Abstract
Severe acute respiratory syndrome coronavirus (SARS-CoV) is a high-risk infectious pathogen. In the proposed model of respiratory failure, SARS-CoV down-regulates its receptor, angiotensin-converting enzyme 2 (ACE2), but the mechanism involved is unknown. We found that the spike protein of SARS-CoV (SARS-S) induced TNF-α-converting enzyme (TACE)-dependent shedding of the ACE2 ectodomain. The modulation of TACE activity by SARS-S depended on the cytoplasmic domain of ACE2, because deletion mutants of ACE2 lacking the carboxyl-terminal region did not induce ACE2 shedding or TNF-α production. In contrast, the spike protein of HNL63-CoV (NL63-S), a CoV that uses ACE2 as a receptor and mainly induces the common cold, caused neither of these cellular responses. Intriguingly, viral infection, judged by real-time RT-PCR analysis of SARS-CoV mRNA expression, was significantly attenuated by deletion of the cytoplasmic tail of ACE2 or knock-down of TACE expression by siRNA. These data suggest that cellular signals triggered by the interaction of SARS-CoV with ACE2 are positively involved in viral entry but lead to tissue damage. These findings may lead to the development of anti-SARS-CoV agents.
Keywords: shedding, cytoplasmic tail, HNL63-CoV, TNF-α
Severe acute respiratory syndrome coronavirus (SARS-CoV) is an infectious pathogen known to have caused acute respiratory distress in >8,000 patients with a mortality rate of ≈10% (1). Although outbreaks of SARS-CoV are now well controlled, the mechanism of severe respiratory failure in infected patients is unknown. Angiotensin-converting enzyme 2 (ACE2), an ACE homolog that functions as a positive regulator of the renin-angiotensin system (RAS) (2, 3), was identified as a receptor of SARS-CoV (4). A possible indicator of a severe clinical outcome, the spike protein of SARS-CoV (SARS-S) was found to down-regulate ACE2 expression (5). ACE2 knockout (KO) mice were also shown to be susceptible to severe respiratory failure after chemical challenge (5, 6), and ACE2 has been shown to moderate ACE-induced intracellular inflammation, suggesting that the mechanism of ACE2 down-regulation may explain the molecular basis of SARS-CoV-related severe respiratory distress.
HNL63-CoV, a CoV (7) that causes the common cold, was recently found to use ACE2 for viral infection (8), and it was further shown that the spike protein of HNL63-CoV (NL63-S) binds ACE2 directly (9). NL63-S and SARS-S show 21% identity (10), whereas that between NL63-S and the spike protein of HCoV-229E, which uses CD13 [a completely different carboxy (C)-peptidase] as a cellular receptor (11), is 55%. It is important to note that, despite their phylogenetically distinct properties and their producing different clinical outcomes, SARS-CoV and HNL63-CoV both use ACE2.
Based on these observations, we hypothesized that the functional modulation of ACE2 may be differentially induced by SARS-S and NL63-S. To test this prediction, we first clarified the mechanism of SARS-S-induced ACE2 down-regulation, and then compared the cellular responses induced by each protein. We found that SARS-S induced TNF-α-converting enzyme (TACE)-dependent shedding of the ectodomain of ACE2, and that the process was coupled with TNF-α production (12). Experiments using deletion mutants of ACE2 revealed that the cytoplasmic tail of ACE2 is required for shedding. Furthermore, viral infection with SARS-CoV was significantly decreased in cells expressing mutant versions of ACE2 lacking the cytoplasmic domain. In addition, siRNAs of TACE and ACE2 blocked viral infection. These observations suggest that TACE activity is modulated by SARS-CoV via the cytoplasmic domain of ACE2, and that it facilitates viral entry but also causes tissue damage through TNF-α production. Together with data showing that NL63-S does not evoke these cellular responses, our results indicate that the cellular components involved in SARS-S-induced ACE2 shedding are good targets for anti-SARS-CoV therapeutics.
Results
SARS-S Induces Shedding of the Ectodomain of ACE2.
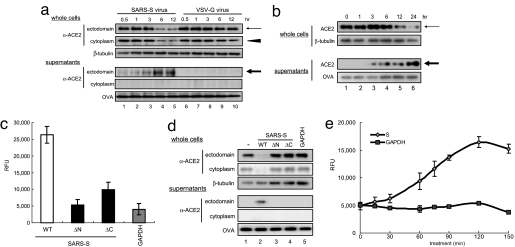
We first examined the mechanism of ACE2 down-regulation after SARS-S binding. In total cellular extracts prepared from Vero E6 cells infected with a pseudotyped virus expressing S protein (S-virus), ACE2 expression was reduced, whereas the expression of an 80-kDa protein in the supernatants was increased (Fig. 1a, lanes 4 and 5). Interestingly, this 80-kDa peptide was not detected by an antibody raised against the cytoplasmic domain of ACE2 (Fig. 1a, lanes 4 and 5, middle row in Lower). Conversely, a control pseudotyped lentivirus expressing vesicular stomatitis virus G protein (VSV-G) did not induce any change in ACE2 expression (Fig. 1a, lanes 6–10). Moreover, recombinant SARS-S showed the same effects (Fig. 1b, lanes 4–6, arrows), whereas binding-defective mutants of the protein (Fig. 1c, ΔC and ΔN) did not (Fig. 1d, lanes 2–4). Consistent with a report showing that the secreted form of ACE2 is the active form (shed form) (2), C-peptidase activity was detected in the supernatants of cultured cells treated with SARS-S (Fig. 1e). Therefore, we concluded that SARS-S down-regulates ACE2 by inducing shedding of the ACE2 ectodomain.
Fig. 1.
Induction of ACE2 shedding by the specific binding of SARS-S and ACE2. (a) ACE2 shedding is induced by SARS-S. After infection with SARS-S, total extracts (Upper) or the supernatants (Lower) of cultured Vero E6 cells were analyzed. The thin arrow and arrowhead indicate the 120-kDa ACE2 protein detected by antibodies against the ectodomain and cytoplasmic domain, respectively. The bold arrow indicates the 80-kDa ACE2 detected by an antibody against the ACE2 cytoplasmic domain (lanes 2–5), which was not generated by control VSV-G virus (lanes 6–10). The proteins present in the culture supernatants were recovered with OVA by TCA precipitation. β-Tubulin was used as a loading control. (b) Recombinant SARS-S induces ACE2 shedding. Cells were treated with 100 μg/ml SARS-S (residues 284–541), and ACE2 in the culture supernatant was examined as described in a. (c) Binding of the SARS-S mutants to ACE2. Two SARS-S mutants, ΔN (residues 332–541) and ΔC (residues 284–490), were prepared according to a report showing the minimum region of SARS-S required for ACE2 binding (27), and their binding activities to ACE2 were examined (see also Materials and Methods). (d) Specific binding to ACE2 is required for ACE2 shedding. After treatment with the ΔN and ΔC SARS-Ss, the amount of ACE2 in the culture supernatant was examined as described in a. (e) C-peptidase activity of the shed ACE2. The C-peptidase activity in the culture supernatant of SARS-S-treated Vero E6 cells was measured (filled diamonds) (see Materials and Methods). GAPDH was used as a negative control (filled squares).
TACE Is a Critical Cellular Factor in SARS-S-Induced ACE2 Shedding.
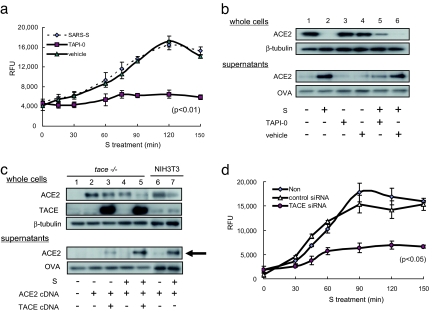
It has been shown that TACE and ADAM17, a member of the ADAM (a disintegrin and metalloprotease) family (12, 13), are required for the induction of ACE2 shedding by phorbol esters [phorbol-12-myristate-13-acetate (PMA)] (14). Therefore, we examined the effects of TAPI-0, an inhibitor of TACE (15), on SARS-S-induced ACE2 shedding. As shown in Fig. 2a, TAPI-0 efficiently attenuated C-peptidase activity in the culture supernatant. Western blot analysis also indicated that TAPI-0 inhibited SARS-S-induced ACE2 shedding (Fig. 2b, lanes 5 and 6). To further determine the involvement of TACE, ACE2 shedding was examined by forced expression of TACE cDNA in transformed mouse fibroblasts derived from tace-KO mouse fibroblasts (tace-KO cells) (16). As shown in Fig. 2c, the tace-KO cells exhibited SARS-S-induced ACE2 shedding when both TACE and ACE2 cDNA was introduced (Fig. 2c, lanes 4 and 5). To obtain direct evidence that TACE plays an important role in ACE2 shedding, TACE siRNA was introduced into Huh-7 cells and SARS-S-induced ACE2 shedding was examined. Huh-7 is a human hepatoma cell line permissive for SARS-CoV infection (17, 18). As shown in Fig. 2d, ACE2 shedding by SARS-S was significantly attenuated by TACE siRNA. As shown in Fig. 5c, TACE siRNA successfully reduced endogenous TACE expression.
Fig. 2.
ACE2 shedding depends on TACE. (a) Inhibitory effects of TAPI-0 on ACE2 shedding. Vero E6 cells were cultured with or without 100 nM TAPI-0, and the amounts of C-peptidase activity in the culture supernatants were measured. SARS-S was used at a concentration of 100 μg/ml. (b) TACE inhibitor blocked SARS-S-induced ACE2 shedding. The experiments described in a were carried out and ACE2 was detected by Western blot analysis. (c) ACE2 shedding depends on TACE activity. tace-KO cells were transfected with cDNAs encoding ACE2 or TACE and then treated with 100 μg/ml SARS-S for 6 h. Shed ACE2 was then detected (bold arrow). (d) The effects of TACE siRNA on ACE2 shedding. After the introduction of TACE or control siRNA, SARS-S-induced ACE2 shedding was examined by measuring the amount of C-peptidase activity in the culture supernatant. Huh-7 cells, a human cell line that expresses endogenous ACE2 and TACE (see Fig. 5c), were analyzed.
Fig. 5.
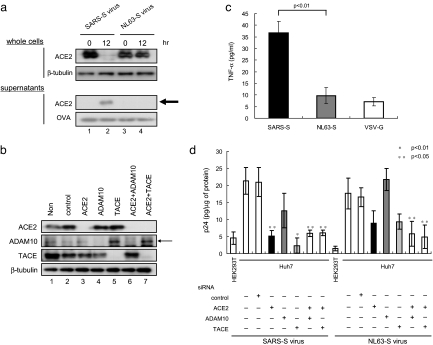
NL63-S does not induce ACE2 shedding. (a) No ACE2 shedding was observed when using NL-63-S. After infection of Vero E6 cells with NL63-S or SARS-S, ACE2 was examined as described in Fig. 1a. (b) NL63-S does not induce TNF-α production. TNF-α was measured in the culture supernatants of ACE2-expressing HEK293T cells. The supernatants were collected 12 h after infection with NL63-S or SARS-S. (c) Effects of siRNAs on the expression of endogenous gene products. Huh-7 cells were transduced with siRNAs against TACE, ACE2, and ADAM10, and the expression of each protein was examined. Lanes 3, 4, and 5 indicate the endogenous expression of ACE2, ADAM10, and TACE, respectively. Lanes 6 and 7 depict the effects of the combined use of siRNAs on protein expression. (d) The differential requirements of TACE for viral entry by NL63-S and SARS-S. Huh-7 cells were infected with SARS-S and NL63-S viruses for 4 h, and the efficiency of viral entry was examined by measuring the intracellular p24 level. Note that the entry of SARS-S was reduced by siRNAs against ACE2 and TACE, whereas NL63-S was not.
The ACE2 Cytoplasmic Domain Is Required for SARS-S-Induced ACE2 Shedding.
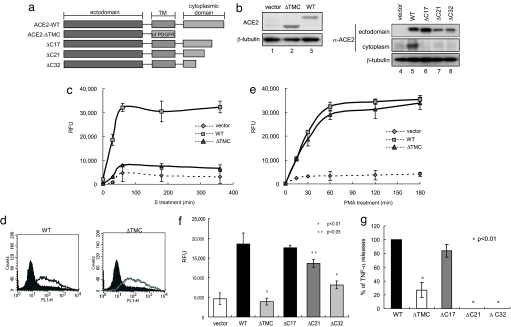
Cellular TACE was activated by the interaction of SARS-S with ACE2, strongly suggesting the involvement of the cytoplasmic domain of ACE2 in this phenomenon. To confirm this suggestion, we prepared several mutants lacking the C-terminal region of ACE2 (Fig. 3a). After confirming the expression of each mutant protein (Fig. 3b), we examined shedding of the ectodomain by SARS-S. SARS-S-induced ACE2 shedding was not observed with a chimeric mutant in which the transmembrane and cytoplasmic domains of ACE2 were eliminated [transmembrane and cytoplasmic domains (ΔTMC) Fig. 3c]. This mutant was able to bind SARS-S (Fig. 3d), and it was susceptible to ectodomain shedding when the cells were stimulated with PMA (Fig. 3e). Experiments with additional deletion mutants revealed that the last 17 aa of the ACE2 C terminus were dispensable for ACE2 shedding (Fig. 3f). Similar to ACE2 shedding, the production of TNF-α was detected in the culture supernatants of cells expressing WT and ΔC17-ACE2 (Fig. 3g). In contrast, no TNF-α production was observed in cells expressing deletion mutants of the cytoplasmic tail of ACE2 (Fig. 3g, P < 0.01).
Fig. 3.
ACE2 shedding by SARS-S requires the cytoplasmic domain of ACE2. (a) Schematic diagrams of the ACE2 cytoplasmic domain mutants. ΔTMC-ACE2 contains only the ectodomain of ACE2 with the transmembrane domain of PDGF receptor. (b) Expression of the ACE2 cytoplasmic domain mutants. The molecular weights of full-length ACE2 (lane 3) and all mutants except ΔTMC-ACE2 (lanes 6–8) were ≈120 kDa. The molecular weight of ΔTMC-ACE2 was ≈95 kDa (lane 2). (c) ΔTMC-ACE2 did not show ectodomain shedding with SARS-S. After treatment with SARS-S, the amounts of C-peptidase activity in the culture supernatant of cells expressing WT (filled sqaures) or ΔTMC-ACE2 (filled diamonds) were examined. (d) Binding of ΔTMC-ACE2 to SARS-S. Recombinant SARS-S was expressed and purified as a chimeric protein with the Fc portion of human IgG (Fc), and bound SARS-S was detected by FACS analysis, using FITC-labeled anti-human IgG (9). Each line indicates the cells that were positive for bound SARS-S. Solid peaks depict cells bound nonspecifically with control IgG-Fc. (e) ΔTMC-ACE2 was competent for ectodomain shedding by PMA. After treatment with PMA for 1 h, the amounts of C-peptidase activity in the culture supernatant of cells expressing WT (filled squares) or ΔTMC-ACE2 (filled diamonds) were examined. (f) The ectodomain shedding of ACE2 depends on the cytoplasmic domain. The levels of C-peptidase activity in the culture supernatant of cells expressing various ACE2 mutants were measured. (g) TACE activation requires the cytoplasmic domain of ACE2. TACE activation, judged by the production of TNF-α in the culture supernatant, was induced by both WT and ΔC17-ACE2. TNF-α was detected 20 h after SARS-S treatment.
The ACE2 Cytoplasmic Domain and TACE Are Positively Involved in Virus Entry.
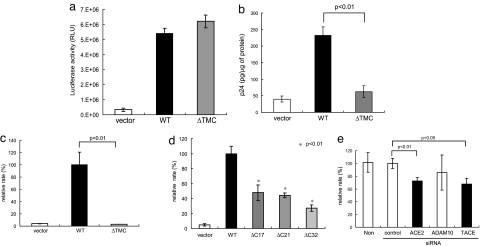
We investigated the role of the cytoplasmic tail of ACE2 and TACE in viral infection. In a reporter assay based on a pseudotyped lentiviral system, no difference in infection was detected between cells expressing WT and ΔTMC-ACE2 (Fig. 4a). Although this observation is consistent with previous reports, we observed a reproducible difference in viral entry based on the intracellular p24 concentration after infection with the same virus (Fig. 4b). To obtain conclusive evidence, we infected the cells with SARS-CoV and quantified SARS-CoV mRNA by real-time RT-PCR. As shown in Fig. 4c, deletion of the cytoplasmic tail greatly reduced viral infection. The efficiency of infection was also markedly reduced in cells infected with ΔC21 or ΔC32-ACE2 (Fig. 4d), even with ΔC17-ACE2.
Fig. 4.
The cytoplasmic tail of ACE2 and TACE promote infection by SARS-CoV. (a) No difference in infection efficiency was detected by reporter assay, using a pseudotyped lentivirus system. After infection of HEK293T cells expressing WT or ΔTMC-ACE2 with a pseudotyped lentivirus encoding luciferase, luciferase activity was measured. Luciferase activity was assayed in triplicate. (b) Detection of differences in viral infection efficiency based on the intracellular p24 level. The experiment in a was followed by measurement of the intracellular p24 level, using an ELISA kit. (c) A marked difference in viral infection efficiency detected by real-time RT-PCR. Each cell was harvested and analyzed 2 h after infection with SARS-CoV. The data were normalized by 18S ribosomal RNA. (d) The cytoplasmic tail is required for efficient viral infection. SARS-CoV was used to infect cells expressing deletion mutants of the cytoplasmic tail of ACE2 (Fig. 3a). Then, real-time RT-PCR was carried out as described in c. (e) TACE is required for efficient viral entry. The efficiency of viral entry into Huh-7 cells was examined before and after the introduction of TACE or ACE2 siRNAs. TACE siRNA [but not ADAM10 (metalloprotease control) siRNA] significantly attenuated viral entry (P < 0.05). ACE2 siRNA also decreased viral infection (P < 0.01).
We next examined the role of TACE in viral infection, using TACE siRNA. As shown in Fig. 4e, viral entry was significantly attenuated (P < 0.05) by the siRNA, whereas control or ADAM10 (control metalloprotease) siRNA did not decrease the incidence of viral infection. It is important to note that ACE2 siRNA significantly decreased the SARS-CoV mRNA copy number (Fig. 4e, P < 0.01), indicating that infection by SARS-CoV via ACE2 was successfully detected by real-time RT-PCR. As shown in Fig. 5c, each siRNA efficiently blocked expression of the target protein. These observations indicate that the cytoplasmic tail of ACE2 and TACE are positively involved in SARS-S-dependent viral infection. In addition, the level of intracellular p24 after infection with pseudotyped lentivirus is a good indicator of viral entry.
HNL63-CoV Spike Protein Does Not Induce ACE2 Shedding or TACE Activation.
To determine the specificity of the ACE2 shedding and TNF-α production induced by SARS-CoV, we investigated the functions of HNL63-CoV. In marked contrast to SARS-S, NL63-S did not induce ACE2 shedding (Fig. 5a, lane 4), although it was able to use ACE2 as a cellular receptor (Fig. 5d). Furthermore, NL63-S did not induce TNF-α production (Fig. 5b). To characterize the differential mode of infection between SARS-S and NL63-S, we examined the effects of siRNAs against ACE2, TACE, and ADAM10, and we examined viral entry by measuring the level of intracellular p24. We began by confirming that each siRNA or combination of siRNAs successfully interfered with expression of the endogenous gene products (Fig. 5c). As shown in Fig. 5d, the ACE2 and TACE siRNAs efficiently blocked viral entry (P < 0.05 and 0.01, respectively), whereas the ADAM10 siRNA did not. However, we detected no definite effect of TACE siRNA on NL63-S infection. The ACE2 and TACE or ADAM10 siRNAs, however, did significantly inhibit NL63-S viral entry.
Taken together, these observations suggest that ACE2 and TACE are differently required for infection by SARS-S and NL63-S and that the functional role of TACE is more prominent for SARS-S than for NL63-S.
Discussion
Down-regulation of ACE2 was proposed to be associated with a severe clinical outcome in cases of SARS-CoV infection, but the mechanism remained to be clarified. In the present study, we found that SARS-S induces ACE2 shedding via a process that is tightly coupled with TNF-α production. In contrast, the spike protein of HNL63-CoV, a CoV and etiological pathogen of the common cold that uses ACE2 as its cellular receptor, did not induce such cellular responses. These observations suggest that ACE2 shedding and its causative cellular signals are attributable to SARS-CoV-induced tissue damage.
As shown in Fig. 3g, the induction of TNF-α production by SARS-S depends on the cytoplasmic tail of ACE2 and TACE activity. Because treatment with SARS-S did not increase the expression of the membrane-bound precursor of TNF-α [supporting information (SI) Fig. S1], it seems that TACE activity is directly affected by the attachment of SARS-S to ACE2. Lambert et al. (14) proposed that PMA-driven ACE2 shedding solely depended on TACE. In addition, the results of our experiments using siRNAs against TACE and ADAM10 revealed that SARS-S-induced ACE2 shedding depends on TACE activity. Interestingly, however, the forced expression of ADAM9 or 10 rendered tace-KO cells competent for SARS-S-dependent viral infection (Fig. S2), implying that TACE may be complemented by the overexpression of other ADAMs. We also observed that rottlerin, a PKC inhibitor (19), partially blocked the phenomenon (Fig. S3). It is likely that functional modulation of the cytoplasmic tail of ACE2 triggers multiple cellular signals involving PKC activation.
Intriguingly, we found that the entry of SARS-CoV depends on the cytoplasmic tail of ACE2 and TACE activity, which was inconsistent with previous reports indicating that the cytoplasmic tail of ACE2 is dispensable for viral infection (18, 20). However, these previous studies were performed by using pseudotyped lentiviruses. As shown in Fig. 4a, we also detected no differences in reporter gene expression in cells expressing WT-ACE2 and ΔTMC-ACE2. However, viral entry, as judged by the amount of p24, a component of the HIV-1-based lentivirus (Fig. 4b), was significantly higher in cells expressing WT-ACE2 than in those expressing ΔTMC-ACE2. Because the replication of SARS-CoV does not require viral integration into the host genome, we assumed that a reporter assay based on a pseudotyped lentivirus was not relevant for evaluating the roles of the factors involved in SARS-CoV infection. Given these expectations, we measured viral entry based on real-time RT-PCR detection of SARS-CoV mRNA. As shown in Fig. 4c, we successfully detected a difference in copy number of the viral mRNA in these cells (P < 0.01). The introduction of an ACE2 siRNA also reduced the copy number of SARS-CoV mRNA (Fig. 4e), indicating that this method actually detects viral infection via ACE2. In addition, TACE siRNA reduced SARS-CoV infection, indicating that TACE plays an important role in the entry of SARS-CoV. Consistent with these data, TAPI-0, a TACE inhibitor, specifically inhibited the entry of SARS-S (Fig. S4). In addition, although ΔTMC-ACE2 was defective in terms of viral infection, it restored receptor function when TACE was activated by addition of PMA (Fig. S5).
It is unclear how TACE facilitates viral entry. Our observations indicated that the TACE requirement is stronger for SARS-S than for NL63-S (Fig. 5d). Recent studies, using peptidases and their inhibitors, suggested that SARS-S is processed in a stepwise fashion (21, 22). Peptidase inhibitors of cathepsin L (23) or Ben·HCl (24) blocked viral infection, whereas the addition of cathepsin L or Factor Xa enhanced viral infection (23–25). The peptidase activity of cathepsin L, which is critical for the fusion of viral particles and cytoplasmic membranes, depends on a low endosomal pH (26). Interestingly, SARS-CoV depends on both cathepsin L (25) and a low endosomal pH (26), whereas HNL63-CoV (25) does not, indicating a possible functional link between TACE and the components required for fusion of viral and plasma membranes.
Because the shed form of ACE2 is catalytically active (Figs. 1–3) and would counteract ACE-induced cellular signals, it is debatable whether ACE2 shedding by itself is a key event leading to tissue damage. It is plausible that prolonged low-level expression of ACE2 is required for the up-regulation of ACE activity in tissue injury. Although it is important to characterize the temporal changes in ACE2 expression after shedding, the results of the present study suggest that the cytoplasmic tail of ACE2 and a cellular response triggered by the interaction between ACE2 and SARS-S are candidate targets for anti-SARS-CoV therapeutics.
Materials and Methods
Cell Culture and Chemicals.
Vero E6, HEK293T, and Huh-7 cells were obtained from the Riken BRC Cell Bank. These cells and NIH 3T3 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% FCS. tace-KO cells (16) were provided by R. A. Black (Amgen) and maintained in Ham's F-12 supplemented with 2% FCS. Phorbol-12-myristate-13-acetate (PMA) (Sigma) was used as a positive control for ACE2 shedding.
Plasmid Construction.
Plasmids encoding full-length hACE2 (4) and residues 12–672 (27) of SARS-S were provided by H. Choe (Harvard Medical School, Boston, MA). Plasmid DNAs encoding the ACE2 mutants or part of SARS-S were constructed by using PCR-based techniques. Each mutant was confirmed by sequencing.
Preparation and Infection of SARS-S and NL63-S.
Pseudotyped lentiviruses were prepared as described in ref. 28. HEK293T cells were cotransfected with HIV-1-based lentiviral DNA and plasmids encoding full-length SARS-S (G. J. Nabel, National Institutes of Health, Bethesda, MA) (26), NL63-S (S. Pöhlmann, Friedrich Alexander University of Erlangen–Nuremberg, Erlangen, Germany) (8, 9), or VSV-G (29). p24 of gag protein was measured by using a Retro-Tek HIV-1 p24 ELISA Kit (ZeptoMetrix) in accordance with the manufacturer's instructions. ACE2-expressing HEK293T cells were infected with 100 ng/ml p24 in SARS-S. After 4 h (before integration of the lentiviral DNA into the host genome) (C. N.-M., unpublished data), the cells were treated with trypsin-EDTA to degrade all unincorporated virus.
siRNA and Protein Analyses.
siRNAs against ACE2, ADAM10, and TACE were purchased from Ambion and transfected by using Oligofectamine Reagent (Invitrogen) in accordance with the manufacturer's instructions. The sequences are summarized in Fig. S7. The cells were assayed 48 h after transfection.
Monoclonal antibodies against the ACE2 ectodomain (R&D Systems) and polyclonal antibodies against the cytoplasmic tail of TACE (Chemicon), ADAM9 (Santa Cruz Biotechnology), ADAM10 (Chemicon), and β-tubulin (loading control) (NeoMarker) were used for Western blot analysis. Rabbit polyclonal antibodies against the ACE2 cytoplasmic domain were produced by MBL Corp. To examine ACE2 shedding, cells were treated with SARS-S, and the culture supernatants were precipitated with trichloroacetic acid (TCA). As a control, 10 μg of ovalbumin (OVA) was added to each sample and the recovered precipitates were probed with anti-OVA antibodies (Chemicon).
Real-Time PCR Analysis of SARS-CoV mRNA Expression.
HEK293T cells or Huh-7 cells in 24-well plates were infected with SARS-CoV FFM-1 strain at an MOI of 1. Four hours after infection, total RNA was purified by ISOGEN (NIPPONGENE). For quantification of SARS-CoV RNA, real time RT-PCR was performed. As a loading control for normalization, 18S ribosomal RNA was quantified. Primers and probes used in the analysis were summarized in Fig. S6. For all of quantification, a one-step RT-PCR kit (Applied Biosystems) was used and the fluorescence intensity generated from the probe was detected by ABI-7700 sequence detector system (Applied Biosystems).
ACE2 Activity Assay.
The enzymatic activity of ACE2 was assayed by using 10 μM 7-methoxycoumarin-YVADAPK (2,4-dinitrophenyl)-OH (R&D Systems) as a fluorogenic substrate. Fluorescence was monitored at an excitation wavelength of 320 nm and an emission wavelength of 450 nm.
FACS Analysis.
SARS-S was expressed as a chimeric protein with IgG-Fc, using pAB61 (S. Pöhlmann). 293FS cells (Invitrogen) were used for protein expression. The recombinant protein was purified by using G-agarose beads (GE Healthcare). Cells expressing WT-ACE2 or ΔTMC-ACE2 were incubated with the Fc-SARS-S protein, and the bound SARS-S was detected by using FITC-labeled α-IgG.
Solid-Phase Assay to Detect the Binding of SARS-S and ACE2.
Recombinant SARS-S and glyceraldehyde dehydrogenase (GAPDH) were expressed and purified by using a pET vector system (Novagen). ACE2 was partially purified from the supernatants of cultured ACE2-expressing 293FS cells by DEAE column chromatography. ELISA plates (Nunc) were coated with viral mutants, and protein and ACE2 was reacted. After incubation α-ACE2 labeled with Alexa Fluor 555 (Molecular Probes), the bound ACE2 was detected by measuring the fluorescence, using Safire 2 (Tecan).
TNF-α Production Assay.
TNF-α production was measured by using a human TNF-α ELISA Kit (R&D Systems) in accordance with the manufacturer's instructions.
Statistical Analysis.
Statistical significance was evaluated by using Student's t test based on triplicate samples unless otherwise stated.
Supplementary Material
Acknowledgments.
We thank Drs. R. A. Black, H. Choe, G. J. Nabel, S. Pöhlmann, E. W. Raines (University of Washington, Seattle), A. Zolkiewska (University of Kansas, Lawrence), and R. Postina (Johannes Gutenberg University, Mainz, Germany) for providing the tace-KO cells and plasmid DNAs encoding ACE2, SARS-S, NL63-S/pAB61, mouse TACE, mouse ADAM9, and bovine ADAM10, respectively. We thank Mr. Y. Okudaira and Ms. S. Nakano for technical assistance. This work was supported by Grants-in-Aid for Research on Emerging and Re-Emerging Infectious Diseases from the Ministry of Health, Labor, and Welfare of Japan and the National Institute of Biomedical Innovation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0711241105/DCSupplemental.
References
- 1.Kaiazek TG, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;15:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 2.Tipnis SR, et al. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J Biol Chem. 2000;275:33238–33243. doi: 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- 3.Nicholls J, Peiris M. Good ACE, bad ACE do battle in lung injury, SARS. Nat Med. 2005;11:821–822. doi: 10.1038/nm0805-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li W, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuba K, et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imai Y, et al. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Hoek L, et al. Identification of a new human coronavirus. Nat Med. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hofmann H, et al. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc Natl Acad Sci USA. 2005;102:7988–7993. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hofmann H, et al. Highly conserved regions within the spike proteins of human coronaviruses 229E and NL63 determine recognition of their respective cellular receptors. J Virol. 2006;80:8639–8652. doi: 10.1128/JVI.00560-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Hoek L, Pyrc K, Berkhout B. Human coronavirus NL63, a new respiratory virus. FEMS Microbiol Rev. 2006;30:760–773. doi: 10.1111/j.1574-6976.2006.00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeager CL, et al. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 1992;357:420–422. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Black RA, et al. A metalloproteinase disintegrin that releases tumour-necrosis factor-α from cells. Nature. 1997;385:729–733. doi: 10.1038/385729a0. [DOI] [PubMed] [Google Scholar]
- 13.Seals DF, Courtneidge SA. The ADAMs family of metalloproteases: Multidomain proteins with multiple functions. Genes Dev. 2003;17:7–30. doi: 10.1101/gad.1039703. [DOI] [PubMed] [Google Scholar]
- 14.Lambert DW, et al. Tumor necrosis factor-α convertase (ADAM17) mediates regulated ectodomain shedding of the severe-acute respiratory syndrome-coronavirus (SARS-CoV) receptor, angiotensin-converting enzyme-2 (ACE2) J Biol Chem. 2005;280:30113–30119. doi: 10.1074/jbc.M505111200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohler KM, et al. Protection against a lethal dose of endotoxin by an inhibitor of tumour necrosis factor processing. Nature. 1994;370:218–220. doi: 10.1038/370218a0. [DOI] [PubMed] [Google Scholar]
- 16.Reddy P, et al. Functional analysis of the domain structure of tumor necrosis factor-alpha converting enzyme. J Biol Chem. 2000;275:14608–14614. doi: 10.1074/jbc.275.19.14608. [DOI] [PubMed] [Google Scholar]
- 17.Simmons G, et al. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc Natl Acad Sci USA. 2004;101:4240–4245. doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofmann H, et al. Susceptibility to SARS coronavirus S protein-driven infection correlates with expression of angiotensin converting enzyme 2 and infection can be blocked by soluble receptor. Biochem Biophyis Res Commun. 2004;319:1216–1221. doi: 10.1016/j.bbrc.2004.05.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gschwendt M, et al. Rottlerin, a novel protein kinase inhibitor. Biochem Biophys Res Commun. 1994;199:93–98. doi: 10.1006/bbrc.1994.1199. [DOI] [PubMed] [Google Scholar]
- 20.Inoue Y, et al. Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted. J Virol. 2007;81:8722–8729. doi: 10.1128/JVI.00253-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu S, et al. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: Implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet. 2004;363:938–947. doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu XD, et al. The spike protein of severe acute respiratory syndrome (SARS) is cleaved in virus infected Vero-E6 cells. Cell Res. 2004;14:400–406. doi: 10.1038/sj.cr.7290240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simmons G, et al. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci USA. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du L, et al. Cleavage of spike protein of SARS coronavirus by protease factor Xa is associated with viral infectivity. Biochem Biophys Res Commun. 2007;359:174–179. doi: 10.1016/j.bbrc.2007.05.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang IC, et al. SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J Biol Chem. 2006;281:3198–3203. doi: 10.1074/jbc.M508381200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang ZY, et al. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J Virol. 2004;78:5642–5650. doi: 10.1128/JVI.78.11.5642-5650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong SK, Li W, Moore MJ, Choe H, Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J Biol Chem. 2004;279:3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakai-Murakami C, et al. HIV-1 Vpr induces ATM-dependent cellular signal with enhanced homologous recombination. Oncogene. 2007;26:477–486. doi: 10.1038/sj.onc.1209831. [DOI] [PubMed] [Google Scholar]
- 29.Tachiwana H, et al. HIV-1 Vpr induces DNA double-strand breaks. Cancer Res. 2006;66:627–631. doi: 10.1158/0008-5472.CAN-05-3144. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.