Abstract
In voltage-gated Na+, K+, and Ca2+ channels, four voltage-sensor domains operate on a central pore domain in response to membrane voltage. In contrast, the voltage-gated proton channel (Hv) contains only a voltage-sensor domain, lacking a separate pore domain. The subunit stoichiometry and organization of Hv has been unknown. Here, we show that human Hv1 forms a dimer in the membrane and define regions that are close to the dimer interface by using cysteine cross-linking. Two dimeric interfaces appear to exist in Hv1, one mediated by S1 and the adjacent extracellular loop, and the other mediated by a putative intracellular coiled-coil domain. It may be significant that Hv1 uses for its dimer interface a surface that corresponds to the interface between the voltage sensor and pore in Kv channels.
Keywords: membrane protein, voltage-dependent ion channel, voltage sensor
Voltage-gated six-transmembrane cation channels (Na+, K+, and Ca2+) contain voltage-sensor and pore domains (1). In this class of ion channels voltage-sensor and pore domains carry out voltage sensing and cation conduction, respectively. Four voltage-sensor domains surround a single, centrally located ion conduction pathway. Each voltage sensor is attached to the pore in a specific manner so that conformational changes within the voltage sensors are transmitted to the pore's gate (2). It was originally thought that voltage-sensor domains existed only in the context of voltage-gated cation channels. However, the cloning of voltage-gated proton (Hv) channels and voltage-sensor phosphatase enzymes revealed that voltage-sensor domains also exist in other contexts, apparently as independent (of a cation pore) membrane proteins (3–5), corroborating the studies of MacKinnon and coworkers (2, 6, 7) and Lu and coworkers (8) that supported the idea that voltage sensors, even in the context of voltage-dependent cation channels, are rather loosely attached to the central pore.
The long-sought molecular identity of Hv1 (9) showed that it contains only a voltage-sensor domain without a separate pore domain in the membrane (3, 4). This observation suggested that in contrast to canonical voltage-dependent cation channels, the voltage-sensor domain of Hv1 is responsible for both H+ conduction and voltage sensing. In this study we evaluate the subunit stoichiometry of the Hv1 channel in cell membranes.
Results
Hv1 Is a Dimer in the Membrane.
To probe the oligomeric state of Hv1, cell membranes isolated from tsA201 cells (HEK293 derivatives) transfected with human Hv1 cDNAs were subjected to the amino-group specific bifunctional cross-linker disuccinimidyl suberate (DSS) and visualized by Western blot analysis using antibodies directed against the Hv1 channel. Amino group-specific cross-linkers have been successful in defining the oligomeric status of several membrane proteins (10–12). Recombinant Hv1 makes functional channels in HEK293 cells (3, 4). In Fig. 1a, human Hv1 migrates at ≈35 kDa in SDS/PAGE under reducing conditions, which is consistent with the molecular mass of a monomer (32 kDa). As the concentration of the cross-linker (DSS) was increased, a band migrating near 73 kDa appeared with a concomitant decrease in the monomer band. Further increase of cross-linker concentration yielded a series of weak bands corresponding to higher oligomers in addition to the strong dimer band. These are probably caused by nonspecific cross-linking with other membrane proteins or cross-linking between the Hv1 dimers. However, the appearance of strong dimer bands at low concentrations of DSS suggests that Hv1 forms a homodimer in the membrane. Dimerization in Hv1 is not a consequence of disulfide bond formation because mutants lacking cysteines show the same DSS-mediated cross-linking pattern as wild type (Fig. 1b).
Fig. 1.
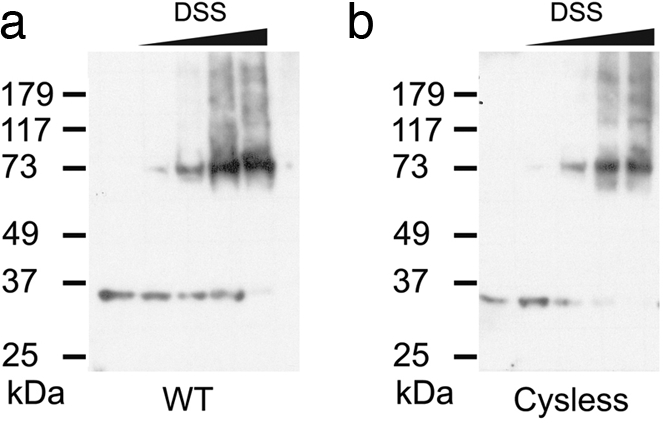
Hv1 is a dimer. Shown are Western blots of membranes from tsA201 cells transfected with WT (a) and Cysless (b) human Hv1 cDNAs and exposed to increasing concentrations of the amino group-specific cross-linker DSS (from left to right: no DSS, 25 μM, 75 μM, 250 μM, 2.5 mM). Membrane samples were run on SDS/PAGE under reducing conditions (see Methods). All of the gels that were used were 12%. Molecular mass markers are shown on the left.
Design of Site-Specific Cross-Links.
The finding of a dimeric subunit stoichiometry for Hv1 leads to the question: how do two voltage sensors arrange relative to each other in the membrane? If the Hv1 homodimer contains a 2-fold rotation axis normal to the membrane, a single cysteine mutation close to the dyad axis should form a cross-linked dimer under oxidizing conditions or in the presence of a bifunctional cross-linker. By sequence analysis Hv1 can be dissected into three subdomains: an N-terminal acid-rich and proline-rich region, a transmembrane voltage-sensor domain, and a C-terminal coiled-coil domain (Fig. 2). Using the voltage sensor of the Kv1.2-Kv2.1 paddle chimera structure as a reference (13), we examined both the two naturally existing cysteine residues and a series of substituted cysteines on a natural-cysteine-free background (Fig. 2): R100 (preceding S1), C107 and L120 (S1), I127 and N132 (S1–S2 loop), T145 and I155 (S2), E164 (S2–S3 turn), L173 and V187 (S3), Q194 (S3–S4 turn), L203 and I212 (S4), T222 (after S4), and C249 (putative coiled coil) (Fig. 2). Limited proteolysis and preliminary biochemical data suggest that the N-terminal region is largely unstructured and does not affect the oligomeric state of the protein (data not shown) and therefore, although the full-length Hv1 was used, the N-terminal region has not been investigated in this cross-linking study.
Fig. 2.
Introduction of cysteine mutations into Hv1. (a) The positions that were mutated to cysteine are indicated by filled circles on the voltage-sensor structure of the Kv 1.2–2.1 paddle chimera (Protein Data Bank ID code 2R9R). The S1–S2 loop from the paddle chimera was replaced with a shorter loop to match the length of Hv1. (b) The amino acid sequence of Hv1 excluding the N-terminal acid-rich and proline-rich region. Predicted transmembrane regions, based on the structure of the paddle chimera and hydropathy, are highlighted in gray. Residues mutated to cysteine are colored red. C107 and C249 are natural cysteines. The region corresponding to the coiled coil was calculated by the program COIL (24).
Dimer Interface of Hv1.
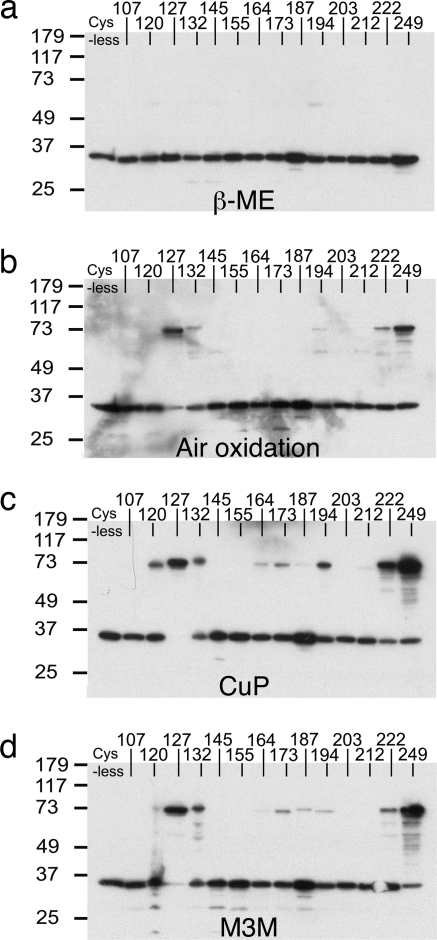
Each of the 15 cysteine mutant cDNAs were transfected into tsA201 cells and membranes isolated from these cells were subjected to cross-linking reactions. The oligomeric state of each mutant was assessed by Western blot analysis. Among the 15 mutants that we chose to test cross-linking, only R100C failed to express by Western blot analysis. In Fig. 3a all mutants migrate as a monomer under reducing conditions on SDS/PAGE. However, several mutants migrate as a dimer under nonreducing conditions (Fig. 3b). I127C, which is located in the loop between S1 and S2, forms an almost complete spontaneous disulfide bond, while C249, which is a natural cysteine located in the predicted coiled-coil domain, forms a significant fraction of spontaneous disulfide bond. N132C and T222C also show some degree of disulfide bond formation. This pattern of disulfide bond formation suggests that S1 and the adjacent loop form a dimer interface on the extracellular side of the cell membrane and that the C terminus, possibly through a coiled coil, forms a dimer interface on the intracellular side of the membrane.
Fig. 3.
Dimer interface of Hv1. Western blots of membranes from tsA201 cells transfected with 14 individual Hv1 cysteine mutant cDNAs that were subjected to reducing reagent [5% (vol/vol) β-mercaptoethanol] (a), air oxidation (b), CuP-induced cross-linking (c), or M3M-mediated cross-linking (d) (see Methods). Cross-linked dimers migrate at ≈73 kDa, whereas monomers migrate at ≈35 kDa. Other weak bands probably correspond to proteolytic fragments of Hv1. A mutant Hv1 in which the native cysteines were removed was included as a control (Cys-less). The numbers represent the amino acid positions of the cysteine residues.
Oligomer formation was also studied under conditions that “force” cross-linking either by the addition of the oxidant CuSO4 and o-phenanthroline (CuP) or the addition of the bifunctional cross-linker 1,3-propanediyl bismethanethiosulfonate (M3M), which has a 13-Å spacer separating the two cysteine-reactive methanethiosulfonate functional groups (Fig. 3 c and d). Both reagents drive cross-links at I127C and C249 nearer to completion. They also produce cross-linked cysteines to a lesser extent at positions near I127 (position 120 within S1 and position 132 in the S1–S2 loop) and near 249 (position 222 within the C terminus).
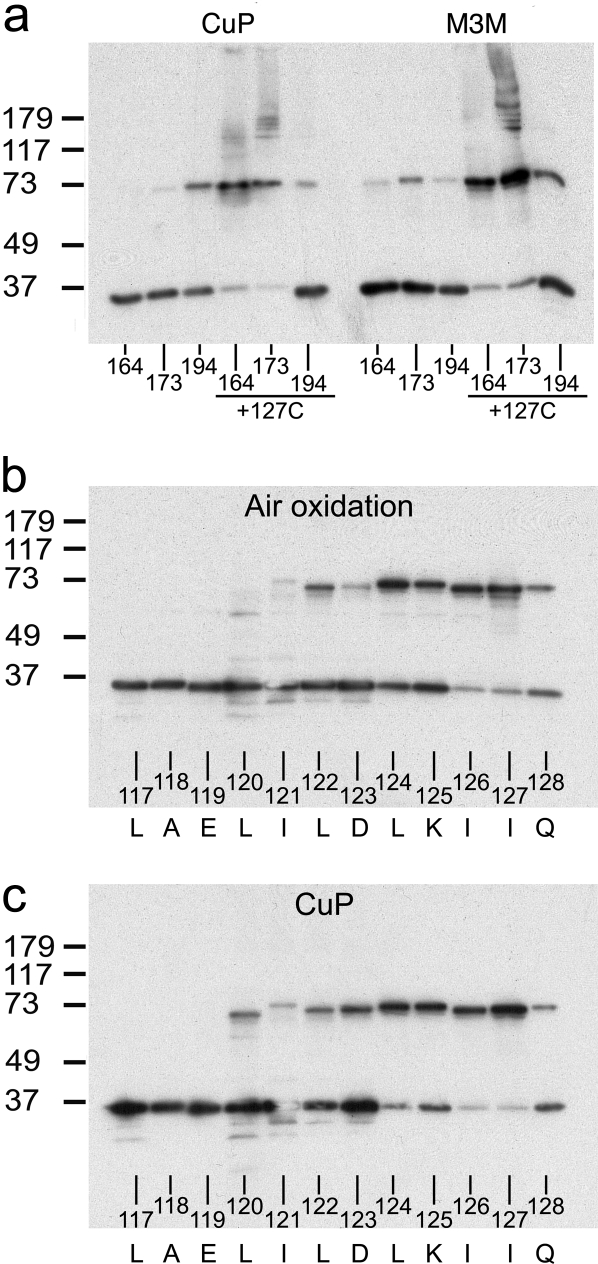
Forcing conditions also result in weak cross-links in two new regions: the S2–S3 turn, which is also known as the membrane interface anchor (positions 164 and 173) (7), and the tip of the voltage sensor paddle (positions 187 and 194) (Fig. 3 c and d) (6). Forcing conditions, by covalently trapping rare conformations or molecular arrangements, can sometimes result in cross-link formation between residues that are only rarely in close proximity to each other. An example of this would be intermolecular cross-links (in this case interdimer cross-bridge formation between pairs of dimers). The blot in Fig. 4a provides evidence for such dimer pairing by cross-linking of cysteines at position 173. When combined with a cysteine at 127, which by itself forms a cross-linked dimer to near completion (Fig. 3b), cysteine at 173 results in the appearance of bands near the size of a tetramer (Fig. 4a). This observation is explicable if cross-links between position 173 cysteines are formed between S2 and S3 loops from separate dimers, because cross-links within a dimer would not produce a tetramer. A similar analysis of a cysteine at position 194 in the background of a cysteine at 127 produces a different, yet equally interesting, result. Cysteine at 194 does not produce tetramers in this setting, but actually reduces the degree to which cysteine at 127 forms a cross-linked dimer (compare Fig. 3 c and d with Fig. 4a). The location of position 194 at the tip of the voltage sensor paddle, which is adjacent to the S1–S2 loop within a single voltage sensor of the dimer (see Fig. 2a), offers a possible explanation for this cross-linking outcome. Cysteine at 194, by forming an intramolecular cross-link with Cys-127 within the same subunit, would compete with Cys-127-mediated dimer cross-linking. These more subtle aspects of the blots suggest that the S2–S3 turn and the voltage-sensor paddle are not actually part of the tight dimer interface, but that under forcing conditions less specific cross-links are formed.
Fig. 4.
Further study of the dimer interface. (a) Distinguishing intradimer from interdimer cross-linking. The numbers at the bottom of each gel indicate the cysteine position. (b and c) Specificity of cross-linking in the S1–S1 dimer interface. Amino acids are shown in single-letter code. Transmembrane region of S1 and the adjacent loop (117 to 128) were subjected to air oxidation (b) and CuP-mediated oxidation (c).
A more extensive inspection of covalent dimer formation via cysteine residues within S1 and the S1–S2 loop show differential propensities to react (Fig. 4 b and c). Even among amino acids that we expect reside outside of the membrane (positions 123–128), positions 126 and 127 react to a greater extent as if they are most optimally positioned (to form a disulfide bridge) relative to their molecular symmetry equivalents. Position 120, which we expect is buried in the membrane, forms a disulfide partially in the presence of CuP. Disulfide formation in a low dielectric environment suggests that position 120 is probably very near its molecular symmetry partner (14, 15).
Discussion
These cross-linking data support the conclusion that Hv1 exists in the cell membrane as a dimer of identical voltage sensor protomers. Two regions of contact between the voltage sensors correspond to the extracellular C-terminal side of S1 and the adjoining S1–S2 loop and intracellular C terminus, which may form a coiled-coil structure. The native cysteine at position 249 may help to stabilize the dimer at the coiled coil, but a disulfide bridge at this position is not essential. We conclude that two voltage sensor protomers of a dimer come into direct contact at both the extracellular and intracellular side of the membrane. A schematic representation of the Hv1 dimer, based on the known crystal structures of Kv channel voltage sensors and the contact surfaces identified in this study, is shown in Fig. 5(2, 6, 13).
Fig. 5.
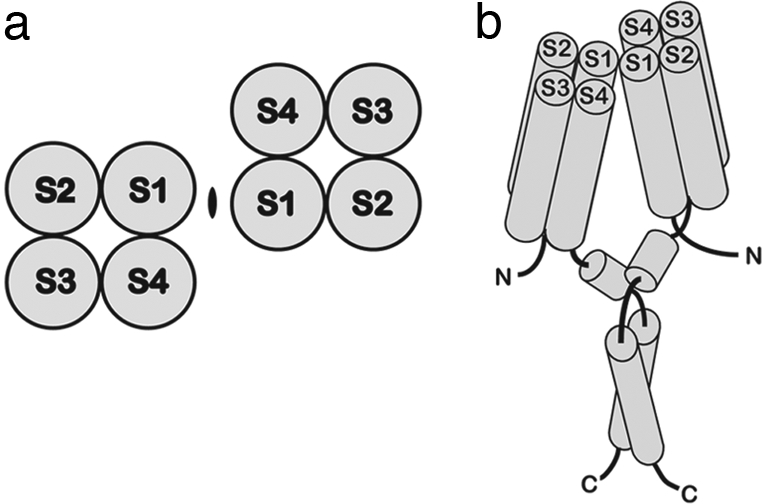
A model of the Hv1 dimer. (a) Topology diagram showing the dimer arrangement of Hv1 transmembrane regions based on the cross-linking data, viewed from the extracellular side. An ellipsoid denotes the 2-fold rotation axis normal to the membrane. (b) A cartoon representation of the Hv1 dimer. Transmembrane helices are labeled. The helical representation of the putative interfacial region (after S4) and the predicted coiled-coil region are also included.
Many membrane proteins, like water-soluble proteins, exist as oligomers of identical or similar subunits. In many cases the necessity of an oligomeric structure is clear. Potassium channels, for example, require four subunits to form a single ion conduction pathway in between the subunits (16). In other membrane proteins the reason for an oligomeric structure is less clear. In aquaporin channels (17) and ClC Cl− channels (18) the water and ion conduction pathways are formed entirely by atoms of a single protomer and yet these transport proteins are tetramers and dimers of identical protomers, respectively.
Into which category does the Hv channel fall? The location of the proton pathway has not yet been established with certainty. Based on electrophysiological studies showing that mutant Kv channel voltage sensors can themselves conduct ions (19) or protons (20) across the membrane and based on atomic structural studies showing that Kv channel voltage sensors contain protonatable chemical groups extending most of the way across the membrane (13), the current thinking is that protons flow through individual voltage sensors in Hv channels. In other words, each sensor is anticipated to contain its own proton conduction pathway.
Sometimes multiple subunits allow active sites to function in a nonindependent manner. Hemoglobin is the most famous example in which an oligomeric structure underlies cooperativity (21), allowing a steep relationship between oxygen saturation and oxygen partial pressure. In ClC Cl− channels one form of gating might arise from its dimeric architecture (22). Thus, oligomeric architectures in both soluble and membrane proteins can allow for much more than the multiplicity of active sites.
In Hv1 a dimeric architecture might have to do with the mechanics of channel gating. It is interesting to note that the location of the extracellular dimer interface in the Hv1 channel corresponds well to the region of contact between voltage sensor and pore in Kv channels (13). In Kv channels the contact between S1 and the pore at the extracellular surface is proposed to serve as a mechanical fixed point so that motions of the voltage-sensor paddle (S3 and S4) are efficiently transmitted to the pore's gate (13) (S.-Y.L., A. Banerjee, and R.M., unpublished work). We wonder whether contacts between S1 helices in the Hv1 dimer serve a similar function by fixing S1 and S2, thus permitting the voltage-sensor paddle to move and open a conduction pathway within each voltage sensor.
Methods
Preparation of Mutations.
Human Hv1 cDNA (GI: 34783431; a gift from David Clapham, Harvard University, Boston) was subcloned into pcDNA4 vector (Invitrogen). Cysteine mutations were generated by using a QuikChange kit (Stratagene). Three additional amino acids (ARG) were introduced into the C terminus as a byproduct of cloning into the expression vector.
Membrane Preparation.
cDNAs encoding human Hv1 were transfected into tsA201 (HEK293 derivatives) cells by using Lipofectamine 2000 (Invitrogen) for 36–48 h. Membranes were prepared as described with modifications (23). Briefly, cells were washed with PBS containing 1 mM EDTA and lysed with a tissue grinder (Wheaton) in the presence of protease inhibitors (1 mM PMSF, 1 μg/ml leupeptin, 1 μg/ml aprotinin, 1 μg/ml pepstatin). Lysed cells were briefly sonicated for 10 s in a bath sonicator and then centrifuged at 900 × g for 15 min. Supernatants were collected and diluted 4-fold with ice-cold buffer [20 mM Hepes (pH 7.5), 5 mM KCl, 150 mM NaCl, 5% glycerol, and 1 mM EDTA] and ultra-centrifuged for 90–120 min at 130,000 × g. After ultracentrifugation, supernatants were discarded and pellets were resuspended in the same buffer and then homogenized with a tissue grinder (Wheaton). Samples were maintained at 4°C at all times.
Cross-Linking.
For nonspecific cross-linking experiments, the amino group-specific bifunctional cross-linker DSS (Pierce) was used. For each reaction, 20-fold concentrated DSS stocks, dissolved in DMSO, were added to prepared membranes and incubated at room temperature for 20 min; reactions were quenched by the addition of Tris·HCl (pH 8.5) to a final concentration of 100 mM. For copper-mediated cross-linking, 6 mM CuSO4 and 1.8 mM o-phenanthroline in water were added to prepared membranes to a final concentration of 300 and 900 μM, respectively and incubated at room temperature for 20 min; reactions were quenched by addition of 20 mM N-ethylmaleimide (NEM) and 50 mM EDTA. For cysteine-directed cross-linking, 1 mM stock of 1,3-propanediyl bismethanethiosulfonate (M3M; Toronto Research Chemicals) in DMSO was added to the prepared membranes to a final concentration of 50 μM and incubated on ice for 1 h; reactions were quenched with 20 mM NEM. Air-oxidized samples, used immediately after membrane preparation, were treated with 10–20 mM NEM to prevent cross-linking during electrophoresis. All of the samples were mixed with equal volume of loading buffers containing 4% (wt/vol) SDS and 10% (vol/vol) β-mercaptoethanol (only for reducing condition), then subjected to SDS/PAGE on 12% gels, transferred onto PVDF membrane, and probed with a mixture of mAbs 25H11 and 9C1. mAbs were raised in mice injected with purified Hv1 protein. Each cross-linking experiment was performed at least twice independently.
Acknowledgments.
We thank J. Butterwick for critical reading. This work was supported by National Institutes of Health Grant GM 43949 (to R.M.). S.-Y.L. was a fellow of the Jane Coffin Childs Memorial Fund. J.A.L. is supported in part by the Natural Science and Engineering Research Council of Canada. R.M. is an Investigator in the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
References
- 1.Hille B. Ion Channels of Excitable Membranes. Sunderland, MA: Sinauer; 2001. [Google Scholar]
- 2.Long SB, Campbell EB, MacKinnon R. Voltage sensor of Kv1.2: Structural basis of electromechanical coupling. Science. 2005;309:903–908. doi: 10.1126/science.1116270. [DOI] [PubMed] [Google Scholar]
- 3.Murata Y, Iwasaki H, Sasaki M, Inaba K, Okamura Y. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature. 2005;435:1239–1243. doi: 10.1038/nature03650. [DOI] [PubMed] [Google Scholar]
- 4.Ramsey IS, Moran MM, Chong JA, Clapham DE. A voltage-gated proton-selective channel lacking the pore domain. Nature. 2006;440:1213–1216. doi: 10.1038/nature04700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sasaki M, Takagi M, Okamura Y. A voltage sensor-domain protein is a voltage-gated proton channel. Science. 2006;312:589–592. doi: 10.1126/science.1122352. [DOI] [PubMed] [Google Scholar]
- 6.Jiang Y, et al. X-ray structure of a voltage-dependent K+ channel. Nature. 2003;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 7.Lee SY, Lee A, Chen J, MacKinnon R. Structure of the KvAP voltage-dependent K+ channel and its dependence on the lipid membrane. Proc Natl Acad Sci USA. 2005;102:15441–15446. doi: 10.1073/pnas.0507651102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu Z, Klem AM, Ramu Y. Ion conduction pore is conserved among potassium channels. Nature. 2001;413:809–813. doi: 10.1038/35101535. [DOI] [PubMed] [Google Scholar]
- 9.Decoursey TE. Voltage-gated proton channels and other proton transfer pathways. Physiol Rev. 2003;83:475–579. doi: 10.1152/physrev.00028.2002. [DOI] [PubMed] [Google Scholar]
- 10.Albright RA, Joh K, Morais-Cabral JH. Probing the structure of the dimeric KtrB membrane protein. J Biol Chem. 2007;282:35046–35055. doi: 10.1074/jbc.M704260200. [DOI] [PubMed] [Google Scholar]
- 11.Aller SG, Eng ET, De Feo CJ, Unger VM. Eukaryotic CTR copper uptake transporters require two faces of the third transmembrane domain for helix packing, oligomerization, and function. J Biol Chem. 2004;279:53435–53441. doi: 10.1074/jbc.M409421200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yernool D, Boudker O, Folta-Stogniew E, Gouaux E. Trimeric subunit stoichiometry of the glutamate transporters from Bacillus caldotenax and Bacillus stearothermophilus. Biochemistry. 2003;42:12981–12988. doi: 10.1021/bi030161q. [DOI] [PubMed] [Google Scholar]
- 13.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 14.Schwem BE, Fillingame RH. Cross-linking between helices within subunit a of Escherichia coli ATP synthase defines the transmembrane packing of a four-helix bundle. J Biol Chem. 2006;281:37861–37867. doi: 10.1074/jbc.M607453200. [DOI] [PubMed] [Google Scholar]
- 15.Yu H, Oprian DD. Tertiary interactions between transmembrane segments 3 and 5 near the cytoplasmic side of rhodopsin. Biochemistry. 1999;38:12033–12040. doi: 10.1021/bi9909492. [DOI] [PubMed] [Google Scholar]
- 16.Doyle DA, et al. The structure of the potassium channel: Molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 17.Murata K, et al. Structural determinants of water permeation through aquaporin-1. Nature. 2000;407:599–605. doi: 10.1038/35036519. [DOI] [PubMed] [Google Scholar]
- 18.Dutzler R, Campbell EB, Cadene M, Chait BT, MacKinnon R. X-ray structure of a ClC chloride channel at 3.0 A reveals the molecular basis of anion selectivity. Nature. 2002;415:287–294. doi: 10.1038/415287a. [DOI] [PubMed] [Google Scholar]
- 19.Tombola F, Pathak MM, Isacoff EY. Voltage-sensing arginines in a potassium channel permeate and occlude cation-selective pores. Neuron. 2005;45:379–388. doi: 10.1016/j.neuron.2004.12.047. [DOI] [PubMed] [Google Scholar]
- 20.Starace DM, Bezanilla F. A proton pore in a potassium channel voltage sensor reveals a focused electric field. Nature. 2004;427:548–553. doi: 10.1038/nature02270. [DOI] [PubMed] [Google Scholar]
- 21.Perutz MF. Mechanisms of cooperativity and allosteric regulation in proteins. Q Rev Biophys. 1989;22:139–237. doi: 10.1017/s0033583500003826. [DOI] [PubMed] [Google Scholar]
- 22.Bykova EA, Zhang XD, Chen TY, Zheng J. Large movement in the C terminus of CLC-0 chloride channel during slow gating. Nat Struct Mol Biol. 2006;13:1115–1119. doi: 10.1038/nsmb1176. [DOI] [PubMed] [Google Scholar]
- 23.Asano S, Tega Y, Konishi K, Fujioka M, Takeguchi N. Functional expression of gastric H+,K+-ATPase and site-directed mutagenesis of the putative cation binding site and catalytic center. J Biol Chem. 1996;271:2740–2745. doi: 10.1074/jbc.271.5.2740. [DOI] [PubMed] [Google Scholar]
- 24.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]