Abstract
Immunization with the highly polymorphic Plasmodium falciparum apical membrane antigen 1 (PfAMA1) induces protection in animals but primarily against parasites that express the same or similar alleles. One strategy to overcome the obstacle of polymorphism is to combine PfAMA1 proteins representing major haplotypes into one vaccine. To determine the minimum number of haplotypes that would confer broad protection, we sequenced the coding region of PfAMA1 from 97 clones from around the world and 61 isolates from Mali, identifying 150 haplotypes for domains 1 to 3 that included previous sequences. A clustering algorithm grouped the 150 haplotypes into six populations that were independent of geographic location. Each of the six populations contained haplotypes predominantly of that population (predominant haplotypes) and haplotypes that were a mixture of haplotypes represented in other populations (admixed haplotypes). To determine the biological relevance of the populations identified through the clustering algorithm, antibodies induced against one predominant haplotype of population 1 (3D7) and one admixed haplotype of population 5 (FVO) were tested for their ability to block parasite invasion of erythrocytes. Parasites expressing PfAMA1s belonging to population 1 were efficiently inhibited by 3D7-specific antibodies, whereas parasites expressing PfAMA1s belonging to other populations were not. For FVO-specific antibodies, we observed growth inhibition against itself as well as isolates belonging to populations 3 and 6. Our data suggests that the inclusion of PfAMA1 sequences from each of the six populations may result in a vaccine that induces protective immunity against a broad range of malaria parasites.
An effective malarial vaccine is urgently needed to reduce disease and death from Plasmodium falciparum in African children. One of the major obstacles to the development of an effective malarial vaccine is the genetic polymorphism of many of the genes in natural parasite populations that otherwise would be promising vaccine candidates. In fact, it may be generally the case that the proteins of pathogens that are the best targets of immune responses are also the most polymorphic because of immune selective pressure (1, 2).
One of the leading malarial vaccine candidates is the highly polymorphic apical membrane antigen 1 (PfAMA1), which is located in the micronemes of P. falciparum merozoites (3), an apical organelle that plays an important role in parasite invasion of erythrocytes during the blood stage of infection. Despite its overall polymorphism, PfAMA1 remains a leading vaccine candidate for several reasons. First, the structure (two Pan domains) and the location of disulfide bridges are highly conserved in all species of Plasmodium (4, 5). Second, targeted disruption of the PfAMA1 gene has not been successful, indicating that the protein is critical for blood-stage survival of the parasite (6). Conversely, introduction of AMA1 from the rodent parasite, Plasmodium berghei, into P. falciparum increases the ability of P. falciparum to invade mouse erythrocytes. Third, vaccination with recombinant AMA1 induces protection against homologous parasite challenges in a number of rodent and primate models. Immunization with AMA1 from P. falciparum (7) and Plasmodium fragile (8) in New World monkeys, from Plasmodium knowlesi in rhesus monkeys (9), and from Plasmodium chabaudi in mice (10) protected the animals against homologous parasite challenge. Moreover, antibodies raised against one parasite haplotype showed greater activity against the homologous haplotype in blocking parasite invasion in vitro (11). Fourth, a study in a coastal region of Kenya showed that the presence of antibodies against the full-length ectodomain of PfAMA1 before the malaria transmission season in children who harbored P. falciparum was associated with subsequent protection from clinical malaria (12). Thus, although it is a promising vaccine candidate, the fact that the immune response to PfAMA1 appears to be strain-specific and that a high degree of polymorphism occurs in field isolates indicates that the polymorphism in this gene must be addressed before development of an effective vaccine based on this protein can be accomplished.
We asked whether the known P. falciparum AMA1 sequences could be grouped into populations such that immunization with any one member of a population would protect against parasites expressing other members of the population. To answer this question, we sequenced the entire coding region of PfAMA1 for a worldwide collection of 97 cloned parasites from Asia, Africa, America, and Papua New Guinea in addition to 210 samples collected from participants in a longitudinal study conducted in a single village in the West African country of Mali. Our analysis showed that >95% of the sequence variation exists within a geographic region compared with minimal variation between regions. By using the clustering algorithm implemented in the STRUCTURE program (13, 14), we show that the full-length PfAMA1 sequences from a worldwide collection of parasite isolates could be best clustered into six populations and that this was also true when only domains 1 to 3 of the PfAMA1 gene were analyzed. Growth inhibition assays (GIA) using antibodies elicited against one member of population 1, P. falciparum 3D7, showed greater inhibition of parasites expressing PfAMA1 haplotypes of population 1 than parasites expressing PfAMA1s in populations 3–6. In contrast, for the admixed haplotype FVO, growth inhibition with FVO-specific antibodies was observed for the homologous parasite and those expressing PfAMA1 haplotypes belonging to populations 3 and 6. Our data show that a highly polymorphic protein such as PfAMA1 can be grouped into immunologically relevant populations, potentially allowing for the identification of the PfAMA1 sequences that can be optimally combined to produce a broadly protective vaccine.
Results
Haplotype Distribution and Genetic Diversity of PfAMA1 Genes.
To address whether PfAMA1 genes could be grouped into immunologically relevant populations, we determined the full-length PfAMA1 sequences from 97 different P. falciparum clones taken from a worldwide collection of parasites (15) and from parasites isolated from residents of the village of Donéguébougou in the West African country of Mali. For the samples collected in Mali, DNA was extracted from a total of 210 filter papers, and those that amplified, by using either PCR or nested PCR as described in Methods, were sequenced. Of the 181 PfAMA1 genes that amplified the target sequence, only 61 contained a single genotype and were included in the analysis [supporting information (SI) Dataset S1]. In total, 158 full-length PfAMA1 sequences were obtained from the 97 parasites clones and the 61 Malian filter paper samples, which comprised 107 unique haplotypes (Dataset S1). Of the 61 PfAMA1 sequences from Mali that were collected at one of the three time points, three sequences were identical to sequences isolated from other parts of the world, and six were identical to sequences from samples collected at one of the other time points in the Malian study. Of the six sequences that were isolated more than once, none was isolated more than once from the same individual. In general, however, the PfAMA1 sequences obtained from samples collected at one time point in Mali did not overlap with sequences of parasites collected at the other two time points.
Although none of the haplotypes could be considered as predominant, there were two high-frequency haplotypes: HapI (FVO clone haplotype) was found in 11 of 158 sequences, mainly from Asia, and Hap 29 was found in 10 of 158 sequences, mainly from Peru and Brazil. All parasites from the worldwide collection with identical PfAMA1 sequences were unique clones, as determined by DNA fingerprint analysis (16).
The sequence diversity of PfAMA1 was analyzed among different geographic samples. The degree of PfAMA1 genetic differentiation across all geographic sampling locations, as indicated by analysis of molecular variance (AMOVA), was low (Fst = 0.0307), with the majority of variation being found within geographic samples (96.93%) and minimal variation between geographic samples (3.07%). For the analysis of PfAMA1 domains 1 to 3, 101 sequences that were reported by Polley and Conway (2, 17) were included, thereby increasing the number of PfAMA1 haplotypes from 107 to 150. Analysis of domains 1 to 3 demonstrated again that sequence variation was primarily within a geographic area (96.33%) with minimal variation between geographic areas (3.67%). Linkage disequilibrium (LD) analysis of the entire coding sequence and of domains 1 to 3 showed a rapid decline of LD with increasing distance between nucleotide sites, as seen in ref. 2.
PfAMA1 Sequences Exhibit Population Structure.
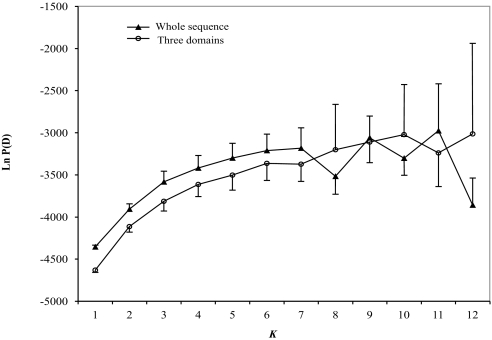
For our analysis, the clustering method (13, 14) attempts to assign individuals to populations on the basis of their genotypes. Generally speaking, this analysis uses departures from Hardy–Weinberg equilibrium to detect population structure. The analysis begins by classifying individuals of unknown origin into a predefined number of populations (Ks), in our case K = 1–12. A Bayesian approach is taken to infer the K value that provides the best fit to the data, as measured by the log-likelihood score. For both the entire coding sequence data (107 haplotypes) and domains 1 to 3 (150 haplotypes), the results were similar (Dataset S2), and we present the findings from domains 1 to 3 here. The estimated log probability of the data [Ln P(D)] plateaued between K = 4–6, and variance around the estimate increased after K = 6 (Fig. 1). Simulation studies have shown that once the real K has been reached, Ln P(D) will typically plateau or continue to increase slightly (18), and this is the case for K = 6 in our sample, indicating that K = 6 provides a good fit to the data (Fig. 1). Although analyses using the second-order rate of change of K (ΔK) suggested only two major populations, inspection of the membership distribution of individual parasites in the inferred populations (Fig. 2A) revealed that populations 1 and 2 remained stable even as K increased to 6, suggesting that these populations were well supported by our data and may be primarily responsible for the ΔK results. In addition, population structure was still evident when the analysis was rerun without populations 1 and 2 (data not shown).
Fig. 1.
Plot of the log probability of the data [LnP(D)] given values for K of 1–12. LnP(D) is shown for sequences of PfAMA1 domains 1 to 3 (amino acids 143–545, circles) and for the entire PfAMA1 coding region (triangles). Results for the two datasets are similar. Bar represents the mean value of variation of 10 replicate runs at each K value.
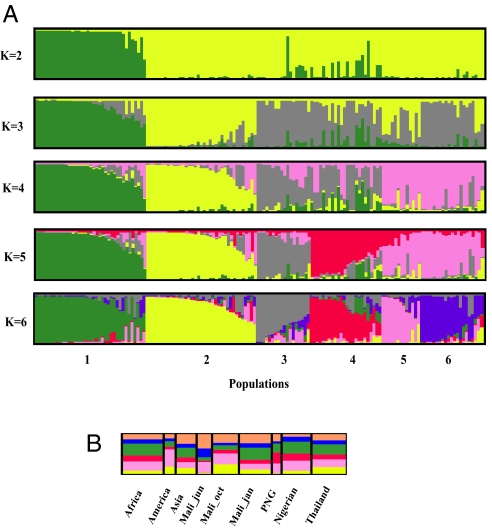
Fig. 2.
Worldwide population structure for P. falciparum sequences of PfAMA1 domains 1 to 3. Graphs of structure results were produced by using the DISTRUCT program. (A) Population clustering for K = 2–6. Each haplotype is represented by a thin vertical line. Each color represents a population, and the color of individual haplotypes represents their proportional membership in the different populations. Shown are population structures for populations 2–6. Populations: green, pop1; yellow, pop2; gray, pop3; red, pop4; pink, pop5; blue, pop6. (B) Distribution of structure-defined populations among geographical locations when K = 6. Membership coefficients for each population were averaged across individuals within a geographical region to create a population Q-matrix. Populations did not segregate based on geographical location or on sampling time points in Mali.
The data are displayed in Fig. 2A. Each haplotype is represented by a vertical line, and each population is shown as a different color. We show clustering results for K values of 2–6. Admixed haplotypes such as PfAMA1-FVO, that is, those with membership coefficients <75% (compared with the homologous members at the left side of each population), could not be assigned with confidence to any one population. The proportion of admixed haplotypes increased only slightly between K = 3–6 (range: 0.33–0.39) and even decreased between K = 4 (0.37) and K = 5 (0.34). Despite uncertainty about the assignment of these haplotypes, six distinct populations could clearly be seen at K = 6 (Fig. 2A). In addition, haplotypes clustered into populations independent of their geographic origin, with all six populations present in each geographical region (Fig. 2B).
Population Structure Defines Immunologically Relevant Groups of PfAMA1 Sequences.
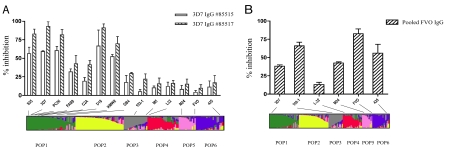
To determine whether the populations defined by our structure analysis represent immunologically relevant groups, we tested whether immunization with one member of a population elicited antibodies capable of inhibiting erythrocyte invasion in vitro by parasites expressing other members of that population and those expressing PfAMA1s belonging to other populations (Fig. 3 and Table 1). To do so, two rabbits were immunized with recombinant PfAMA1 based on the 3D7 sequence belonging to population 1. IgGs isolated from the rabbit sera showed significantly higher inhibition of invasion for eight parasites from population 1 than for parasites from other populations (Fig. 3A and Table 1). Within population 1, 3D7-specific antibodies showed greater inhibition of erythrocyte invasion for parasites expressing PfAMA1s that were more similar to 3D7 than more distantly related sequences (e.g., GB4, Fig. 3 and Table 1). Association analysis for anti-3D7 antibodies in population 1 identified the mutation of 1 aa (E to K at amino acid residue 187) as having the greatest effect on the efficiency of parasite invasion as showed in GIA experiments using the FAB9 and C2A clones (Table 1).
Fig. 3.
In vitro growth inhibition of different P. falciparum parasites by IgG from rabbits immunized with the 3D7 or FVO form of PfAMA1. (A) IgGs from two different rabbits (85515 and 85517) elicited against the 3D7 form of PfAMA1 were tested at a concentration of 0.75 mg/ml for their ability to inhibit the growth of 14 different strains of P. falciparum. Parasites from different populations included: Population 1: S35 (Mali), 3D7 (The Netherlands), PC26 (Peru), FAB9 (South Africa), C2A (Thailand), D10 (Papua New Guinea), KMWII (Kenya), and GB4 (Ghana); population 3: 102/1 (Sudan) and M5 (Mali); population 4: L32 (Liberia); population 5: FVO (Asia) and M24 (Kenya); and population 6: 425 (Gambia). Percentage inhibition values are presented as the mean ± SE from three (for population 1) or two (for other populations) independent experiments. (B) IgGs (1.5 mg/ml) from a pool of rabbit sera elicited against the FVO form of PfAMA1 were tested for their ability to inhibit the growth of six different strains of P. falciparum. Parasites in different groups included: population 1: 3D7 (The Netherlands); population 3: 102/1 (Sudan); population 4: L32 (Liberia); population 5: FVO (Asia) and M24 (Kenya); and population 6: 425 (Gambia). Percentage inhibition values are presented as the mean ± SE from two independent experiments.
Table 1.
Amino acid polymorphisms in the AMA-1 coding region of 14 P. falciparum parasites used in GIA
| Parasite G-POP* | GIA, % inhibition |
Amino acid position in whole sequence |
||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IgGs 85515 | IgGs 85517 | Domain 1 (27) |
Domain 2 (11) |
Domain 3 (11) |
||||||||||||||||||||||||||||||||||||||||||||||||
| 167 | 172 | 174 | 175 | 187 | 189 | 190 | 196 | 197 | 200 | 201 | 204 | 206 | 207 | 224 | 225 | 230 | 242 | 243 | 244 | 267 | 269 | 282 | 283 | 285 | 296 | 300 | 308 | 325 | 330 | 332 | 393 | 395 | 404 | 405 | 407 | 435 | 439 | 448 | 451 | 485 | 493 | 496 | 498 | 503 | 505 | 512 | 526 | 544 | ||||
| 3D7 | pop1 | 59.0 ± 0.6 | 93.0 ± 3.6 | T | G | Q | Y | E | L | M | D | E | H | F | D | K | Y | M | I | K | D | K | D | E | K | I | S | Q | D | K | Q | H | P | I | H | K | T | E | Q | I | N | D | M | K | D | M | S | R | F | R | E | K |
| D10 | pop1 | 66.7 ± 12.3 | 91.0 ± 3 | T | G | Q | Y | E | L | M | D | E | H | F | D | K | Y | M | I | K | D | K | D | E | K | I | S | Q | D | K | Q | H | S | I | H | K | T | E | Q | I | H | N | M | K | D | I | S | N | F | R | E | K |
| S35 | pop1 | 56.3 ± 4.9 | 82.6 ± 3.7 | T | G | Q | Y | E | L | M | D | E | H | F | D | K | Y | M | I | K | D | K | D | E | K | I | S | Q | D | K | Q | D | S | N | H | K | T | E | H | I | N | N | M | K | D | M | S | R | F | R | E | K |
| PC26 | pop1 | 60.3 ± 3.2 | 81.7 ± 4.1 | T | G | Q | Y | E | L | M | D | E | H | F | D | K | Y | M | I | K | D | K | D | E | K | I | S | Q | D | K | Q | H | S | I | H | K | T | E | Q | I | N | N | M | K | D | I | S | N | F | K | E | K |
| KMWII | pop1 | 52.3 ± 1.8 | 69.7 ± 5.6 | T | E | Q | D | E | L | M | D | Q | H | F | D | K | Y | M | I | K | D | K | D | Q | I | I | S | Q | D | K | K | H | P | I | H | K | T | E | Q | I | N | D | K | I | A | M | S | R | F | K | E | K |
| FAB9 | pop1 | 31.7 ± 2.3 | 42.7 ± 6.4 | T | E | Q | D | K | L | M | D | Q | H | F | D | K | Y | M | I | K | D | K | D | E | K | I | S | Q | D | K | Q | H | P | I | H | K | R | E | Q | I | N | N | M | K | D | I | S | N | F | K | E | K |
| C2A | pop1 | 19.0 ± 2.9 | 41.3 ± 3.2 | T | E | Q | D | K | L | M | D | Q | H | F | D | K | Y | M | I | K | D | K | D | E | K | I | S | Q | D | K | Q | H | S | N | H | K | T | E | H | I | N | D | M | K | D | I | S | N | F | K | E | K |
| GB4 | pop1 | 17.3 ± 5.5 | 29.3 ± 0.9 | T | E | Q | D | K | L | M | D | Q | H | F | D | K | Y | M | I | K | Y | E | D | E | K | K | L | E | D | E | E | H | S | N | H | K | R | E | Q | I | H | N | M | K | D | I | S | N | F | K | E | K |
| 102–1 | pop3 | 5.0 ± 2 | 22.0 ± 5 | T | E | K | D | E | L | M | D | D | R | F | D | E | Y | M | N | K | Y | N | D | Q | K | K | L | E | D | E | E | H | S | N | H | K | R | E | Q | N | H | D | K | I | D | I | S | N | F | K | E | K |
| M5 | pop3 | 10.0 ± 2 | 15.5 ± 5.5 | T | E | Q | D | N | P | M | N | G | D | L | N | E | Y | M | N | K | Y | E | D | Q | K | K | S | Q | D | K | E | H | S | N | H | R | R | K | Q | N | H | D | K | I | A | I | S | N | F | K | E | K |
| L32 | pop4 | 12.0 ± 4 | 16.0 ± 3 | T | E | Q | D | E | L | M | D | R | D | F | N | E | D | I | N | K | D | K | N | Q | K | K | S | Q | D | E | E | H | P | I | H | R | T | K | Q | I | H | D | K | I | A | I | T | N | F | K | E | K |
| M24 | pop5 | 8.0 ± 4 | 15.0 ± 5 | T | E | Q | D | K | L | M | D | Q | D | F | N | E | Y | M | N | E | Y | K | D | Q | K | K | L | E | H | K | Q | H | S | N | R | K | T | K | Q | I | H | D | K | I | D | M | S | R | Y | R | E | N |
| FVO | pop5 | 4.0 ± 2 | 9.5 ± 3.5 | K | G | Q | D | N | L | I | N | G | D | F | N | E | Y | M | N | K | Y | N | D | Q | K | K | L | E | D | E | E | H | S | N | H | K | R | E | Q | I | H | D | K | K | D | M | S | R | F | R | E | N |
| 425 | pop6 | 11.0 ± 6 | 17.0 ± 7 | K | G | Q | D | N | L | I | D | H | D | F | N | E | Y | M | N | K | Y | E | D | Q | K | K | L | Q | H | E | Q | D | S | N | H | K | T | E | H | I | N | N | M | K | D | M | S | R | F | R | E | K |
Only anti-3D7 GIA data were shown in table; G-POP: the defined populations; 85515 and 85517 are rabbit ID numbers; numbers in parentheses of each domain indicate the total mutations in the region.
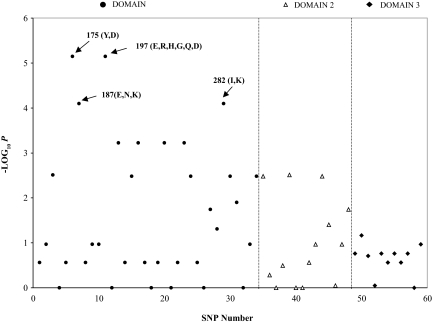
In the analysis of parasites from all of the populations, amino acid residues 175, 197, and 282 also showed a significant impact on inhibition of erythrocyte invasion (Fig. 4). All residues that impacted variation in erythrocyte invasion were found in AMA1 domain 1, with residue 197 being the most polymorphic. These data are only for anti-3D7 antibodies and would presumably be different for antibodies raised against single sequences from different populations.
Fig. 4.
Association analysis of GIA and SNPs across the three domains of PfAMA1. The analysis, using only anti-3D7 GIA data, is based on single-marker association using the χ2 test. SNPs (total of 59 in our analysis) located in the PfAMA1 gene are depicted as different symbols for the different domains: domain 1 circles, domain 2 triangles, and domain 3 diamonds. The arrows indicate the sites with the most significant P values.
To test the protection potential of PfAMA1 sequences with admixed ancestry, the FVO PfAMA1 sequence was selected to generate antibodies in rabbits, and a GIA experiment was performed by using six parasites belonging to five of the six different PfAMA1 populations. Interestingly, FVO-specific antibodies showed high cross-population growth inhibition of parasites from PfAMA1 populations 3 and 6 (Fig. 3B) in contrast to 3D7-specific antibodies, showing that antibodies to the PfAMA1 molecule with admixed sequences protect against parasites with PfAMA1 carrying homogeneous sequences from multiple populations.
Discussion
After decades of research and development, we still do not have an effective vaccine to control malaria. The emergence of drug-resistant parasites, particularly to chloroquine, has increased the need for such a vaccine. One of the obstacles to malarial vaccine development is P. falciparum's ability to escape host immune responses to a parasite antigen by altering key amino acids of that antigen (1, 2). Many known malaria antigens (e.g., PfAMA1) identified so far are highly polymorphic (19). Various studies have shown that antisera against PfAMA1 can block parasite invasion of erythrocytes in vitro if the parasite has the same haplotype as the protein used for vaccination but not parasites carrying heterologous PfAMA1 haplotypes (11). This presents a significant challenge for vaccine design, which can be surmounted either by using a conserved antigen, which may not exist, or by using a vaccine that combines all of the potential haplotypes that might be found in malaria-endemic areas.
For a highly polymorphic antigen like PfAMA1, a vaccine including all of the identified haplotypes is not feasible. Our analyses have included 150 PfAMA1 haplotypes from around the world. Undoubtedly, the number of haplotypes will increase as more samples are analyzed. In our effort to find a way to improve vaccine design, we asked whether it is possible to develop a vaccine containing a limited number of PfAMA1 haplotypes that could protect against the majority of parasites in the field. Although PfAMA1 undergoes mutation when subjected to host immune pressure, it also plays an important role in erythrocyte invasion by the parasite (6) that may functionally constrain against certain modifications to the protein. Because mutations in protein coding regions are often deleterious, these mutants may not be able to compete with parasites in the field carrying mutations that do not affect protein function. Thus, the PfAMA1 sequences obtained from parasites in the field result from forces of immune selection and constraints imposed by its function.
Here, we show that a worldwide collection of PfAMA1 haplotypes could be clustered into six populations. Moreover, these clusters were present in all of the geographical regions that were sampled. This finding was in contrast to the results of previous studies where geography was an important determinant in the chromosome-wide analysis of P. falciparum SNPs (20) and in HIV (21). The P. falciparum isolates that were analyzed in our study were derived from distinct and widely dispersed geographic locations and over a long time period, indicating a relatively stable population structure of PfAMA1. This suggests that a PfAMA1 vaccine containing alleles representative of each of the six identified populations may be effective against nearly all haplotypes.
To test the biological relevance of defined PfAMA1 populations for vaccine development, we immunized rabbits against one PfAMA1 sequence belonging to population 1 and tested the ability of purified IgGs to block erythrocyte invasion in vitro of multiple members of population 1 and of parasites expressing PfAMA1 from the other populations. The IgGs blocked invasion of parasites expressing PfAMA1s of population 1 more effectively than parasites from other populations, suggesting that the population structure grouping was immunologically relevant. This protection, however, was far less effective against a parasite with an admixed PfAMA1 haplotype (P. falciparum GB4 strain) with ancestry primarily from populations 1 and 6.
The evidence that a vaccine containing six PfAMA1 alleles representing each of the different populations may not select for new variants comes from studies of antibodies induced against the FVO clone of PfAMA1. The FVO PfAMA1 haplotype displays admixture compared with the homogeneous population 5 isolates (e.g., M24) at the left side of Fig. 2A. Antibodies against FVO PfAMA1 inhibited invasion of two parasites expressing PfAMA1s from other populations. This result suggests that including these six different PfAMA1s in a vaccine may provide protection against parasites expressing these admixed PfAMA1 sequences.
As far as we are aware, this application of population structure analysis to a large number of P. falciparum PfAMA1 sequences for the purpose of rationalizing vaccine design has not previously been tested. Testing whether antibodies to one PfAMA1 sequence from each population at K = 4, K = 5, or K = 6 can block invasion of parasites expressing the entire spectrum of sequences, including admixtures, remains to be done. These studies will define the biological relevance of the population structure prediction and determine its utility in guiding vaccine design.
Methods
DNA Sample Collection.
Genomic DNA from 97 P. falciparum clones that were collected from around the world (34 from Africa, 31 from Asia, 21 from America, and 11 from Papua New Guinea) has been described (15, 22). A multicopy microsatellite marker (PfRRM) was used to genotype the parasites to ensure that the same clone was used in the PfAMA1 sequencing step and the GIA experiment (16). Field samples were collected from participants of a study conducted in Donéguébougou, Mali, a rural village of ≈1,300 inhabitants located 30 km northeast of the Malian capital, Bamako. Individuals between the ages of 6 months and 45 years were randomly selected from the village census and invited to participate in the study. Two hundred nine participants were enrolled in July 2002, at the beginning of the malaria transmission season, and were sampled at three visits: in July 2002, October 2002, and January 2003, corresponding to the start, the midpoint, and 1 month after the end of the malaria transmission season, respectively. Capillary blood was obtained by finger prick using sterile lancets and was collected onto blood-collection paper (Schleicher & Schuell) and microscope slides for thick-film microscopy. Blood collection papers were air-dried and stored at room temperature in individually sealed plastic bags containing desiccant packs until DNA extraction was performed. A random sample of 210 filter papers, drawn equally from the three different sampling time points, was analyzed as part of the study. Of these 210 samples, 181 amplified the target sequence, and of these, only 61 contained a single genotype and were included in the analysis (Dataset 1).
Community permission for the study in Mali was obtained from village elders, along with approval from the ethical review committees of the Faculty of Medicine, Pharmacy, and Dentistry at the University of Bamako (Mali) and the National Institutes of Allergy and Infectious Diseases (Bethesda). Individual written informed consent was obtained from all participants or their guardians, and human experimentation guidelines of the U.S. Department of Health and Human Services and those of the participating institutions were followed in conducting this research.
Amplification and Sequencing of the PfAMA1 Coding Region.
DNA from laboratory-adapted P. falciparum clones and blood-collection papers was extracted according to the protocols and reagents of the QIAamp DNA mini kit (Qiagen), as recommended by the manufacturer. PCR primer pair (F2, 5′-GTACTTGTTATAAATTGTACA-3′; R8, 5′-TTTTAGCATAAAAGAGAAGC-3′); and nested PCR primer pair (N1, 5′-ATGAGAAAATTATACTGCGT-3′; N2, 5′-TGATTATATCAGACGTTGAA-3′) were used to amplify the entire coding region sequence of the PfAMA1 gene. PCR conditions and set-up were the same as described (23). Negative and positive controls were included in each PCR. After treatment with ExoSAP-it (U.S. Biochemical), PCR products were used directly in sequence reactions and analyzed on an ABI 3730XL automatic sequencer as recommended by the manufacturer (Applied Biosystems).
GIA.
Fourteen parasite clones (3D7, D10, S35, PC26, KMWII, FAB9, C2A, GB4, FVO, 102/1, M5, M24, L32, and 425) from different populations were selected for testing in the in vitro GIA (Fig. 4 and Table 1). The GIA method used was the same as described in ref. 11. IgGs were purified from the sera (11) of two rabbits that had been vaccinated with recombinant PfAMA1 that corresponded to the sequence of the 3D7 P. falciparum clone. Additionally, anti-PfAMA1 IgGs were purified from a pool of sera obtained from rabbits vaccinated with the PfAMA1-FVO recombinant protein. Purified anti-PfAMA1 IgGs (0.75 or 1.5 mg/ml for anti-PfAMA1–3D7 or FVO, respectively) were incubated with cultured P. falciparum-parasitized erythrocytes, and the parasite growth after 40 h of culture was determined by a biochemical assay specific for parasite lactate dehydrogenase. The results of the GIA experiments were expressed as percentage inhibition, which was calculated as follows: 100 − [(OD650 of infected RBCs with tested IgG − OD650 of normal RBCs only)/(OD650 of infected RBCs without any IgG − OD650 of normal RBCs only) × 100].
Data Analysis.
SNP identification and verification.
DNA sequences were aligned by using SEQUENCHER 4.5 (Gene Codes). The quality of the DNA sequences and all SNPs was assessed by visual inspection.
Haplotype construction and genetic diversity analysis.
Each unique PfAMA1 sequence was treated as a separate haplotype, and the haplotype files were created by using DNASP 4.1 (24). LD across the PfAMA1 gene was analyzed by using the same software. ARLEQUIN was used to perform the AMOVA test to evaluate the genetic variance present both between and among PfAMA1 sequences from different geographic regions.
Cluster (population) analysis.
To determine whether our samples could be grouped into genetic clusters and to infer the number of clusters that best fit the data, we used the Bayesian clustering method implemented in the STRUCTURE program (13, 14). The log-probability of the data, given a certain value of K [LnP(D∣K)] was calculated and compared across a range of K values to determine which one provided the best fit to the data. Markov Chain Monte Carlo searches consisted of 50,000 “burn-in” steps, followed by 100,000 iterations. We performed 10 replicate runs at each K from 1 to 12 under the admixture model with correlated allele frequencies.
Statistical analysis.
The association between SNPs and GIA activity was assessed by using the χ2 test including a permutation test of 1,000 iterations as described (15).
Supplementary Material
Acknowledgments.
We thank Dr. Susan K. Pierce for suggestions on the manuscript; Mr. Ababacar Diouf for parasite culture and GIA analysis; and the volunteers of Donéguébougou, Mali, the Malaria Research and Training Center Field and Laboratory staff, and the local authorities. This work was supported by the Intramural Program of the National Institute of Allergy and Infectious Diseases of the National Institutes of Health. Support for GIA analysis came from the PATH–Malaria Vaccine Initiative, Bethesda.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802328105/DCSupplemental.
References
- 1.Coley AM, et al. The most polymorphic residue on Plasmodium falciparum apical membrane antigen 1 determines binding of an invasion-inhibitory antibody. Infect Immun. 2006;74:2628–2636. doi: 10.1128/IAI.74.5.2628-2636.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Polley SD, Conway DJ. Strong diversifying selection on domains of the Plasmodium falciparum apical membrane antigen 1 gene. Genetics. 2001;158:1505–1512. doi: 10.1093/genetics/158.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Healer J, Crawford S, Ralph S, McFadden G, Cowman AF. Independent translocation of two micronemal proteins in developing Plasmodium falciparum merozoites. Infect Immun. 2002;70:5751–5758. doi: 10.1128/IAI.70.10.5751-5758.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bai T, et al. Structure of AMA1 from Plasmodium falciparum reveals a clustering of polymorphisms that surround a conserved hydrophobic pocket. Proc Natl Acad Sci USA. 2005;102:12736–12741. doi: 10.1073/pnas.0501808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pizarro JC, et al. Crystal structure of the malaria vaccine candidate apical membrane antigen 1. Science. 2005;308:408–411. doi: 10.1126/science.1107449. [DOI] [PubMed] [Google Scholar]
- 6.Triglia T, et al. Apical membrane antigen 1 plays a central role in erythrocyte invasion by Plasmodium species. Mol Microbiol. 2000;38:706–718. doi: 10.1046/j.1365-2958.2000.02175.x. [DOI] [PubMed] [Google Scholar]
- 7.Stowers AW, et al. Vaccination of monkeys with recombinant Plasmodium falciparum apical membrane antigen 1 confers protection against blood-stage malaria. Infect Immun. 2002;70:6961–6967. doi: 10.1128/IAI.70.12.6961-6967.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins WE, et al. Protective immunity induced in squirrel monkeys with recombinant apical membrane antigen-1 of Plasmodium fragile. Am J Trop Med Hyg. 1994;51:711–719. doi: 10.4269/ajtmh.1994.51.711. [DOI] [PubMed] [Google Scholar]
- 9.Deans JA, et al. Vaccination trials in rhesus monkeys with a minor, invariant, Plasmodium knowlesi 66 kD merozoite antigen. Parasite Immunol. 1988;10:535–552. doi: 10.1111/j.1365-3024.1988.tb00241.x. [DOI] [PubMed] [Google Scholar]
- 10.Anders RF, et al. Immunisation with recombinant AMA-1 protects mice against infection with Plasmodium chabaudi. Vaccine. 1998;16:240–247. doi: 10.1016/s0264-410x(97)88331-4. [DOI] [PubMed] [Google Scholar]
- 11.Kennedy MC, et al. In vitro studies with recombinant Plasmodium falciparum apical membrane antigen 1 (AMA1): production and activity of an AMA1 vaccine and generation of a multiallelic response. Infect Immun. 2002;70:6948–6960. doi: 10.1128/IAI.70.12.6948-6960.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polley SD, et al. Human antibodies to recombinant protein constructs of Plasmodium falciparum Apical Membrane Antigen 1 (AMA1) and their associations with protection from malaria. Vaccine. 2004;23:718–728. doi: 10.1016/j.vaccine.2004.05.031. [DOI] [PubMed] [Google Scholar]
- 13.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mu J, et al. Multiple transporters associated with malaria parasite responses to chloroquine and quinine. Mol Microbiol. 2003;49:977–989. doi: 10.1046/j.1365-2958.2003.03627.x. [DOI] [PubMed] [Google Scholar]
- 16.Su XZ, Carucci DJ, Wellems TE. Plasmodium falciparum: Parasite typing using a multicopy microsatellite marker, PfRRM. Exp Parasitol. 1998;89:262–265. doi: 10.1006/expr.1998.4299. [DOI] [PubMed] [Google Scholar]
- 17.Polley SD, Chokejindachai W, Conway DJ. Allele frequency-based analyses robustly map sequence sites under balancing selection in a malaria vaccine candidate antigen. Genetics. 2003;165:555–561. doi: 10.1093/genetics/165.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 19.Mu J, et al. Genome-wide variation and identification of vaccine targets in the Plasmodium falciparum genome. Nat Genet. 2007;39:126–130. doi: 10.1038/ng1924. [DOI] [PubMed] [Google Scholar]
- 20.Mu J, et al. Recombination hotspots and population structure in Plasmodium falciparum. PLoS Biol. 2005;3:e335. doi: 10.1371/journal.pbio.0030335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCutchan FE, Salminen MO, Carr JK, Burke DS. HIV-1 genetic diversity. AIDS. 1996;10(Suppl 3):S13–S20. [PubMed] [Google Scholar]
- 22.Wootton JC, et al. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature. 2002;418:320–323. doi: 10.1038/nature00813. [DOI] [PubMed] [Google Scholar]
- 23.Wang X, et al. Decreased prevalence of the Plasmodium falciparum chloroquine resistance transporter 76T marker associated with cessation of chloroquine use against P. falciparum malaria in Hainan, People's Republic of China. Am J Trop Med Hyg. 2005;72:410–414. [PubMed] [Google Scholar]
- 24.Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DNASP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.