Abstract
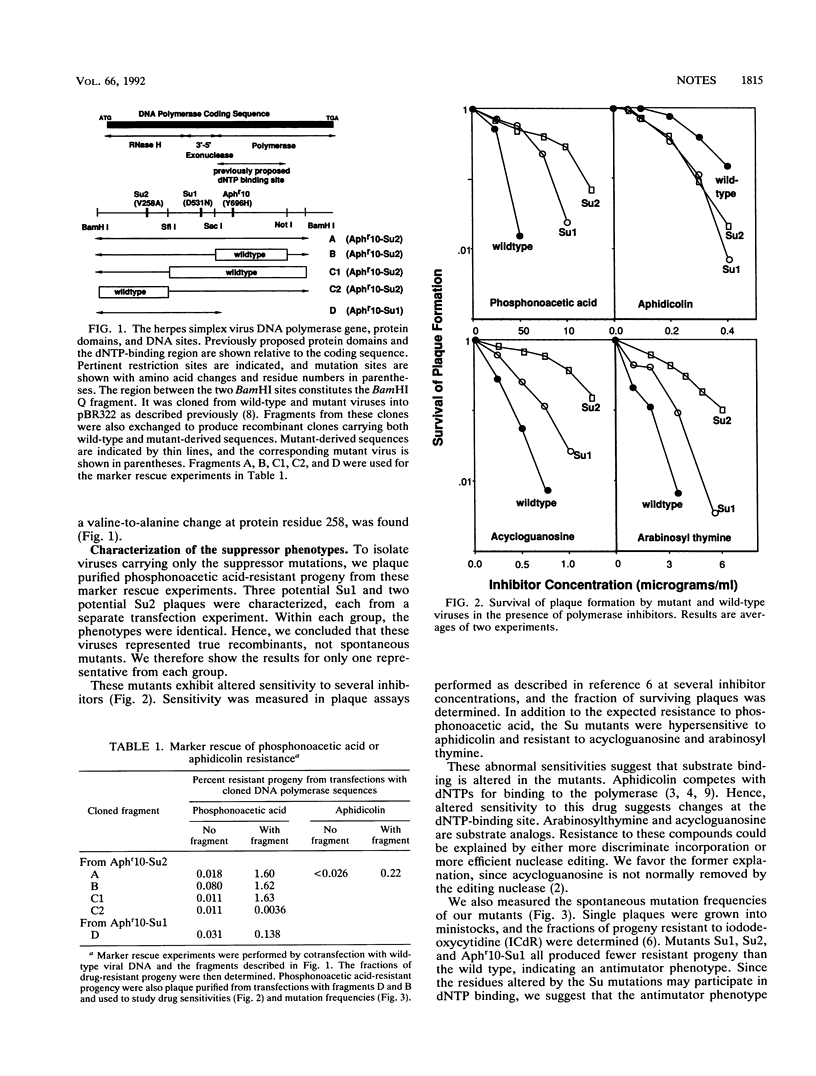
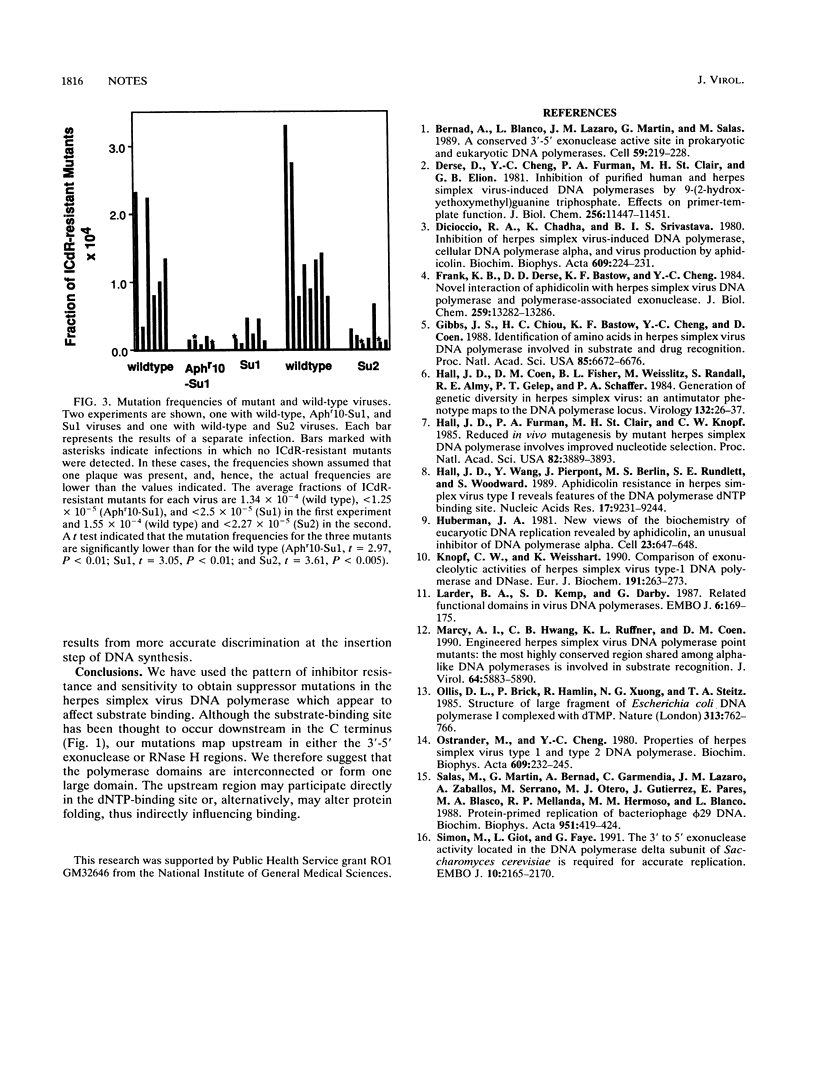
Herpes simplex virus DNA polymerase mutations which map in the N-terminal part of the protein and appear to alter deoxynucleoside triphosphate (dNTP) substrate specificity are described. These mutations suppress a drug hypersensitivity associated with the downstream mutation, Aphr10. We suggest that the mutant residues form part of the dNTP-binding site, a site previously thought to be confined to the C terminus.
Full text
PDF
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bernad A., Blanco L., Lázaro J. M., Martín G., Salas M. A conserved 3'----5' exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell. 1989 Oct 6;59(1):219–228. doi: 10.1016/0092-8674(89)90883-0. [DOI] [PubMed] [Google Scholar]
- Derse D., Cheng Y. C., Furman P. A., St Clair M. H., Elion G. B. Inhibition of purified human and herpes simplex virus-induced DNA polymerases by 9-(2-hydroxyethoxymethyl)guanine triphosphate. Effects on primer-template function. J Biol Chem. 1981 Nov 25;256(22):11447–11451. [PubMed] [Google Scholar]
- Dicioccio R. A., Chadha K., Sahai Srivastava B. I. Inhibition of herpes simplex virus-induced DNA polymerase, cellular DNA polymerase alpha, and virus production by aphidicolin. Biochim Biophys Acta. 1980 Sep 19;609(2):224–231. doi: 10.1016/0005-2787(80)90233-6. [DOI] [PubMed] [Google Scholar]
- Frank K. B., Derse D. D., Bastow K. F., Cheng Y. C. Novel interaction of aphidicolin with herpes simplex virus DNA polymerase and polymerase-associated exonuclease. J Biol Chem. 1984 Nov 10;259(21):13282–13286. [PubMed] [Google Scholar]
- Gibbs J. S., Chiou H. C., Bastow K. F., Cheng Y. C., Coen D. M. Identification of amino acids in herpes simplex virus DNA polymerase involved in substrate and drug recognition. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6672–6676. doi: 10.1073/pnas.85.18.6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J. D., Coen D. M., Fisher B. L., Weisslitz M., Randall S., Almy R. E., Gelep P. T., Schaffer P. A. Generation of genetic diversity in herpes simplex virus: an antimutator phenotype maps to the DNA polymerase locus. Virology. 1984 Jan 15;132(1):26–37. doi: 10.1016/0042-6822(84)90088-6. [DOI] [PubMed] [Google Scholar]
- Hall J. D., Furman P. A., St Clair M. H., Knopf C. W. Reduced in vivo mutagenesis by mutant herpes simplex DNA polymerase involves improved nucleotide selection. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3889–3893. doi: 10.1073/pnas.82.11.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J. D., Wang Y. S., Pierpont J., Berlin M. S., Rundlett S. E., Woodward S. Aphidicolin resistance in herpes simplex virus type I reveals features of the DNA polymerase dNTP binding site. Nucleic Acids Res. 1989 Nov 25;17(22):9231–9244. doi: 10.1093/nar/17.22.9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman J. A. New views of the biochemistry of eucaryotic DNA replication revealed by aphidicolin, an unusual inhibitor of DNA polymerase alpha. Cell. 1981 Mar;23(3):647–648. doi: 10.1016/0092-8674(81)90426-8. [DOI] [PubMed] [Google Scholar]
- Knopf C. W., Weisshart K. Comparison of exonucleolytic activities of herpes simplex virus type-1 DNA polymerase and DNase. Eur J Biochem. 1990 Jul 31;191(2):263–273. doi: 10.1111/j.1432-1033.1990.tb19119.x. [DOI] [PubMed] [Google Scholar]
- Larder B. A., Kemp S. D., Darby G. Related functional domains in virus DNA polymerases. EMBO J. 1987 Jan;6(1):169–175. doi: 10.1002/j.1460-2075.1987.tb04735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcy A. I., Hwang C. B., Ruffner K. L., Coen D. M. Engineered herpes simplex virus DNA polymerase point mutants: the most highly conserved region shared among alpha-like DNA polymerases is involved in substrate recognition. J Virol. 1990 Dec;64(12):5883–5890. doi: 10.1128/jvi.64.12.5883-5890.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollis D. L., Brick P., Hamlin R., Xuong N. G., Steitz T. A. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. 1985 Feb 28-Mar 6Nature. 313(6005):762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- Ostrander M., Cheng Y. C. Properties of herpes simplex virus type 1 and type 2 DNA polymerase. Biochim Biophys Acta. 1980 Sep 19;609(2):232–245. doi: 10.1016/0005-2787(80)90234-8. [DOI] [PubMed] [Google Scholar]
- Salas M., Martín G., Bernad A., Garmendia C., Lázaro J. M., Zaballos A., Serrano M., Otero M. J., Gutiérrez J., Parés E. Protein-primed replication of bacteriophage phi 29 DNA. Biochim Biophys Acta. 1988 Dec 20;951(2-3):419–424. doi: 10.1016/0167-4781(88)90115-7. [DOI] [PubMed] [Google Scholar]
- Simon M., Giot L., Faye G. The 3' to 5' exonuclease activity located in the DNA polymerase delta subunit of Saccharomyces cerevisiae is required for accurate replication. EMBO J. 1991 Aug;10(8):2165–2170. doi: 10.1002/j.1460-2075.1991.tb07751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]



