isolation of the Flagellar Basal-Body.
Full-length MotY, MotY-N, and MotY-C were expressed in the ΔmotY strain TH2 (19) from plasmids pAS101, pAS102, and pAS103, respectively. The strain TH2 was derived from multipolar-flagellated mutant KK148 (31) so that we can obtain high yields of flagellar basal bodies from multiple polar flagella. The cells were grown in the VPg medium exponentially, and isolation of the basal bodies from each strain was carried out as described in ref. 19. Isolated basal body samples were analyzed by SDS/PAGE followed by immunoblotting. For detection of the P-ring protein FlgI, we used anti-Salmonella FlgI antibody that cross-reacts with FlgI of V. alginolyticus.
Motility Assay.
GRF2 cells were transformed with the respective plasmids, and fresh colonies were inoculated onto the VPG soft agar plates (1% bactotryptone, 0.4% K2HPO4, 3% NaCl, 0.5% glycerol, and 0.25% bact-agar) containing kanamycin 100 μg/ml and incubated at 30°C for 5 h. To measure the fraction of swimming cells, cells of each construct were grown exponentially in the VPg medium at 30°C, harvested and suspended in TMN300 medium [50 mM Tris·HCl (pH 7.5), 5 mM MgCl2, 5 mM glucose, and 300 mM NaCl]. Then, a small amount of the cell suspension was diluted 10-fold into the fresh TMN300 medium, motility of the cells was observed at room temperature under a dark-field microscope, and images were recorded on videotapes. The fraction of swimming cells was determined by counting the ratio of motile to total cells among the total cells in one video image.
Supplementary Material
Acknowledgments.
We thank I. Kawagishi for invaluable discussions, S. Nagashima for help with machine setup for modeling and refinement; S. Tatematsu for technical support; N. Shimizu, M. Kawamoto, K. Hasegawa, and M. Yamamoto at SPring-8 for technical help in use of beamlines; F. Oosawa, S. Asakura, and S. Hayashi for continuous support and encouragement during this work; K. Kutsukake (Okayama University, Okayama, Japan) for providing anti-Salmonella FlgI antibody. This work was supported in part by Ministry of Education, Science and Culture of Japan Grants-in-Aid for Scientific Research 18074006 (to K.I.) and 16087207 (to K.N.), the National Project on Protein Structural and Functional Analyses (K.I. and M.H.), and the Soft Nano-Machine Project of the Japan Science and Technology Agency (A.S., T.Y., and M.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in Protein Data Bank, www.pdb.org (PDB ID code 2ZF8).
This article contains supporting information online at www.pnas.org/cgi/content/full/0800308105/DCSupplemental.
References
- 1.Berg HC. The rotary motor of bacterial flagella. Annu Rev Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 2.Kojima S, Blair DF. The bacterial flagellar motor: Structure and function of a complex molecular machine. Int Rev Cytol. 2004;233:93–134. doi: 10.1016/S0074-7696(04)33003-2. [DOI] [PubMed] [Google Scholar]
- 3.Yorimitsu T, Homma M. Na+-driven flagellar motor of Vibrio. Biochim Biophys Acta. 2001;1505:82–93. doi: 10.1016/s0005-2728(00)00279-6. [DOI] [PubMed] [Google Scholar]
- 4.Ryu WS, Berry RM, Berg HC. Torque-generating units of the flagellar motor of Escherichia coli have a high duty ratio. Nature. 2000;403:444–447. doi: 10.1038/35000233. [DOI] [PubMed] [Google Scholar]
- 5.Sowa Y, et al. Direct observation of steps in rotation of the bacterial flagellar motor. Nature. 2005;437:916–919. doi: 10.1038/nature04003. [DOI] [PubMed] [Google Scholar]
- 6.Asai Y, et al. Putative channel components for the fast-rotating sodium-driven flagellar motor of a maine bacterium. J Bacteriol. 1997;179:5104–5110. doi: 10.1128/jb.179.16.5104-5110.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okunishi I, Kawagishi I, Homma M. Cloning and characterization of motY, a gene coding for a component of the sodium-driven flagellar motor in Vibrio alginolyticus. J Bacteriol. 1996;178:2409–2415. doi: 10.1128/jb.178.8.2409-2415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okabe M, Yakushi T, Asai Y, Homma M. Cloning and characterization of motX, a Vibrio alginolyticus sodium-driven flagellar motor gene. J Biochem (Tokyo) 2001;130:879–884. doi: 10.1093/oxfordjournals.jbchem.a003061. [DOI] [PubMed] [Google Scholar]
- 9.Sato K, Homma M. Functional reconstitution of the Na+-driven polar flagellar motor component of Vibrio alginolyticus. J Biol Chem. 2000;275:5718–5722. doi: 10.1074/jbc.275.8.5718. [DOI] [PubMed] [Google Scholar]
- 10.Sato K, Homma M. Multimeric structure of PomA, the Na+-driven polar flagellar motor component of Vibrio alginolyticus. J Biol Chem. 2000;275:20223–20228. doi: 10.1074/jbc.M002236200. [DOI] [PubMed] [Google Scholar]
- 11.Pautsch A, Schulz GE. Structure of the outer membrane protein A transmembrane domain. Nat Struct Biol. 1998;5:1013–1017. doi: 10.1038/2983. [DOI] [PubMed] [Google Scholar]
- 12.Cascales E, Lloubes R. Deletion analysis of the peptidoglycan-associated lipoprotein Pal reveals three independent binding sequences including a TolA box. Mol Microbiol. 2004;51:873–885. doi: 10.1046/j.1365-2958.2003.03881.x. [DOI] [PubMed] [Google Scholar]
- 13.Grizot S, Buchanan SK. Structure of the OmpA-like domain of RmpM from Neisseria meningitidis. Mol Microbiol. 2004;51:1027–1037. doi: 10.1111/j.1365-2958.2003.03903.x. [DOI] [PubMed] [Google Scholar]
- 14.De Mot R, Vanderleyden J. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both gram-positive and gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol Microbiol. 1994;12:333–334. doi: 10.1111/j.1365-2958.1994.tb01021.x. [DOI] [PubMed] [Google Scholar]
- 15.Koebnik R. Proposal for a peptidoglycan-associating alpha-helical motif in the C-terminal regions of some bacterial cell-surface proteins. Mol Microbiol. 1995;16:1269–1270. doi: 10.1111/j.1365-2958.1995.tb02348.x. [DOI] [PubMed] [Google Scholar]
- 16.Yagasaki J, Okabe M, Kurebayashi R, Yakushi T, Homma M. Roles of the intramolecular disulfide bridge in MotX and MotY, the specific proteins for sodium-driven motors in Vibrio spp. J Bacteriol. 2006;188:5308–5314. doi: 10.1128/JB.00187-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okabe M, Yakushi T, Kojima M, Homma M. MotX and MotY, specific components of the sodium-driven flagellar motor, colocalize to the outer membrane in Vibrio alginolyticus. Mol Microbiol. 2002;46:125–134. doi: 10.1046/j.1365-2958.2002.03142.x. [DOI] [PubMed] [Google Scholar]
- 18.Okabe M, Yakushi T, Homma M. Interactions of MotX with MotY and with the PomA/PomB sodium ion channel complex of the Vibrio alginolyticus polar flagellum. J Biol Chem. 2005;280:25659–25664. doi: 10.1074/jbc.M500263200. [DOI] [PubMed] [Google Scholar]
- 19.Terashima H, Fukuoka H, Yakushi T, Kojima S, Homma M. The Vibrio motor proteins, MotX and MotY, are associated with the basal body of Na+-driven flagella and required for stator formation. Mol Microbiol. 2006;62:1170–1180. doi: 10.1111/j.1365-2958.2006.05435.x. [DOI] [PubMed] [Google Scholar]
- 20.Shinohara A, et al. Crystallization and preliminary X-ray analysis of MotY, a stator component of the Vibrio alginolyticus polar flagellar motor. Acta Crystallogr F. 2007;63:89–92. doi: 10.1107/S1744309106055850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parsons LM, Lin F, Orban J. Peptidoglycan recognition by Pal, an outer membrane lipoprotein. Biochemistry. 2006;45:2122–2128. doi: 10.1021/bi052227i. [DOI] [PubMed] [Google Scholar]
- 22.Gosink KK, Häse CC. Requirements for conversion of the Na+-driven flagellar motor of Vibrio cholerae to the H+-driven motor of Escherichia coli. J Bacteriol. 2000;182:4234–4240. doi: 10.1128/jb.182.15.4234-4240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Asai Y, Yakushi T, Kawagishi I, Homma M. Ion-coupling determinants of Na+-driven and H+-driven flagellar motors. J Mol Biol. 2003;327:453–463. doi: 10.1016/s0022-2836(03)00096-2. [DOI] [PubMed] [Google Scholar]
- 24.Doyle TB, Hawkins AC, McCarter LL. The complex flagellar torque generator of Pseudomonas aeruginosa. J Bacteriol. 2004;186:6341–6350. doi: 10.1128/JB.186.19.6341-6350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leslie AGW. CCP4+ESF-EACMB Newslett. Protein Crystallogr. 1992;26:27–33. [Google Scholar]
- 26.Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 27.Terwilliger TC, Berendzen J. Automated structure solution for MIR and MAD. Acta Crystallogr D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 29.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 30.Kawagishi I, Okunishi I, Homma M, Imae Y. Removal of the periplasmic DNase before electroporation enhances efficiency of transformation in a marine bacterium Vibrio alginolyticus. Microbiology. 1994;140:2355–2361. [Google Scholar]
- 31.Kusumoto A, et al. Regulation of polar flagellar number by the flhF and flhG genes in Vibrio alginolyticus. J Biochem (Tokyo) 2006;139:113–121. doi: 10.1093/jb/mvj010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.