Abstract
Dimeric myosins V and VI travel long distances in opposite directions along actin filaments in cells, taking multiple steps in a “hand-over-hand” fashion. The catalytic cycles of both myosins are limited by ADP dissociation, which is considered a key step in the walking mechanism of these motors. Here, we demonstrate that external loads applied to individual actomyosin V or VI bonds asymmetrically affect ADP affinity, such that ADP binds weaker under loads assisting motility. Model-based analysis reveals that forward and backward loads modulate the kinetics of ADP binding to both myosins, although the effect is less pronounced for myosin VI. ADP dissociation is modestly accelerated by forward loads and inhibited by backward loads. Loads applied in either direction slow ADP binding to myosin V but accelerate binding to myosin VI. We calculate that the intramolecular load generated during processive stepping is ≈2 pN for both myosin V and myosin VI. The distinct load dependence of ADP binding allows these motors to perform different cellular functions.
Keywords: gating mechanism, intramolecular load, molecular motors, processivity
Processive dimeric myosins V and VI “walk” along individual actin filaments via a “hand-over-hand” mechanism, in which the two catalytic subunits, or “heads,” alternately switch leading and trailing positions (1–3). Models of processive myosin V and myosin VI motility implicate a steady-state walking intermediate with both heads, supposedly bound to ADP, attached to actin (4–12). Such an intermediate would require some form of communication between the heads such that rate-limiting ADP dissociation and subsequent ATP binding would occur in the trailing head with a higher probability than in the leading head. Electron microscopy and structural modeling (13) suggest that during double-headed binding to actin, the leading head is pulled back by the trailing head, which, in turn, is pulled forward by the leading head. The resulting intramolecular load affects the ATPase cycle kinetics of at least one head (5, 7, 14) and is hypothesized to create the asymmetry responsible for effective, directional processive stepping (4, 5, 7, 13, 14). The hypothesis that the intramolecular load increases the efficiency of the processive movement by modulating biochemical kinetics is supported by mechanical measurements of individual single-headed myosin V molecules (15, 16). However, these studies yield partly conflicting conclusions regarding the kinetic basis of how the load-induced asymmetry between the two ADP-bound heads arises, and it remains unclear whether head-head communication through the intramolecular load results solely in inhibition of ADP dissociation from the leading head, or, additionally, acceleration of ADP dissociation from the trailing head.
ADP binding to myosins V and VI is coupled to a rotation of the “lever” (17, 18) and should, therefore, be sensitive to load (19). Biochemical solution studies (5, 7, 20), mechanical measurements of individual motor molecules (6, 8, 15, 16), and modeling of processive run lengths (9) are consistent with this prediction. However, direct experimental evidence demonstrating load-dependent modulation of ADP affinity of individual myosin V or VI molecules is lacking. In addition, the load dependence of ADP dissociation and binding to single-headed myosin VI remains uncharacterized, and studies (15, 16) do not identify the load dependence of ADP binding to myosin V.
In this report, we use optical nanometry to examine the effect of applied external loads on individual actomyosin V and VI bonds to test the hypothesis that loads modulate biochemical processes within the motor domain, namely the ADP affinity, in an asymmetric manner, with the effect of loads applied toward the barbed end of an actin filament differing from that of loads applied toward the pointed end. We use a model analysis to identify the kinetic basis of the load-dependent affinity. Quantitative knowledge of the load-dependent ADP binding parameters allows us to calculate the intramolecular load generated during processive motility of myosins V and VI.
Results and Discussion
Experimental Measurement of the Actomyosin Unbinding Force.
Individual single-headed myosin V or myosin VI molecules were attached to an actin filament, and a significant external load was applied in either a forward or backward direction of the examined myosin's motility, mimicking the proposed effect on the trailing or leading head, respectively, as we have done for the kinesin–microtubule interaction (21). A bead with a single bound myosin molecule was held by optical tweezers and brought close to an actin filament that had been immobilized on the glass surface, and the stage was slowly displaced with a constant speed along the actin filament axis (Fig. 1A). The unbinding forces of individual actomyosin complexes were measured in the absence of nucleotides (rigor state) or in the presence of various ADP concentrations (Fig. 1B).
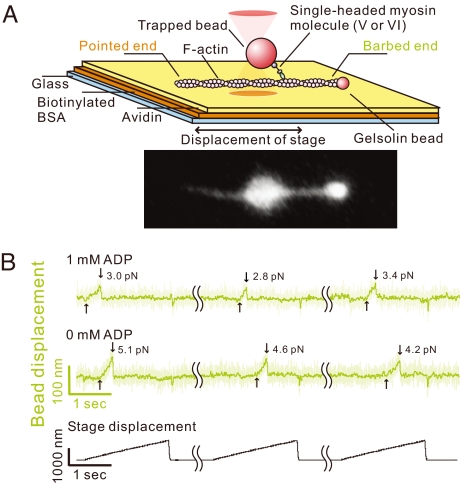
Fig. 1.
Myosins V and VI unbinding force measurements. (A) Schematic drawing of the experimental setup and the fluorescence image of a bead–actin filament system. Polarity of a bead-tailed actin filament can be clearly determined. (B) Typical traces showing bead movement during pulling–unbinding of individual single-headed myosin V–actin complex under forward (barbed-end directed) load. Traces at each ADP concentration (1 and 0 mM) were obtained with the same bead; the futile stage displacements, during which no binding was detected, and waiting time (5–10 sec between consecutive stage displacements) are not shown (wavy lines indicate elipsed data). Light green trace indicates the raw data (collected at 10 kHz), and dark green trace is the low-pass filtered data (50 Hz). The moment at which the external load began to be imposed on the actomyosin bond, and the moment of the unbinding are indicated by the upward and downward arrows, respectively. The trace jumps slightly when the stage quickly returns to the initial position. Representative traces for myosin VI are shown in Fig. S4.
Effect of ADP on the Actomyosin V Unbinding Force.
The unbinding force distributions (Fig. 2A) of single-headed myosin V having all six IQ domains (MV-6IQ) indicate that the actomyosin V-6IQ bond (±ADP) is more stable and dissociates less readily under backward pointed-end loads than under forward barbed-end loads. The absolute values of the actomyosin V unbinding forces in the nucleotide-free, or rigor, state are 5.1 ± 0.1 pN and 4.6 ± 0.2 pN for backward and forward loading, respectively (Table 1). Saturating ADP (1 mM) weakens the actomyosin bond and decreases the unbinding force to 4.0 ± 0.1 pN and 3.1 ± 0.1 pN for backward and forward loads, respectively. Such small separation of the peaks is due to only 10-fold difference in the lifetimes of the nucleotide-free and ADP-bound states on actin in the absence of load, contrary to 150-fold difference in case of kinesin (22). However, the t test confirms that for both loading directions the two peaks are statistically distinguishable (two-tailed t test; P < 0.05). The nucleotide-dependent unbinding force indicates that ADP binding induces conformational rearrangement of the actin-myosin binding interface (23).
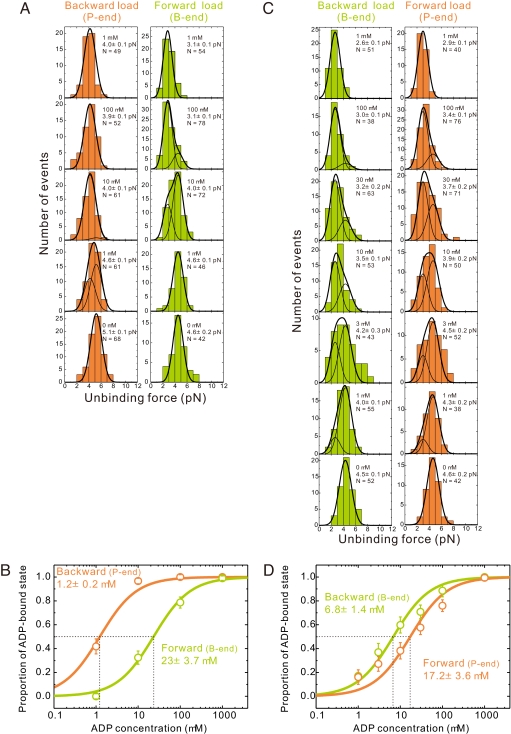
Fig. 2.
ADP affinity of myosins V and VI depends on loading direction. (A) Unbinding force distributions of individual myosin V–actin complexes at various ADP concentrations (0–1 mM). ADP concentration and total number of data, N, are shown in the upper right corners. Slender curves are the results of globally fitting histograms to two Gaussian fits [corresponding to the detachment in stronger (rigor) and weaker (ADP) binding states], separately for the backward (pointed-end directed) and forward (barbed-end directed) loading. The bold curves are the sum of two slender curves. (B) Estimation of the apparent ADP dissociation constants of actomyosin V under backward or forward load. Proportion of the ADP state is calculated as the ratio of the area of the left peak to the total area of the unbinding force distribution obtained by a global fit. Error bars represent the SEM, calculated as (p·(1 − p)/N)1/2, where p is the existence probability of the ADP-bound state and N is the total number of data at each ADP concentration. (C and D) Similar analysis for individual myosin VI–actin complexes. In all images, data obtained under barbed (B)- and pointed (P)-end-directed loading are green and orange, respectively.
Table 1.
Unbinding forces and ADP affinities of individual actomyosin V or VI complexes under directional loading
| Myosin | Unbinding force, pN |
ADP affinity, μM |
||||
|---|---|---|---|---|---|---|
| ADP state |
Rigor state |
|||||
| P end | B end | P end | B end | P end | B end | |
| V | 5.1 ± 0.1 (5.2) | 4.6 ± 0.2 (4.5) | 4.0 ± 0.1 (4.2) | 3.1 ± 0.1 (2.8) | 1.2 ± 0.2 | 23 ± 3.7 |
| VI | 4.6 ± 0.2 (4.6) | 4.5 ± 0.1 (4.3) | 2.9 ± 0.1 (3.0) | 2.6 ± 0.1 (2.6) | 17.2 ± 3.6 | 6.8 ± 1.4 |
Unbinding force values are given as average ± SEM for the rigor state (0 mM ADP) and the ADP state (1 mM ADP). Values in parentheses were obtained from global fit.
Directional Load Induces Strong Asymmetry of the Apparent Actomyosin V ADP Affinities.
The unbinding force distributions at every [ADP] are composed of the unbinding events in the stronger (rigor) and weaker (ADP) actomyosin binding states, and the confirmed statistical difference between the peaks allows for reliable fitting to a bimodal distribution. The fraction of rigor and ADP-bound states changes with [ADP] (Fig. 2B), permitting the affinity for ADP under load to be determined from the [ADP] dependence of the relative population of the two states by globally fitting the unbinding force distributions. The apparent ADP dissociation constant (Kd) (ADP affinity) under barbed-end (forward) loading and pointed-end (backward) loading differs ≈20-fold (23 μM and 1.2 μM, respectively), indicating that load strongly affects the ADP affinity of actomyosin V in an asymmetric direction-dependent manner such that the ADP affinity of the trailing head is lower than that of the leading head.
The unbinding force distributions with other binning parameters (0.75 pN and 0.5 pN) show no significant difference with 1 pN bins [supporting information (SI) Figs. S1 A and B and S2 A and B), which confirms that the results are independent of the binning size. Additionally, because the bimodal distribution is much less evident than in the case of kinesin, the ADP affinities were estimated from the average values of the unbinding forces (Fig. S3; see Methods for details), which also gave similar results as when fitting the distributions to double Gaussian functions.
Load Dependence of ADP Dissociation and Binding Rates in Actomyosin V.
The ADP dissociation constant for single-headed myosin V-6IQ bound to actin in the absence of load, obtained from solution studies, is 1.2 μM (Table S1). The apparent ADP dissociation constants (Kd) determined in this study (23 μM and 1.2 μM for the trailing and the leading head, respectively) appear to indicate that load modulates ADP binding to the trailing head, whereas the leading head is unaffected. However, because Kd = kADP−/kADP+, load could equally modulate ADP binding and dissociation so that the overall affinity of the leading head is not affected. To determine the load dependence of ADP binding and dissociation kinetics in both heads, we performed model-based analysis on the experimentally obtained unbinding force distributions.
The populations of the ADP-bound and the nucleotide-free states at each [ADP] are defined by the load-dependent rates of ADP dissociation and binding (Fig. 3). Using the kinetic rate constants of ADP dissociation and binding (kADP,0− and kADP,0+, respectively) for actomyosin V and the lifetimes (τD,0 and τφ,0, respectively) of the actomyosin bond in the ADP-bound and nucleotide-free states in the absence of load, obtained from solution studies, as the initial parameters (Table S1), the characteristic distances for ADP dissociation and binding (Table S2) and, therefore, load dependence of ADP dissociation and binding rates could be determined (see Methods for details), as summarized in Fig. 4 A and B. Note here that in our model neither dADP− no dADP+ were constrained to be symmetrical under forward and backward loads, which allowed us to detect the asymmetrical effects of load on the dissociation and binding in the two heads.
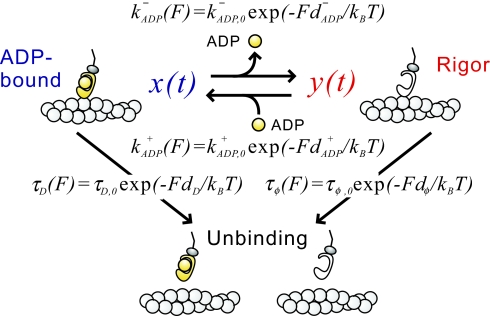
Fig. 3.
Schematic representation of the model. kADP− and kADP+ are the rates of ADP dissociation and binding, respectively; dD and dφ are the characteristic distances for the interaction between actin and myosin in the ADP-bound and the rigor state, respectively; dADP− and dADP+ are the characteristic distances for the ADP dissociation and binding, respectively; and τD and τφ are the lifetimes of actomyosin bond in the ADP-bound and the rigor states, respectively. Index 0 denotes corresponding parameters obtained in the absence of load by the solution studies.
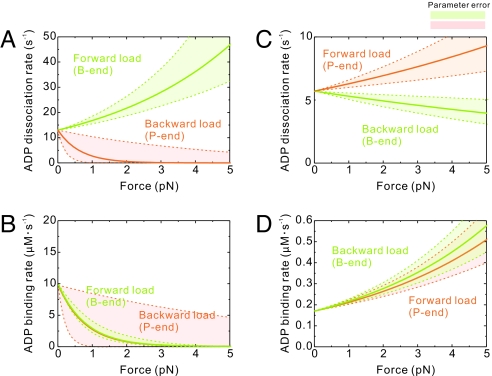
Fig. 4.
Loads modulate rates of ADP dissociation and binding in myosins V and VI. Dependence of the ADP dissociation (A and C) and binding (B and D) rates on load is plotted for myosins V (A and B) and VI (C and D). In all panels data obtained under barbed (B)- and pointed (P)-end directed loading are colored green and orange, respectively.
Load Inhibits ADP Dissociation from the Leading Head in Myosin V.
ADP dissociation from myosin V (Fig. 4 A and B) is only modestly accelerated (≤2-fold) by forward load (trailing head) in the range of loads up to 3 pN and inhibited >20-fold by ≈2 pN backward load (leading head). These results favor models (6, 7, 15) in which preferential forward stepping of myosin V is achieved due to a significant inhibition of ADP dissociation from the leading head. This load-induced asymmetry between the heads leads to the preferential release of ADP from the trailing head, providing an efficient gating mechanism that contributes to processive stepping.
To estimate the intramolecular load exerted by the heads of myosin V during double-headed binding to actin, we used the determined characteristic distances to compare the lifetime of bound ADP to the time that the actomyosin bond can sustain the applied load under various loads (Table S3). The hand-over-hand mechanism predicts that ADP dissociates from the trailing head before the detachment of myosin-ADP from actin, which implies that the intramolecular load is ≤2.6 pN. ADP binding to both heads is slowed >10-fold to 0.34 μM−1·s−1 (leading head) and 0.39 μM−1·s−1 (trailing head), under a 2.6-pN load. However, the experimental observation that ADP is a potent inhibitor of myosin V motility (4, 6) indicates that the intramolecular load generated during processive stepping is not sufficient to completely inhibit ADP binding. The stepping velocity in the presence of 1 mM ATP was reduced by 50% with 200 μM ADP. At these concentrations, the observed rate constants of ATP and ADP binding are therefore comparable, indicating that during processive stepping the ADP binding rate is ≈5-fold faster (≈4.5 μM−1·s−1) than that of ATP (0.9 μM−1·s−1) (4). This observation suggests that intramolecular load generated when ADP is released from the trailing head is actually as small as ≈1 pN (Fig. 4B), although it should be larger when both heads have ADP bound, because the ADP dissociation is accompanied by the rotation of the lever in the powerstroke direction (17, 19), which eases the load on the cross-bridge. ADP dissociation from the trailing head during processive stepping is reported to be only weakly accelerated (≤2-fold) (7), if at all (4, 24), which implies (upper curve in Fig. 4A) that the intramolecular load is <2 pN even when ADP is bound to both actin-attached heads.
The load-dependent ADP binding affinity and kinetics determined here are consistent with a rotation of the myosin V lever arm toward the pointed end of actin on ADP binding (17), because loads acting in the forward direction should facilitate ADP dissociation, whereas the opposing loads stabilize the ADP-bound conformation (19). At the same time, both forward and backward loads inhibit ADP binding to actomyosin V (Fig. 4B), but probably for different reasons: A forward barbed end load inhibits the lever arm rotation that is coupled to ADP binding (7, 19, 20, 23); a backward pointed end load blocks ADP binding by keeping the nucleotide binding site inaccessible (19).
Load-Induced Asymmetry of ADP Affinities Is Smaller in Myosin VI.
The unbinding force distributions of single-headed myosin VI molecules with bound ADP (Fig. 2C) are shifted to the weaker forces compared with the rigor state, similarly to myosin V. However, the actomyosin VI-ADP bond is more stable under forward load than under backward load (unbinding force is 2.9 ± 0.1 pN and 2.6 ± 0.1 pN for forward and backward loading, respectively), whereas in the absence of ADP the strength of the actomyosin VI bond hardly depends on the loading direction (unbinding force is 4.6 ± 0.2 pN and 4.5 ± 0.1 pN for forward and backward loading, respectively) (Table 1). Similarly to myosin V, the ADP affinity is lower under forward load than under backward load (17.2 μM and 6.8 μM, respectively) (Fig. 2D), indicating that, despite oppositely directed motility, ADP binds more weakly to the trailing head than to the leading head, introducing chemical asymmetry between the two actin-bound heads of both myosins V and VI. However, such difference between the two heads in the case of myosin VI is significantly smaller than in myosin V, which may contribute to the lower processivity of myosin VI at low [ADP] (14).
Insights into the Load-Induced Gating in Myosin VI.
Myosin VI differs from myosin V in that both forward (5) and backward (8) loads accelerate ADP binding, and weakly affect ADP dissociation (5, 8) (Fig. 4 C and D). This behavior is most likely mediated by two unique insertions in the myosin VI motor domain: one located between the converter and IQ domain, which is solely responsible for the reversed movement of the lever arm (25), and the other located near the active site, which blocks nucleotide binding (5, 14, 20, 26). Under forward loads <5 pN, ADP dissociation from the trailing head is marginally accelerated ≈1.5-fold, consistent with solution (5, 14) and single-molecule (8) studies of dimeric myosin VI. Under backward loads, ADP dissociation is only slightly inhibited, raising the possibility that the leading head will release ADP and bind ATP, resulting in a backward step or termination of a processive run. The frequent backward stepping (≈4% of all steps) observed during processive runs of dimeric myosin VI in the presence (8) and absence of external load (2) is consistent with this interpretation; myosin V does not step back until external loads approach stall (4, 6, 27). Estimation of the intramolecular load in myosin VI, performed similarly to myosin V, gives the maximum value of 2.2 pN (Table S3).
The accelerated ADP binding to myosin VI under forward and backward load is consistent with myosin VI playing an anchoring role when external (8) or intramolecular (5) loads exist. An accelerated ADP binding under load would contribute to an increased efficiency of myosin VI processivity at physiological [ADP], which is supported by recent run length measurements (14). However, dimeric myosin VI is processive even in the absence of high concentrations of exogenous ADP (2, 3, 8, 28, 29), which indicates that there must exist a factor other than load-dependent kinetics of ADP binding regulating directional, processive stepping of myosin VI. This hypothesis has gained support from solution measurements (5, 14) demonstrating the acceleration of ATP binding (≈3- to 10-fold) by forward load and argues that asymmetric rates of ATP binding may contribute to the efficient mechanism of myosin VI gating (5, 14, 20).
Common Mechanochemistry in Myosins and Kinesins.
These results and our data on the kinesin-microtubule interaction (21, 30) strongly suggest that the intramolecular load that arises from double-headed binding of a motor to the lattice track is a major regulator of the mechanochemical kinetics, which coordinates the enzymatic cycles in the two heads and controls the unidirectional stepping of dimeric ATP-driven processive motors, regardless of whether the lattice is an actin filament or a microtubule. However, the different characteristics of load dependence of ADP binding/dissociation kinetics between myosins V and VI suggest that this regulatory mechanism is adapted to the particular cellular functions of each molecular motor.
Methods
Proteins.
Actin was purified from rabbit skeletal muscle and biotinylated (6). Myosin constructs, myc-tagged at the C terminus, were coexpressed with calmodulin in Sf9 insect cells and purified by coprecipitation with actin, followed by FLAG- or His-affinity chromatography. The T406A mutant was used for the experiments with myosin VI. Before the measurements myosin molecules were attached to 1-μm polystyrene beads coated with anti-myc antibody (Invitrogen) and rhodamine-labeled BSA. The number of functional myosin molecules per bead was estimated by the statistical methods established in ref. 31 to be ≤1. Polarity of actin filaments was determined (32) by using gelsolin-coated 200-nm yellow-green fluorescent polystyrene beads (Molecular Probes).
Measurements of the Unbinding Force.
The experiments were performed in assay buffer containing 20 mM Hepes-KOH (pH 7.8), 25 mM KCl, 5 mM MgCl2, and 1 mM EGTA. The experimental system was created by consecutive infusion of the following solutions into a flow cell in the following order: 3.5 mg·ml−1 biotinylated BSA (incubated for 4 min), 0.5 mg·ml−1 streptavidin (incubated for 2 min), and fluorescent and polarity-marked 10%-biotinylated F-actin (incubated for 10 min). The flow cell was washed with two volumes of the assay buffer after each step and, finally, filled with the solution containing myosin-bound beads; an oxygen scavenging system (4.5 mg·ml−1 glucose, 50 units·ml−1 catalase, 50 units·ml−1 glucose oxidase, and 10 mM DTT); and, when necessary, ADP to a final concentration of 1 μM to 1 mM. Nucleotide-free solutions did not contain apyrase, because our probe measurements showed that its presence did not affect the unbinding force distributions. For measurements in the presence of ADP, 1 unit·ml−1 hexokinase was added. The experiments were performed at 25 ± 1°C.
To induce external load on the actomyosin bond, a piezo substage (P-611 NanoCube; Physik Instrumente) was repeatedly displaced for 1 μm with a constant speed (500 nm·sec−1), using a function generator (FG-300; Yokogawa) and rapidly returned to the initial position in the end of each cycle (Fig. 1B). The position of the optically trapped (using a 1,064-nm laser; Spectra-Physics) myosin-bound bead was tracked by projecting its bright-field image onto a quadrant photodiode (Sentech). A piezo substage-function generator unit is the improvement to the apparatus used in our studies in refs. 21, 30, and 33. The binding events were repeatedly detected with the same bead and, presumably, the same myosin molecule during the subsequent stage displacements, confirming that the unbinding does not lead to the denaturation of a myosin molecule. On average, 10 unbinding events per bead were recorded, which means that 4–8 different beads per a histogram were tested. The unbinding force was calculated as bead displacement × trap stiffness (0.076 pN·nm−1). Data recording and analysis were performed with the PowerLab software (ADInstruments).
Global Fitting.
The global fit of the unbinding force distributions was performed with OriginLab software. The two sets of data at various [ADP] were fitted separately for the barbed- and pointed-end loading. For each loading direction, the positions of the two Gaussian peaks and their width were set to be common for the distributions at all [ADP] but were not constrained to match those obtained in the absence of ADP (rigor state) or in the presence of saturating ADP (ADP state). The width of the two peaks was independent of each other. The proportion of the unbinding events in the ADP state (the fraction of the weaker binding peak in the total distribution area) was plotted against [ADP] and fit to a hyperbola, from which the apparent ADP dissociation constant (Kd) was determined.
Analysis Using the Average Forces.
To estimate the apparent ADP affinity without fitting the unbinding force distributions with the double Gaussians, the population of the ADP state, pADP, was determined from the values of average forces at each [ADP] as pADP = (Fφ − F̄)/(Fφ − FADP), where Fφ and FADP denote the average unbinding force for the distributions at 0 and 1 mM ADP, respectively, and F̄ is the average unbinding force for any given distribution.
Model Analysis.
We applied the Bell equation (34, 35) to model the load dependence of binding and dissociation kinetics of actin-myosin and myosin-ADP (Eq. 1):
|
|
where kADP− and kADP+ are the rates of ADP dissociation and binding; dADP− and dADP+ are the according characteristic distances; τφ and τD are the lifetimes of actomyosin bond in the rigor and the ADP states, respectively; and dφ and dD are the characteristic distances for actomyosin bond in the rigor and the ADP states, respectively. Index 0 denotes corresponding parameters obtained in the absence of load by the solution studies. Neither dADP− nor dADP+ are constrained to change symmetrically under loads applied in the opposite directions; their load dependence is determined separately for the backward and forward loads.
Consequently, the populations of the ADP and the nucleotide-free states under load (x(t) and y(t), respectively) are defined by the following equations (Fig. 3):
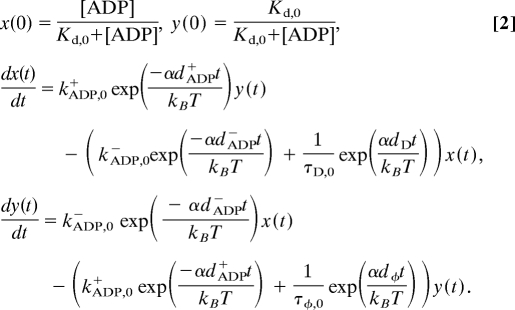 |
α, the loading rate, was fixed to be 15 pN·s−1, determined from the average slope of a bead displacement in raw data records.
First, dφ and dD were obtained by solving Eq. 2 separately for the detachment of a myosin head in the rigor or the ADP-bound state (Fig. 3 and Table S2). Next, dADP− and dADP+ were determined by separately fitting the corresponding experimentally obtained proportions at each [ADP] with the normalized modeled curves of ADP and rigor states. To this end, Eq. 2 was solved by using the determined values for dφ and dD and substituting various parameters for dADP− and dADP+ to obtain the best correlation between the modeled distributions and the experimentally obtained proportions of ADP and rigor states at each [ADP]. dx(t)/dt and dy(t)/dt, obtained by solving Eq. 2, correspond to the proportion of the ADP and the rigor states, respectively, and the sum corresponds to the unbinding force distribution. The restraining condition for each pair of dADP− and dADP+ is that the ratio of the according ADP dissociation and binding rates should be equal to the experimentally obtained apparent dissociation constant, Kdobserved = kADP−/kADP+, that is,
 |
where tmax is the longest time between the moment load is applied to an actomyosin bond and the moment it unbinds, that is, the time corresponding to the largest load exerted on an actomyosin bond. The values of dADP− and dADP+ were then determined. The experimentally obtained unbinding force distributions were fitted with the modeled curves in Fig. S5 A and C, and the proportion of the ADP state at each [ADP], determined from the modeled curves, was plotted against [ADP] in Fig. S5 B and D. Using these values, the dependence of kADP− and kADP+ on the applied load was determined (Fig. 4). The error of the defined characteristic distances was estimated from the χ2 distribution of the residuals between the predicted (modeled) values of the proportion of ADP and rigor states and the experimentally obtained values at each [ADP]. Best-fit parameters are summarized in Table S2.
The calculations presented do not consider the hyperbolic [ADP] dependence of ADP binding rates (20). In the case of actomyosin VI, the observed rate constant of ADP binding depends linearly on the [ADP] over the range examined in this study (0–1 mM). However, the observed rate constant of ADP binding to actomyosin V significantly deviates from the linear dependence as [ADP] approaches 1 mM (≈900 s−1 compared with 9,800 s−1 predicted from the linear dependence). Therefore, the actual ADP binding rates for myosin V at each [ADP] were constrained in Eq. 2 to the experimentally determined values (20). The resulting parameters are summarized in Table S2. Comparison of these values with the according values obtained without considering the nonlinear dependence of ADP binding rates on [ADP] confirms that this nonlinearity does not significantly alter the conclusions obtained from the simplified model within the experimental uncertainties.
The results of the additional calculations when dADP− and dADP+ were constrained to be symmetrical under forward and backward loads, as in ref. 16, are shown in Table S4 and Figs. S6 and S7, revealing that imposing such constraints results in large deviation from the experimental data. Calculations were performed by using Mathematica for Windows.
Supplementary Material
Acknowledgments.
We thank Dr. Jim Sellers for providing the myosin V construct. This research was supported in part by Grants-in-Aid for Specially Promoted Research, Scientific Research (A), The 21st Century COE Program (Physics of Self-Organization Systems), the “Academic Frontier” Project, and “Establishment of Consolidated Research Institute for Advanced Science and Medical Care” from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to S. I.); National Institutes of Health Grant GM071688, National Science Foundation Grant MCB-0546353, and American Heart Association Grant 0655849T (to E.M.D.L.C.); grants from The 21st Century COE Program (Physics of Self-Organization Systems), Waseda University (to Y.O., S.V.M. and T.O.); and National Institutes of Health Predoctoral Fellowship F31AR051614 (to A.O.O.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0800564105/DCSupplemental.
References
- 1.Yildiz A, et al. Myosin V walks hand-over-hand: Single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 2.Yildiz A, et al. Myosin VI steps via a hand-over-hand mechanism with its lever arm undergoing fluctuations when attached to actin. J Biol Chem. 2004;279:37223–37226. doi: 10.1074/jbc.C400252200. [DOI] [PubMed] [Google Scholar]
- 3.Okten Z, Churchman LS, Rock RS, Spudich JA. Myosin VI walks hand-over-hand along actin. Nat Struct Mol Biol. 2004;11:884–887. doi: 10.1038/nsmb815. [DOI] [PubMed] [Google Scholar]
- 4.Rief M, et al. Myosin-V stepping kinetics: A molecular model for processivity. Proc Natl Acad Sci USA. 2000;97:9482–9486. doi: 10.1073/pnas.97.17.9482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robblee JP, Olivares AO, De La Cruz EM. Mechanism of nucleotide binding to actomyosin VI: Evidence for allosteric head-head communication. J Biol Chem. 2004;279:38608–38617. doi: 10.1074/jbc.M403504200. [DOI] [PubMed] [Google Scholar]
- 6.Uemura S, Higuchi H, Olivares AO, De La Cruz EM, Ishiwata S. Mechanochemical coupling of two substeps in a single myosin V motor. Nat Struct Mol Biol. 2004;11:877–883. doi: 10.1038/nsmb806. [DOI] [PubMed] [Google Scholar]
- 7.Rosenfeld SS, Sweeney HL. A model of myosin V processivity. J Biol Chem. 2004;279:40100–40111. doi: 10.1074/jbc.M402583200. [DOI] [PubMed] [Google Scholar]
- 8.Altman D, Sweeney HL, Spudich JA. The mechanism of myosin VI translocation and its load-induced anchoring. Cell. 2004;116:737–749. doi: 10.1016/s0092-8674(04)00211-9. [DOI] [PubMed] [Google Scholar]
- 9.Baker JE, et al. Myosin V processivity: Multiple kinetic pathways for head-to-head coordination. Proc Natl Acad Sci USA. 2004;101:5542–5546. doi: 10.1073/pnas.0307247101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olivares AO, Chang W, Mooseker MS, Hackney DD, De La Cruz EM. The tail domain of myosin Va modulates actin binding to one head. J Biol Chem. 2006;281:31326–31336. doi: 10.1074/jbc.M603898200. [DOI] [PubMed] [Google Scholar]
- 11.Shiroguchi K, Kinosita K., Jr Myosin V walks by lever action and Brownian motion. Science. 2007;316:1208–1212. doi: 10.1126/science.1140468. [DOI] [PubMed] [Google Scholar]
- 12.Dunn AR, Spudich JA. Dynamics of the unbound head during myosin V processive translocation. Nat Struct Mol Biol. 2007;14:246–248. doi: 10.1038/nsmb1206. [DOI] [PubMed] [Google Scholar]
- 13.Burgess S, et al. The prepower stroke conformation of myosin V. J Cell Biol. 2002;159:983–991. doi: 10.1083/jcb.200208172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sweeney HL, et al. How myosin VI coordinates its heads during processive movement. EMBO J. 2007;26:2682–2692. doi: 10.1038/sj.emboj.7601720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purcell TJ, Sweeney HL, Spudich JA. A force-dependent state controls the coordination of processive myosin V. Proc Natl Acad Sci USA. 2005;102:13873–13878. doi: 10.1073/pnas.0506441102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Veigel C, Schmitz S, Wang F, Sellers JR. Load-dependent kinetics of myosin-V can explain its high processivity. Nat Cell Biol. 2005;7:861–869. doi: 10.1038/ncb1287. [DOI] [PubMed] [Google Scholar]
- 17.Volkmann N, et al. The structural basis of myosin V processive movement as revealed by electron cryomicroscopy. Mol Cell. 2005;19:595–605. doi: 10.1016/j.molcel.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 18.Wells AL, et al. Myosin VI is an actin-based motor that moves backwards. Nature. 1999;401:505–508. doi: 10.1038/46835. [DOI] [PubMed] [Google Scholar]
- 19.Nyitrai M, Geeves M. Adenosine diphosphate and strain sensitivity in myosin motors. Phil Trans R Soc Lond B. 2004;359:1867–1877. doi: 10.1098/rstb.2004.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robblee JP, Cao W, Henn A, Hannemann DE, De La Cruz EM. Thermodynamics of nucleotide binding to actomyosin V and VI: A positive heat capacity change accompanies strong ADP binding. Biochemistry. 2005;44:10238–10249. doi: 10.1021/bi050232g. [DOI] [PubMed] [Google Scholar]
- 21.Uemura S, Ishiwata S. Loading direction regulates the affinity of ADP for kinesin. Nat Struct Mol Biol. 2003;10:308–311. doi: 10.1038/nsb911. [DOI] [PubMed] [Google Scholar]
- 22.Uemura S, et al. Kinesin-microtubule binding is dependent on both nucleotide state and loading direction. Proc Natl Acad Sci USA. 2002;99:5977–5981. doi: 10.1073/pnas.092546199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hannemann DE, Cao W, Olivares AO, Robblee JP, De La Cruz EM. Magnesium, ADP, and actin binding linkage of myosin V: Evidence for multiple myosin V-ADP and actomyosin V-ADP states. Biochemistry. 2005;44:8826–8840. doi: 10.1021/bi0473509. [DOI] [PubMed] [Google Scholar]
- 24.De La Cruz EM, Wells AL, Rosenfeld SS, Ostap EM, Sweeney HL. The kinetic mechanism of myosin V. Proc Natl Acad Sci USA. 1999;96:13726–13731. doi: 10.1073/pnas.96.24.13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bryant Z, Altman D, Spudich JA. The power stroke of myosin VI and the basis of reverse directionality. Proc Natl Acad Sci USA. 2007;104:772–777. doi: 10.1073/pnas.0610144104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ménétrey J, et al. The structure of the myosin VI motor reveals the mechanism of directionality reversal. Nature. 2005;435:779–785. doi: 10.1038/nature03592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mehta AD, et al. Myosin-V is a processive actin-based motor. Nature. 1999;400:590–593. doi: 10.1038/23072. [DOI] [PubMed] [Google Scholar]
- 28.Nishikawa S, et al. Class VI myosin moves processively along actin filaments backward with large steps. Biochem Biophys Res Commun. 2002;290:311–317. doi: 10.1006/bbrc.2001.6142. [DOI] [PubMed] [Google Scholar]
- 29.Rock RS, et al. Myosin VI is a processive motor with a large step size. Proc Natl Acad Sci USA. 2001;98:13655–13659. doi: 10.1073/pnas.191512398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawaguchi K, Ishiwata S. Nucleotide-dependent single- to double-headed binding of kinesin. Science. 2001;291:667–669. doi: 10.1126/science.291.5504.667. [DOI] [PubMed] [Google Scholar]
- 31.Svoboda K, Schmidt CF, Schnapp BJ, Block M. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 1993;365:721–727. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- 32.Nishizaka T, Miyata H, Yoshikawa H, Ishiwata S, Kinosita K., Jr Unbinding force of a single motor molecule of muscle measured using optical tweezers. Nature. 1995;377:251–254. doi: 10.1038/377251a0. [DOI] [PubMed] [Google Scholar]
- 33.Kawaguchi K, Uemura S, Ishiwata S. Equilibrium and transition between single- and double-headed binding of kinesin as revealed by single-molecule mechanics. Biophys J. 2003;84:1103–1113. doi: 10.1016/S0006-3495(03)74926-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bell GI. Models for the specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 35.Nishizaka T, Seo R, Tadakuma H, Kinosita K, Jr, Ishiwata S. Characterization of single actomyosin rigor bonds: Load dependence of lifetime and mechanical properties. Biophys J. 2000;79:962–974. doi: 10.1016/S0006-3495(00)76350-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.